Background: Constitutive activation of NFκB has been found in various cancers, causing resistance to chemotherapeutic drugs.
Results: Curcumin pretreatment alleviates p65NFκB activation and hence tailors p65NFκB-p300 cross-talk in favor of p53-p300 in drug-resistant cells.
Conclusion: This preclinical study suggests curcumin as a potent chemo-sensitizer to improve the therapeutic index.
Significance: These results suggest that curcumin can be developed into an adjuvant chemotherapeutic drug.
Keywords: Apoptosis, DNA Damage, Drug Resistance, NFκB, p300, p53, Tumor, SMAR1, Curcumin, Doxorubicin
Abstract
Breast cancer cells often develop multiple mechanisms of drug resistance during tumor progression, which is the major reason for the failure of breast cancer therapy. High constitutive activation of NFκB has been found in different cancers, creating an environment conducive for chemotherapeutic resistance. Here we report that doxorubicin-induced SMAR1-dependent transcriptional repression and SMAR1-independent degradation of IkBα resulted in nuclear translocation of p65NFκB and its association with p300 histone acetylase and subsequent transcription of Bcl-2 to impart protective response in drug-resistant cells. Consistently SMAR1-silenced drug-resistant cells exhibited IkBα-mediated inhibition of p65NFκB and induction of p53-dependent apoptosis. Interestingly, curcumin pretreatment of drug-resistant cells alleviated SMAR1-mediated p65NFκB activation and hence restored doxorubicin sensitivity. Under such anti-survival condition, induction of p53-p300 cross-talk enhanced the transcriptional activity of p53 and intrinsic death cascade. Importantly, promyelocyte leukemia-mediated SMAR1 sequestration that relieved the repression of apoptosis-inducing genes was indispensable for such chemo-sensitizing ability of curcumin. A simultaneous decrease in drug-induced systemic toxicity by curcumin might also have enhanced the efficacy of doxorubicin by improving the intrinsic defense machineries of the tumor-bearer. Overall, the findings of this preclinical study clearly demonstrate the effectiveness of curcumin to combat doxorubicin-resistance. We, therefore, suggest curcumin as a potent chemo-sensitizer to improve the therapeutic index of this widely used anti-cancer drug. Taken together, these results suggest that curcumin can be developed into an adjuvant chemotherapeutic drug.
Introduction
Although chemotherapy plays an important role in the treatment of breast cancer, the high percentage of non-responders and of failures after initial responses highlights the critical role played by drug resistance mechanisms in breast cancer management (1). Mechanistically, the resistance phenomena may be explained by (i) mutation or overexpression of drug target proteins and/or (ii) inactivation of drugs by a reduction in uptake or enhanced detoxification and removal of drugs. Doxorubicin, an anthracycline antibiotic, is one of the commonly prescribed chemotherapeutic agents against a wide-spectrum of cancers including breast cancer (2). However, the clinical efficacy and usefulness of doxorubicin-based treatment regimens is still limited because of dose-limiting toxicity and induction of drug resistance (3). Therefore, there is an urgent need to develop new sensitizing agents that enhance the efficacy of doxorubicin and circumvent chemoresistance.
Resistance to the apoptotic effect of doxorubicin is speculated to be multifactorial, involving the activation of nuclear factor κB in cancer cells (4). Coincidently NFκB3 is constitutively active in human breast cancer tissues and breast cancer cell lines (5). Moreover, it is proposed to be one of the early events in breast oncogenesis, as shown by early NFκB DNA binding in neoplastic transformation of mammary cells. Consequently in studies by Montagut et al. (6) activation of NFκB in breast cancer pre-chemotherapy specimens was found to be a predictive factor of chemoresistance. It has been shown that activation of the NFκB pathway renders many types of tumor cells more resistant to chemotherapy presumably via induction of anti-apoptotic proteins (7). Therefore, inhibition of the NFκB has been extensively exploited as a novel approach to sensitize cancers to chemotherapy but has achieved mixed results (7). Therefore, further studies are urgently needed to gain a better understanding of how manipulation of the NFκB pathway regulates breