Abstract
Recent studies have shown that autophagy is essential for proper β-cell function and survival. However, it is yet unclear under what pathogenic conditions autophagy is inhibited in β-cells. Here, we report that long term exposure to fatty acids and glucose block autophagic flux in β-cells, contributing to their toxic effect. INS1 cells expressing GFP-LC3 (an autophagosome marker) were treated with 0.4 mm palmitate, 0.4 mm oleate, and various concentrations of glucose for 22 h. Kinetics of the effect of fatty acids on autophagy showed a biphasic response. During the second phase of autophagy, the size of autophagosomes and the content of autophagosome substrates (GFP-LC3, p62) and endogenous LC3 was increased. During the same phase, fatty acids suppressed autophagic degradation of long lived protein in both INS1 cells and islets. In INS1 cells, palmitate induced a 3-fold decrease in the number and the acidity of Acidic Vesicular Organelles. This decrease was associated with a suppression of hydrolase activity, suppression of endocytosis, and suppression of oxidative phosphorylation. The combination of fatty acids with glucose synergistically suppressed autophagic turnover, concomitantly suppressing insulin secretion. Rapamycin treatment resulted in partial reversal of the inhibition of autophagic flux, the inhibition of insulin secretion, and the increase in cell death. Our results indicate that excess nutrient could impair autophagy in the long term, hence contributing to nutrient-induced β-cell dysfunction. This may provide a novel mechanism that connects diet-induced obesity and diabetes.
Keywords: Autophagy, Diabetes, Fatty Acid, Glucose, Pancreatic Islets
Introduction
Macroautophagy (hereafter named autophagy) is the main mechanism the cell uses to degrade damaged and redundant organelles. It involves the formation of a double-membrane structure called the phagophore, which evolves into the autophagosome (AP),2 an organelle that sequesters cytoplasmic material such as mitochondria, peroxisomes, endoplasmic reticulum, protein aggregates, and lipids. Upon acidification (1), the AP fuses with the lysosome to form the autolysosome, which degrades its content (2).
The main approach to study autophagy is by tracking APs using LC3 (microtubule-associated protein 1 light chain 3), a cytosolic protein that upon stimulation of autophagy is lipidated and recruited to the AP membrane. LC3 remains bound to the AP until released to the cytosol or degraded by lysosomal enzymes (3).
Stimulators of autophagy are known to increase the number of APs. However, the quantification of APs to assess autophagy can be misleading, APs being but one component in the chain constituting autophagic degradation (3). Thus, for example, in the case of various neuronal diseases, the increase in the number of APs was originally falsely interpreted as an increase in autophagic turnover, although it is now known to be the result of a decrease in autophagic turnover downstream to AP formation (4, 5).
Type 2 diabetes is a disease in which glucose homeostasis is impaired due to peripheral insulin resistance accompanied by a decrease in pancreatic β-cell function and a decrease in β-cell mass. One of the causes for the deterioration of β-cells during type 2 diabetes appears to be the toxicity of nutrient overload on β-cells. High concentrations of free fatty acids (FFAs) (“lipotoxicity”), high concentrations of glucose (“glucotoxicity”), and the combination of FFAs and glucose (“glucolipotoxicity”) impair insulin secretion and induce cell death by apoptosis (6–8).
Autophagy has been shown to be important for β-cell viability and function both in the whole animal (9–11) and in INS1 cells, a commonly used β-cell secreting line (12, 13). Whether any of the diabetogenic factors such as nutrition and obesity impair autophagy is yet unclear. In various animal models (including mice fed high fat diet) (9, 14) and cellular models of diabetes (including glucotoxicity and lipotoxicity) (9, 13–16), the number of APs has been reported to increase. Whether this increase reflects an increase or a decrease in autophagic turnover is unresolved. Hints to the effect of FFAs on turnover were provided; those, however, are somewhat contradictory. In INS1 cells AP formation is found to be increased under long term exposure to FFAs, when autophagosome degradation is blocked (9). In those same cells, short term palmitate exposure increases protein degradation mediated by autophagy (17). In contrast to these findings, Masini et al. (18) reported an accumulation of overloaded APs in β-cells from pancreata of human diabetic cadavers and in dispersed β-cells exposed to FFAs, suggesting an impairment of AP maturation.
In this study, we use a battery of approaches to study the effect of nutrient overload on autophagic turnover in β-cells. We show that although in the long term, FFAs can increase the formation of APs, they suppress acidification and autophagic degradation. Glucose suppresses autophagy as well and can synergize with FFAs.
EXPERIMENTAL PROCEDURES
Reagents
The following reagents were used: rapamycin (Sigma), palmitate (Sigma), oleate (Sigma), bafilomycin (LC Laboratories), [14C]phenylalanine (PerkinElmer Life Sciences), and leupeptin (Sigma).
Animals and Islet Isolation
Nine- to 10-week-old C57Bl6 male mice were used for islet isolation. They were housed in accordance with the Boston University Institutional Guidelines for Animal Care (IACUC no. 1104) in compliance with United States Public Health Service Regulation. Animals were fed standard chow and kept at normal housing conditions (19–22 °C and a 14:10-h light-dark cycle) until death by CO2 asphyxiation and cervical dislocation.
Islets of Langerhans were isolated as described previously (19). Shortly, the pancreata were inflated with Hanks' buffer containing collagenase (Roche Applied Science). After 30 min of digestion at 37 °C, the islets were isolated using Ficoll gradient.
Cell Culture
INS1 832/13 cells were cultured in RPMI 1640 media supplemented with 10% FBS, 10 mm HEPES buffer, 1 mm pyruvate, 50 μm 2-β-mercaptoethanol, 50 units/ml penicillin, and 50 μg/ml streptomycin.
Palmitate and Oleate Complexed to BSA
Palmitate and oleate were dissolved in DMSO to a final concentration of 0.4 m and then dissolved at 56 °C in RPMI 1640 media containing 5% fatty acid-free BSA (Calbiochem) to make a 10× stock. For control, we used RPMI 1640 media containing 5% BSA and 1% DMSO. The day of the experiment, the stocks were added to RPMI 1640 media containing 1% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin and glucose at either 10 mm or as designated concentration.
Protein Degradation Assay
The basics of protein degradation were reported previously (20). Shortly, cells were incubated in plain media supplemented with [14C]phenylalanine. After 24 h, the media were switched to media containing 2 mm nonradioactive phenylalanine for 24 h. The cells were then incubated in either control media or media containing 0.4 mm palmitate or 0.4 mm oleate, in the presence or absence of bafilomycin. Media samples were collected at different time points, and proteins were precipitated in TCA. At the end of the experiment, the cells were lysed using 1% SDS in PBS. Cellular proteins were isolated by TCA precipitation. Disintegrations/min (dpm) in the nonprecipitable fraction of the media and in precipitable fraction of the cells were measured with a β-counter. Protein degradation rate was calculated by the ratio of the nonprecipitable C14 in the media to the cell precipitable C14 divided by time of incubation after chase.
Fluorescence-activated Cell Sorting (FACS) Analysis
INS1 cells were infected with lentivirus encoding either for GFP-LC3 or DsRed targeted to the mitochondria (12). The cells expressing GFP-LC3 were then sorted according to GFP intensity using MoFlo Cell Sorter. FACS analyses of the cells after treatment was done with ×620 FACScan. FACS data analysis was performed using FACScalibur (Beckman Coulter). Cell debris was excluded by gating on the forward and side scatter plot.
Cells were stained with 1 μm Acridine Orange (Sigma) for 15 min and washed twice with PBS. The cells were then trypsinized, washed twice with PBS by centrifugation, and then subjected to flow cytometry. Cells incubated for 4 h in KRB or bafilomycin were used as positive and negative controls, respectively.
LysoTracker, LysoSensor, MR-Cathepsin L, pHrodo-dextran Staining, and Immunostaining
Cells were stained with 50 nm LysoTracker red (Molecular Probes) for 30 min, then washed three times with PBS before being fixed with 4% paraformaldehyde for 15 min, and washed again three times with PBS and mounted with Mowiol.
Cells were stained with 1 μm LysoSensor yellow/blue for 5 min followed by confocal imaging using a 360 nm excitation and collecting images at the yellow wavelength range (510–641 nm) and at the range of blue wavelength (404–456 nm). The ratio between yellow and blue was calculated using metamorph software. Cells were stained with 10 μg/ml Magic red cathepsin L (MR-cathepsin L; Immunochemistry Technologies) for 1 h. The cells were then washed three times with RPMI 1640 media and observed under a confocal microscope.
For p62 immunostaining, the cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, permeabilized in 0.5% Triton X-100 for 15 min, washed, and then blocked with 1% BSA/PBS for 15 min. The fixed cells were incubated with primary antibody against p62 anti-mouse (Abnova), diluted 1:200, washed, incubated with the secondary antibody Alexa 660 anti mouse (Invitrogen), diluted 1:500 for 1 h, washed with PBS, and mounted with Mowiol (Calbiochem) mounting media. The Images were taken at ×63 magnification using Zeiss LSM 510 Meta confocal microscope, and analyzed with MetaMorph software.
Western Blot
Samples were prepared as described previously (21). They were loaded in 12% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane using a semidry transfer machine. LC3 (Sigma), β-actin (Sigma), p62 (Abnova), GAPDH (Santa Cruz Biotechnology), GFP (Abcam), COX-IV subunit 1 (Invitrogen), and COX-IV subunit 4 (Santa Cruz Biotechnology) antibodies were used according to the manufacturer's instructions.
O2 Consumption
Oxygen consumption in INS1 was measured by an XF24 oxygen consumption assay (Seahorse Bioscience, Billerica, MA) as described previously (22).
Mitochondrial Membrane Potential
Mitochondrial membrane potential was analyzed as described previously (23). Briefly, INS1 cells were stained with 5 nm tetramethylrhodamine ethyl ester perchlorate and 100 nm MitoTracker Green (Molecular Probes) for 45 min. The cells were washed and incubated with RPMI 1640 media containing tetramethylrhodamine ethyl ester perchlorate alone and observed under a 710 Zeiss confocal microscope.
WIPI-1 Measurement
INS1 cells were transfected with WIPI-1-GFP vector (a generous gift from Dr. Tassula Proikas-Cezanne) using TransIT-LT1 Reagent (Mirus) according to the manufacturer's recommendations. After 24 h cells were treated with control media or palmitate for 14 h and imaged with confocal microscope. Cells containing several distinct puncta were counted.
ATP Measurement
ATP was measured using a bioluminescence kit (Roche Applied Science) according to manufacturer's protocol, after extracting the ATP using boiled deionized water as described previously (24).
Statistics
Unless stated otherwise, error bars indicate means ± S.E., and an unpaired t test was used to validate statistical differences.
RESULTS
Palmitate Induces Accumulation of Overloaded Autophagosomes in INS1 Cells
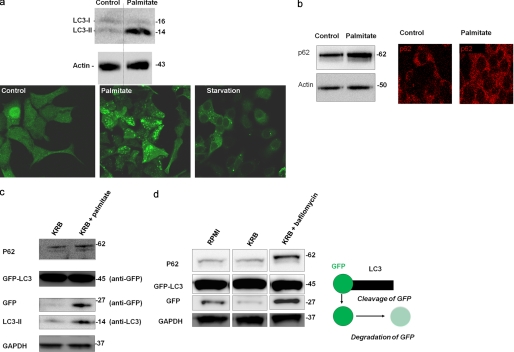
It has previously been shown that type 2 diabetes patients accumulate large autophagosomes in their pancreatic β-cells, suggesting impairment in autophagy (18). We tested whether a similar phenomenon could be reproduced in a cellular model of β-cell lipotoxicity. INS1 cells stably expressing GFP-LC3 were exposed for 6 h to either 0.4 mm palmitate bound to BSA or to amino acid-free medium (KRB) or to control media containing BSA (Fig. 1a). Although both amino acid starvation and palmitate increased the number of autophagosomes, under palmitate, autophagosomes were larger, and GFP fluorescence intensity (FI) was brighter than under KRB, suggesting an impairment in AP degradation. Consistently, p62, a protein known to be degraded exclusively by autophagy, was accumulated with palmitate treatment (Fig. 1b). Absence of amino acids in the media did not prevent the accumulation of substrates of autophagosomes in the cell. Cells expressing GFP-LC3 and treated for 14 h with palmitate in KRB showed an increase in p62 (Fig. 1c). Likewise, palmitate increased unprocessed free GFP (Fig. 1c), as did bafilomycin, a blocker of the lysosomal H+ pump (Fig. 1d).
FIGURE 1.
Palmitate increases the number and size of the APs and the content of AP substrates. INS1 cells were cultured in the presence or absence of 0.4 mm palmitate for 14 h. a, representative experiment (n = 6) Western blot analysis of LC3 and images of INS1 cells expressing GFP-LC3 and treated for 14 h with palmitate or KRB. Two bands were obtained, the cytosolic LC3-I and the autophagosomal LC3-II. Actin was used as a loading control. Below are confocal images of INS1 cells expressing GFP-LC3 and treated with control, palmitate, or KRB (starvation) media. b, Western blot analysis and immunostaining of p62 in INS1 cells incubated in the presence or absence of palmitate in full RPMI 1640 media for 24 h. c and d, Western blot analysis of p62, GFP, LC3 in cells cultured in amino acid-free medium (KRB) alone or in amino acid-free medium in the presence of 0.4 mm palmitate (c) or in the presence of 200 nm bafilomycin (d) after 14 h of incubation. GAPDH was used as a loading control. Two bands were obtained with anti-GFP antibody as follows: a heavy one (45 kDa) and a lighter one (27 kDa) corresponding to the chimeric GFP-LC3 and to free GFP, respectively. Free GFP represents partial degradation of GFP-LC3 as indicated by its accumulation upon exposure to bafilomycin.
Palmitate Increases AP Formation but Autophagy Does Not Reach Steady State
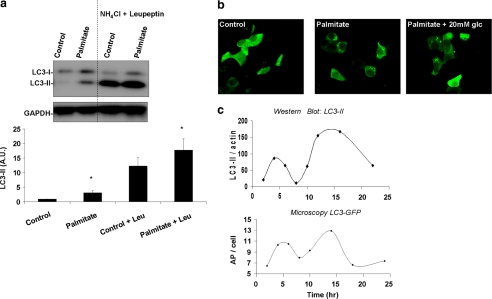
Previous studies undertaken in hepatocytes have shown that AP formation is suppressed by FFAs (25). In contrast, in INS1 cells, prolonged exposure to FFAs increase AP formation (9). To verify the latter, we measured LC3 after 14 h of exposure to 0.4 mm palmitate in the presence or absence of protease inhibitors for the last 2 h. As shown in Fig. 2a, the lipidated form of LC3 (LC3-II) was significantly increased in the presence of palmitate alone, indicating an increase in the number of APs. Blockage of autophagy by the combination of ammonium chloride and leupeptin increased LC3-II expression. Palmitate further increased LC3-II, indicating that the accumulation observed in AP number reflects, at least partially, an increase in the rate of AP formation. To further confirm increased AP formation, we measured the effect of palmitate on accumulation of WIPI-1 puncta, a marker of phagophores (Fig. 2b) (26, 27). INS1 cells transfected with WIPI1-GFP showed a significant (61%) increase in WIPI-1 puncta in the presence of palmitate as compared with control after 14 h of exposure. Although under control conditions only 26 ± 11% of the cells showed multiple WIPI-1 puncta, treatment with palmitate increased this value to 42 ± 14% (n = 3, p < 0.05, paired t test). In the presence of high glucose (20 mm), palmitate had a milder effect on abundance of WIPI-1 puncta (33 ± 9% of the cells having multiple WIPI-1 puncta).
FIGURE 2.
Palmitate increases AP formation without reaching steady state. a, Western blot analysis of LC3. INS1 cells were incubated for 14 h with or without 0.4 mm palmitate, in the presence or absence of 20 mm ammonium chloride (NH4Cl) and 100 μm leupeptin (Leu) during the last 3 h of incubation. Below is the densitometry of LC3-II signal normalized against GAPDH. Note that palmitate increased LC3-II both in the absence and presence of ammonium chloride and leupeptin. *, p < 0.05 (n = 6); error bars, S.E. b, confocal images of INS1 cells expressing WIPI-GFP and incubated for 14 h in control media (containing 10 mm glucose), in media containing 0.4 mm palmitate along with 10 mm glucose or along with 20 mm glucose. c, representative curve (n = 3) of LC3-II protein levels over time as determined by densitometry of Western blot analyses. LC3-II signal was normalized against actin. Below is the average AP number per cell over time as determined from confocal images of cells expressing GFP-LC3. Between 40 and 50 cells were analyzed per condition.
Unless APs are in steady state (rate of formation = rate of degradation), an increase in AP formation does not necessarily reflect an increase in their degradation. To examine whether under palmitate autophagosomes reach steady state, we followed the change in AP number with time. In agreement with previous publications (13, 17), autophagosome number showed a fluctuation with time (Fig. 2c). Both Western blot densitometry for endogenous LC3-II in noninfected INS1 cells and the number of APs in INS1 cells transduced with GFP-LC3 lentivirus displayed a biphasic pattern with the first peak emerging after 8 h of exposure and the second peak emerging after 14 h of exposure (Fig. 2c). Given that autophagosomes do not reach steady state, we had to rely on other tools to study autophagic degradation.
FFAs Suppress Autophagic Turnover as Reflected by GFP-LC3 Intensity
Autophagic turnover depends on the fusion between the lysosomes and the APs to form the acidic autolysosomes. Given that GFP fluorescence decreases within an acidic environment, total GFP FI in cells expressing GFP targeted to the APs (GFP-LC3) can be used to estimate the rate of fusion between autophagosomes and lysosomes. We employed an approach in which the turnover of GFP-LC3 is measured by flow cytometry (28, 29). In this technique, the total FI of GFP in cells expressing GFP-LC3 decreases when autophagic turnover increases. Thus, under stimulation of autophagic turnover, the increase in fusion between APs and lysosomes is expected to lead to a decrease in GFP fluorescence, whereas blockage of autophagic turnover is expected to lead to an accumulation of GFP-LC3 in the APs and in the cytosol and thus to an increase in the number of cells with high GFP FI.
We compared FI of cells expressing GFP-LC3 under various conditions known to affect autophagy (Fig. 3a). As shown previously (29), amino acid starvation and treatment with rapamycin (an mTOR inhibitor) decreased GFP FI relative to control cells, indicating an increase in autophagic turnover. Palmitate and oleate, however, significantly increased the average GFP FI by 23 ± 6 and 39 ± 9%, respectively (Fig. 3a), as compared with control media. Fatty acyl-induced increase in GFP FI was found to be significantly smaller than that obtained with bafilomycin (217 ± 25% of control cells), a strong inhibitor of acidification, suggesting a partial blockage of autophagic flux by fatty acids.
FIGURE 3.
Exposure to fatty acids decreases the turnover of GFP-LC3 while not affecting its synthesis. a, summary of flow cytometry data from GFP-LC3 expressing cells presented as average GFP FI normalized to control. The cells were exposed for 14 h to one of the following conditions: Krebs ringer bicarbonate (starvation), rapamycin (200 nm), palmitate (400 μm), or oleate (400 μm). Cells were also exposed to bafilomycin (200 nm) for 3 h to inhibit autophagy. GFP-LC3 intensity was inversely correlated with the autophagosome flux. Thus blockage of autophagosome flux with bafilomycin showed an increase in FI, whereas induction of flux with KRB and rapamycin decreased FI. The data are expressed as average percentage increase or decrease intensity compared with no treatment. Error bars, S.E. b, fatty acids induce an early and transient increase in autophagic flux followed by two phases of flux inhibition. Chart shows average GFP intensities of INS1 cells expressing GFP-LC3 and treated with oleate or palmitate and subjected to flow cytometry after 0, 2, 4, 6, 8, 10, and 14 h. The FI was normalized to the respective control cells at each time point. Average of four experiments; error bars, S.E. c, palmitate does not change synthesis rates of transduced proteins under CMV promoter. Bar graph shows the average DsRed intensity (A.U., arbitrary units) as determined by confocal microscopy. Error bars, S.D. d, lack of additive effects between bafilomycin and palmitate. Flow cytometry histogram of INS1 cells expressing GFP-LC3 and treated for 14 h in the presence or absence of palmitate or oleate with bafilomycin added the last 3 h.
To investigate whether the effect of FFAs on autophagic flux is time-dependent, we followed the change in GFP FI with time in INS1 cells expressing GFP-LC3 and exposed to 0.4 mm palmitate or 0.4 mm oleate (Fig. 3b). Cell exposure to both palmitate and oleate induced two waves of increased GFP intensity compared with control levels (with peaks at the 6th and 10th hours). The first period of attenuated flux was preceded by an additional early phase, which occurred during the first 4 h of palmitate exposure. This early phase was characterized by a decrease in the average intensity as compared with control, suggesting an increase in autophagic turnover during that period of time.
FFAs are known to suppress protein translation (30, 31). However, to rule out the possibility that the increase in GFP FI by palmitate is mediated by an effect other than autophagy, we examined FI in conditions that are not expected to be influenced by autophagy.
First, we tested whether palmitate was able to increase the content of a fluorescent protein not targeted to the autophagosomes. INS1 cells transduced with DsRed targeted to mitochondria under the same promoter as the one used for GFP-LC3 were treated with palmitate for 14 h. The average fluorescence intensity of DsRed was similar in cells exposed to palmitate and in control cells (Fig. 3c). Next, we examined whether FFAs can increase GFP FI in INS1 cells expressing GFP-LC3 after autophagy has been completely suppressed with bafilomycin. As shown in Fig. 3d, in the presence of bafilomycin, palmitate and oleate showed a similar GFP FI as compared with bafilomycin alone (Fig. 3d). These results indicate that the increase in the intensity of GFP is mediated by autophagic degradation.
FFAs Decrease Acidification and Impair Lysosomal Function
Because the increase in GFP FI was not mediated by an increase in synthesis, we examined whether it reflects a decrease in acidification.
INS1 cells were exposed to palmitate for 14 h and stained with LysoTracker red, a basic dye that stains acidic vesicular organelles (AVOs). Palmitate decreased the average LysoTracker red staining intensity as measured by flow cytometry (Fig. 4a). A similar result was obtained with acridine orange, another indicator of AVOs (data not shown). Imaging of cells stained with LysoTracker red, showed a 63% decrease in the number of AVOs per cell in the presence of palmitate as compared with control (Fig. 4b). Although LysoTracker red and acridine orange are reliable dyes to quantify AVOs, they are not sensitive enough for pH measurement. LysoSensor yellow/blue is a pH-sensitive probe that emits predominantly yellow fluorescence in acidic organelles, although in less acidic organelles it emits blue fluorescence. Treatment of INS1 cells with palmitate significantly decreased the yellow/blue ratio in the AVOs reflecting an increase in pH from 4.6 to 5.1 (Fig. 4c). Interestingly, this decrease in acidification was concomitant with a 2-fold increase in AVO size (Fig. 4d), further indicating impairment in lysosomal function.
FIGURE 4.
Palmitate decreases lysosomal acidification and cathepsin L activity. INS1 cells were stained with either the probe for acidic vesicular organelles LysoTracker red, or the pH indicator LysoSensor yellow/blue, or the cathepsin L activity indicator magic red after treatment with or without palmitate for 14 h. a, flow cytometry intensity histogram of cells stained with LysoTracker red. b, confocal microscopy analysis of INS1 cells expressing GFP-LC3 (green) and stained with LysoTracker red (red). Nuclei were co-stained with Hoechst (purple). Bar, 10 μm. Quantification of the number of AVOs. Error bars, S.D., *, p < 0.05. Note that LysoTracker red and GFP-LC3 did not co-localize, reflecting the sensitivity of GFP to the acidic pH of the lysosomes and autolysosomes. c, palmitate decreases AVO acidification. Images show INS1 cells stained with LysoSensor yellow/blue and imaged using two filter ranges, 404–456 nm (blue) and 510–641 nm (yellow). Subcellular organelles with lower pH are identified as pixels with increased yellow/blue ratio. Error bars, S.E., n = 7. d, average AVO size measured from LysoSensor images. e, palmitate suppresses cathepsin L activity. Staining for MR-cathepsin L. The graph expresses the average red intensity. Error bars, S.E., n = 6, **, p < 0.01. f, pHrodo-dextran fluorescence intensity as a measure for endosome acidity. After 14 h in the presence or absence of palmitate, the cells were incubated with pHrodo-dextran for 1 h and pHrodo intensity was measured; Error bars, S.E., n = 3, **, p < 0.01.
Acidification of the lysosomes is required for the activity of lysosomal hydrolases. To test the functional significance of palmitate-induced alkalinization of lysosomes, we assessed the functionality of the hydrolases. Cells were stained with MR-cathepsin L, a substrate-based assay for the activity of the lysosomal protease cathepsin L. Although both control and palmitate-treated cells showed active cathepsin L, the cells treated with palmitate displayed a significantly lower intensity of MR-cathepsin L. The intensity of MR-cathepsin L was 36% lower than for controls indicating impairment in cathepsin L activity (Fig. 4e). Palmitate impaired other lysosomal functions beside autophagy, as shown by pHrodo-dextran staining, a pH-sensitive probe that enters the cells via endocytosis. Treatment of INS1 cells with palmitate induced a robust decrease in the intensity and number of pHrodo puncta as compared with control (Fig. 4f).
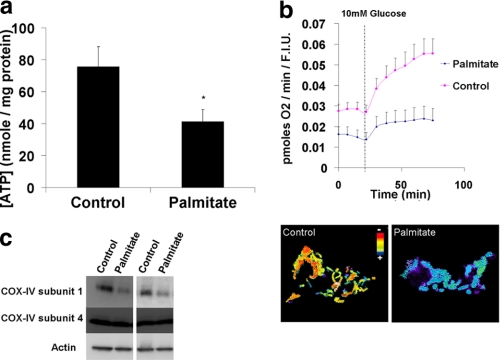
FFAs Decrease Cellular ATP Levels and Oxygen Consumption Rate and Impair Mitochondrial Function
Because lysosomal acidification is dependent on ATP hydrolysis by the H+ pump, we measured cellular ATP levels (Fig. 5a). Palmitate strongly decreased cellular ATP contents as compared with control (41 ± 7 and 75 ± 12 nm/mg protein, respectively). This decrease was associated with a suppression of cellular oxygen consumption as well as with mitochondrial depolarization. This was also accompanied by fragmentation of mitochondrial network (Fig. 5b) (as previously shown in Refs. 23, 32, 33). The combination of depolarization and reduced oxygen consumption suggested a respiratory chain defect. To further assess this possibility, we examined the level respiratory chain proteins. Mitochondrial respiration is influenced, among others, by the levels of respiratory complex subunits encoded by the nucleus and by the mitochondria. Palmitate did not affect the expression of COX-IV subunit 4, a protein encoded by the nuclear DNA, but it decreased the expression of COX-IV subunit 1, a protein encoded by the mitochondrial DNA (Fig. 5c).
FIGURE 5.
Palmitate suppresses cellular bioenergetics and mitochondrial function. a, cellular ATP levels. b, cellular oxygen consumption and mitochondrial potential as measured by the ratio of tetramethylrhodamine ethyl ester perchlorate to MitoTracker green intensities. c, two blots from two separate experiments showing the effect of palmitate on COX-IV subunit 1 (encoded by the mitochondria) and COX-IV subunit 4 (encoded by the nucleus).
FFAs Suppress Autophagic Degradation of Long Lived Proteins
Long lived proteins tend to be degraded by autophagy. We examined the effect of FFAs on long lived protein degradation by performing a pulse-chase experiment with [14C]phenylalanine. The protein degradation rate was calculated by measuring [14C]phenylalanine released into control media or into media containing FFAs. Under control conditions, the degradation rate of long lived proteins was about 5 ± 0.6% per h in INS1 cells (Fig. 6a). Bafilomycin blocked ∼50% of protein degradation indicating that half of the proteins are degraded by autophagy. Palmitate suppressed 27% of INS1 protein degradation (Fig. 6a). The decrease in [14C]phenylalanine release by palmitate was the same whether bafilomycin was present or absent in the media, indicating that the suppression of protein degradation by palmitate was mediated by autophagy.
FIGURE 6.
FFAs suppress long lived protein degradation. Rate of degradation of proteins with long half-life in INS1 cells (a) and in islets (b) treated with or without 0.4 mm palmitate or 0.4 mm oleate for 24 h in the presence or absence of bafilomycin. INS1 cells were pulsed with [14C]phenylalanine for 24 h and then washed and chased in media containing 2 mm nonradioactive phenylalanine for 24 h. The cells were then incubated in either control media or media containing 0.4 mm palmitate or 0.4 mm oleate, in the presence or absence of bafilomycin. The radioactivity was measured in the free amino acids that were in the media as well as in the cellular proteins. Note that in INS1 cells, palmitate just partially suppressed protein degradation mediated by autophagy, although in islets it decreased the rate of degradation to the level obtained with bafilomycin. Error bars, S.E. *, p < 0.05. c, glucose synergizes with fatty acids in decreasing autophagic flux. INS1 cells expressing GFP-LC3 were treated with or without 0.4 mm palmitate or 0.4 mm oleate for 22 h in the presence of 2.5, 5, 10, or 20 mm glucose. After flow cytometry cells were lysed and Western blot for GFP-LC3 was performed (bottom panel). Mean ± S.E., n = 3. d, degradation rate of long lived protein in medium containing 0.4 mm palmitate with either 10 or 20 mm glucose. Error bars, S.E. *, p < 0.05.
To test if suppression of autophagy occurs also in primary β-cells, we examined the effect of FFAs on long lived protein degradation in islets retrieved from mice. Under control conditions, long lived protein degradation in islets was similar to that observed in INS1 cells (5 ± 0.3% per h). Addition of bafilomycin showed that 40% of this degradation is mediated by autophagy. In islets, like in INS1 cells, FFAs significantly suppressed autophagy. Palmitate and oleate decreased protein degradation by 33% after 24 h of incubation (Fig. 6b). Bafilomycin did not further decrease protein degradation in the presence of palmitate, indicating a complete blockage of autophagy by palmitate in primary cells. The combination of oleate and bafilomycin, however, showed a synergistic effect on protein degradation. When combined, oleate and bafilomycin led to a stronger decrease in protein degradation than under oleate or bafilomycin alone, suggesting the blockage of additional degradative pathways besides autophagy.
Glucose Synergizes with Palmitate or Oleate in the Reduction of Autophagic Flux
β-Cells are known to be particularly sensitive to glucose and to the combination of palmitate and glucose, a phenomenon known as glucolipotoxicity, where the two nutrients synergize in the induction of cell death (6). Because accumulation of APs is associated with cell death, we tested if this synergy applies to the impairment of autophagic turnover. INS1 cells expressing GFP-LC3 were treated for 22 h with different concentrations of glucose (2.5, 5, 10, and 20 mm) in the presence or absence of palmitate or oleate and then were subjected to flow cytometry. As shown in Fig. 6c, glucose alone increased the average GFP-LC3 FI in a dose-dependent manner. The average GFP intensity under 20 mm glucose was 50% higher than under 2.5 mm glucose. Palmitate and oleate further increased the intensity of GFP, adding to the effect of glucose. The addition of palmitate amplified GFP intensity under all glucose concentrations but 20 mm, where the intensity was similar to glucose alone. Because palmitate is particularly toxic to the cells when combined with 20 mm glucose (glucolipotoxicity), we assumed that the apparent loss of synergism observed in GFP-LC3 FI assay could have reflected the inclusion of dead cells in the measurement. We therefore measured long lived protein degradation (Fig. 6d), which spare the dead cells, thereby avoiding this drawback. Indeed, protein degradation was strongly decreased in 20 mm glucose with palmitate as compared with 10 mm glucose with palmitate.
Like palmitate, oleate showed an amplification of GFP signal. The GFP FI was higher (up to 40% above the control at 10 and 20 mm glucose) in the presence of oleate at all glucose concentrations as compared with the concomitant glucose concentration without fatty acids. Neither WIPI puncta (see Fig. 2b) nor GFP-LC3 total expression (Fig. 6c) were affected by the addition of glucose, indicating that the increase in GFP-LC3 FI is not due to its content but possibly to the low acidic environment.
Rapamycin Reduces the Blockage of Autophagic Turnover and Rescues the Cells from Glucolipotoxicity
Palmitate does not affect mTOR activity (31), implying that palmitate-induced inhibition of autophagy is mediated via a different pathway than that of rapamycin. We thus hypothesized that rapamycin could alleviate the suppressive effect of palmitate by suppressing the mTOR pathway. As shown in Fig. 7a, rapamycin prevented the effect of palmitate on autophagic flux. When combined with rapamycin, palmitate did not elevate GFP FI as compared with control.
FIGURE 7.
Rapamycin prevents the decrease in autophagic flux and protects from lipotoxicity. a, rapamycin partially reverses palmitate-induced inhibition of autophagic flux. Flow cytometry of cellular GFP intensity in INS1-expressing GFP-LC3 normalized to control. Mean ± S.E., n = 4. b, rapamycin protects cells from palmitate-induced cell death. Flow cytometry analysis of dead cells with propidium iodide (PI); above are images of the cells taken by a Celigo plate imager. Blue stains for the nuclei with Hoechst, and red represents propidium iodide staining; S.E., n = 6. c, rapamycin protects from suppression of glucose-stimulated insulin secretion. Insulin secretion in response to 2 mm (black columns) and 11 mm glucose (white columns) of cells exposed for 18 h to 0.4 mm palmitate in the presence or absence of rapamycin. Error bars, S.E. *, p < 0.05; **, p < 0.01.
Rapamycin induced stimulation of autophagic flux was associated with a significant decrease in cell death under lipotoxicity. When combined with rapamycin, treatment with palmitate reduced cell death by 40% (Fig. 7b).
In addition to their cytotoxic effect, FFAs also suppress glucose-stimulated insulin secretion. We tested whether rapamycin could alleviate this suppression. Exposure to palmitate for 24 h induced a significant decrease in glucose-stimulated insulin secretion (Fig. 7c). Rapamycin alone did not affect insulin secretion significantly. When added to palmitate, rapamycin prevented the decrease in glucose-stimulated insulin secretion observed with palmitate alone.
DISCUSSION
Autophagy has been shown to be essential for β-cell function and survival (9, 10, 12). However, thus far a link between diet-induced diabetes and impaired autophagy has not been demonstrated.
Treatment of β-cells with FFAs and glucose has been previously associated with an increase in AP number (9, 13, 15), which was attributed to an increase in autophagic flux. Although recent advances have provided some evidence questioning this interpretation (18), this study is the first attempt to focus on the long term effect of FFAs on autophagic turnover. Autophagy serves as an essential quality maintenance mechanism, a function that is of particular importance in the β-cell, which is frequently exposed to nutrient-induced oxidative stress while harboring a relatively minor antioxidant machinery (34). Under these circumstances, inhibition of autophagic flux may be of crucial consequences.
In this study, we show that nutrient abundance, which has been associated with various pathologies, suppresses autophagic turnover in the long term, contributing to its toxic effect. The reason for the discrepancy in autophagic flux measurements is likely due to the different approaches employed in the study of autophagic turnover. Autophagic turnover is commonly assessed by treating cells with protease inhibitors while measuring the effect on the levels of LC3-II. Although this approach efficiently uncovers stimulation of AP formation, it cannot serve as a reliable indicator of autophagic degradation unless the system is under steady state during the measurements. Although our results confirm that after 14 h of incubation, palmitate increases AP formation (Fig. 1a), they indicate that autophagic steady state is not reached during the first 24 h of incubation (Fig. 2b).
We provide a series of evidence indicating a long term blockage of autophagy by FFAs as follows: 1) increased cellular GFP-LC3 FI; 2) accumulation of substrates of autophagy, and 3) a decrease in long lived protein degradation. This blockage appears to be due to an impairment in lysosomal acidification and hydrolytic capacity.
Although the kinetics of GFP-LC3 FI in response to palmitate and oleate was remarkably similar (Fig. 3b), there were differences in the effects of the two FFAs on protein degradation, with oleate-blocking protein degradation also in presence of bafilomycin. It is thus possible that unlike palmitate oleate impairs the proteasome system as well as autophagy. This hypothetically could contribute to the higher endoplasmic reticulum stress and higher toxicity observed under palmitate compared with oleate (6, 30).
Palmitate-induced decrease in autophagic flux was associated with accumulation of large APs (Fig. 1a) and large AVOs (Fig. 4d). This finding was reported in β-cells of diabetic patients (18).
A potential explanation for the increase in the number of APs, the decrease of autophagic turnover, and the decrease in endocytosis is the suppression in lysosomal acidification that we observed. Indeed, the staining for acridine orange, LysoTracker red, LysoSensor blue/yellow, and pHrodo-dextran, all pH-sensitive probes, was decreased under palmitate. We have quantified the AVO pH and found it to be increased with palmitate from 4.6 to 5.1. A pH change within this range was also recently reported in cellular models of Alzheimer where it appears to cause accumulation of inactive lysosomes and unfused Aps, which contribute to the pathology (35).
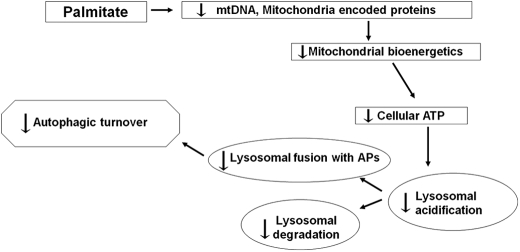
Alkalinization of the lysosomes could account for the decrease in the fusion of the APs with lysosomes, as well as for the suppression of lysosomal hydrolase activity. Although the cause of the impairment in lysosomal acidification has not been addressed directly, a plausible explanation is the observed decrease in cellular ATP. Indeed H+ pump activity is highly sensitive to a decrease in ATP concentrations (36) and so is the capacity of the lysosomes to fuse with APs (37). Reduction in ATP levels may be the result respiratory chain dysfunction reflected by the suppression of oxygen consumption as well as by mitochondrial depolarization. We have previously reported that palmitate induces the arrest of mitochondrial fusion and fragmentation in INS1 cells (23). Others have shown that inhibition of mitochondrial fusion leads to a decrease in mitochondrial DNA levels (38). Furthermore, FFAs have been reported to induce mitochondrial DNA damage in β-cells (39). These abnormalities could account for the observed decrease in mitochondrion-encoded proteins that we report here, which could contribute to reduced oxygen consumption (Fig. 8).
FIGURE 8.
Model for the impairment of autophagy by palmitate. By suppressing mitochondrial bioenergetics, palmitate impairs cellular ATP levels, thus decreasing the lysosomal H+ pump capacity to acidify its lumen, a process required for hydrolase activity and for the fusion of the AP with the lysosome.
Our data suggest that inhibition of autophagic turnover was accompanied by induction of AP formation. This is evidenced by the increase in WIPI-1 puncta and the increase in LC3 in the presence of lysosomal blockers, which is supported by previous studies (9).
Although the combination of increased AP formation along with suppression of autophagic turnover could appear paradoxical, it is plausible that the first is a response to the second. Indeed, in a recent study undertaken on melanoma cells, it was shown that pharmacological inhibition of the lysosomal proton pump induces the formation of APs as a survival mechanism to cope with the stress (40). It is also possible that the inhibition of the lysosomal function by FFAs suppresses nutrient processing, which in turn could trigger AP formation.
Interestingly, we found that both palmitate and oleate decreased GFP intensity during the first 4 h of incubation, suggesting an increase in autophagic flux during that stage. This result supports the recent report that in the short term FFAs induce autophagic protein degradation (17). It is possible that this induction is beneficial to the cell for coping with lipotoxicity. Further study on this effect is required.
The long term effect of palmitate as well as the effect of oleate on GFP intensity in GFP-LC3-expressing cells was found to be synergistic with glucose. Such synergy with glucose is also well known with respect to β-cell death (glucolipotoxicity) (6). The suppression of autophagic turnover as reflected by GFP-LC3 could be alleviated using rapamycin, an mTOR inhibitor; this was accompanied by a protection from lipotoxicity indicating that mTOR could be a target for treating palmitate-induced suppression of autophagy. The impairment of autophagy by nutrient overload described here marks AP maturation as an important impairment contributing to compromised cell function and viability. Inhibition of AP maturation may therefore be playing a role in the pathophysiology of diabetes and may serve as a target for treatment of metabolic diseases.
Acknowledgments
We thank Drs. Barbara Corkey, Jude Deeney, and Andrea Havasi for their helpful advice. We thank Dr. Dani Dagan for this helpful comments on the manuscript.
This work was supported by the Evans Center and the Mitochondria-Affinity Research Collaborative (mtARC) and by National Institutes of Health Grants R01 HL071629-03 and R01 DK074778 and DK035914 (to O. S. S. and B. C. Corkey).
- AP
- autophagosome
- FFA
- free fatty acid
- mTOR
- mammalian target of rapamycin
- FI
- fluorescence intensity
- KRB
- Krebs ringer bicarbonate
- AVO
- acidic vesicular organelle.
REFERENCES
- 1. Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. (1998) Cell Struct. Funct. 23, 33–42 [DOI] [PubMed] [Google Scholar]
- 2. He C., Klionsky D. J. (2009) Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kundu M., Thompson C. B. (2008) Annu. Rev. Pathol. 3, 427–455 [DOI] [PubMed] [Google Scholar]
- 5. Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El-Assaad W., Buteau J., Peyot M. L., Nolan C., Roduit R., Hardy S., Joly E., Dbaibo G., Rosenberg L., Prentki M. (2003) Endocrinology 144, 4154–4163 [DOI] [PubMed] [Google Scholar]
- 7. Zhou Y. P., Grill V. E. (1994) J. Clin. Invest. 93, 870–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poitout V., Robertson R. P. (2008) Endocr. Rev. 29, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., Watada H. (2008) Cell Metab. 8, 325–332 [DOI] [PubMed] [Google Scholar]
- 10. Jung H. S., Chung K. W., Won Kim J., Kim J., Komatsu M., Tanaka K., Nguyen Y. H., Kang T. M., Yoon K. H., Kim J. W., Jeong Y. T., Han M. S., Lee M. K., Kim K. W., Shin J., Lee M. S. (2008) Cell Metab. 8, 318–324 [DOI] [PubMed] [Google Scholar]
- 11. Wu J. J., Quijano C., Chen E., Liu H., Cao L., Fergusson M. M., Rovira I. I., Gutkind S., Daniels M. P., Komatsu M., Finkel T. (2009) Aging 1, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi S. E., Lee S. M., Lee Y. J., Li L. J., Lee S. J., Lee J. H., Kim Y., Jun H. S., Lee K. W., Kang Y. (2009) Endocrinology 150, 126–134 [DOI] [PubMed] [Google Scholar]
- 14. Fujimoto K., Hanson P. T., Tran H., Ford E. L., Han Z., Johnson J. D., Schmidt R. E., Green K. G., Wice B. M., Polonsky K. S. (2009) J. Biol. Chem. 284, 27664–27673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaniuk N. A., Kiraly M., Bates H., Vranic M., Volchuk A., Brumell J. H. (2007) Diabetes 56, 930–939 [DOI] [PubMed] [Google Scholar]
- 16. Lupi R., Dotta F., Marselli L., Del Guerra S., Masini M., Santangelo C., Patané G., Boggi U., Piro S., Anello M., Bergamini E., Mosca F., Di Mario U., Del Prato S., Marchetti P. (2002) Diabetes 51, 1437–1442 [DOI] [PubMed] [Google Scholar]
- 17. Komiya K., Uchida T., Ueno T., Koike M., Abe H., Hirose T., Kawamori R., Uchiyama Y., Kominami E., Fujitani Y., Watada H. (2010) Biochem. Biophys. Res. Commun. 401, 561–567 [DOI] [PubMed] [Google Scholar]
- 18. Masini M., Bugliani M., Lupi R., del Guerra S., Boggi U., Filipponi F., Marselli L., Masiello P., Marchetti P. (2009) Diabetologia 52, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 19. Wikstrom J. D., Katzman S. M., Mohamed H., Twig G., Graf S. A., Heart E., Molina A. J., Corkey B. E., de Vargas L. M., Danial N. N., Collins S., Shirihai O. S. (2007) Diabetes 56, 2569–2578 [DOI] [PubMed] [Google Scholar]
- 20. Bauvy C., Meijer A. J., Codogno P. (2009) Methods Enzymol. 452, 47–61 [DOI] [PubMed] [Google Scholar]
- 21. Kimura S., Fujita N., Noda T., Yoshimori T. (2009) Methods Enzymol. 452, 1–12 [DOI] [PubMed] [Google Scholar]
- 22. Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., Chomicz S., Ferrick D. A. (2007) Am. J. Physiol. Cell Physiol. 292, C125–C136 [DOI] [PubMed] [Google Scholar]
- 23. Molina A. J., Wikstrom J. D., Stiles L., Las G., Mohamed H., Elorza A., Walzer G., Twig G., Katz S., Corkey B. E., Shirihai O. S. (2009) Diabetes 58, 2303–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang N. C., Ho W. M., Chen Y. H., Hu M. L. (2002) Anal. Biochem. 306, 323–327 [DOI] [PubMed] [Google Scholar]
- 25. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Proikas-Cezanne T., Pfisterer S. G. (2009) Methods Enzymol. 452, 247–260 [DOI] [PubMed] [Google Scholar]
- 27. Proikas-Cezanne T., Ruckerbauer S., Stierhof Y. D., Berg C., Nordheim A. (2007) FEBS Lett. 581, 3396–3404 [DOI] [PubMed] [Google Scholar]
- 28. Shvets E., Elazar Z. (2009) Methods Enzymol. 452, 131–141 [DOI] [PubMed] [Google Scholar]
- 29. Shvets E., Fass E., Elazar Z. (2008) Autophagy 4, 621–628 [DOI] [PubMed] [Google Scholar]
- 30. Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. (2006) Endocrinology 147, 3398–3407 [DOI] [PubMed] [Google Scholar]
- 31. Bachar E., Ariav Y., Ketzinel-Gilad M., Cerasi E., Kaiser N., Leibowitz G. (2009) PLoS One 4, e4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koshkin V., Dai F. F., Robson-Doucette C. A., Chan C. B., Wheeler M. B. (2008) J. Biol. Chem. 283, 7936–7948 [DOI] [PubMed] [Google Scholar]
- 33. Koshkin V., Wang X., Scherer P. E., Chan C. B., Wheeler M. B. (2003) J. Biol. Chem. 278, 19709–19715 [DOI] [PubMed] [Google Scholar]
- 34. Lenzen S., Drinkgern J., Tiedge M. (1996) Free Radic. Biol. Med. 20, 463–466 [DOI] [PubMed] [Google Scholar]
- 35. Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., Peterhoff C. M., Wolfe D. M., Martinez-Vicente M., Massey A. C., Sovak G., Uchiyama Y., Westaway D., Cuervo A. M., Nixon R. A. (2010) Cell 141, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamashiro D. J., Fluss S. R., Maxfield F. R. (1983) J. Cell Biol. 97, 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koga H., Kaushik S., Cuervo A. M. (2010) FASEB J. 24, 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen H., Vermulst M., Wang Y. E., Chomyn A., Prolla T. A., McCaffery J. M., Chan D. C. (2010) Cell 141, 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grishko V., Rachek L., Musiyenko S., Ledoux S. P., Wilson G. L. (2005) Free Radic. Biol. Med. 38, 755–762 [DOI] [PubMed] [Google Scholar]
- 40. Marino M. L., Fais S., Djavaheri-Mergny M., Villa A., Meschini S., Lozupone F., Venturi G., Della Mina P., Pattingre S., Rivoltini L., Codogno P., De Milito A. (2010) Cell Death Dis. 1, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]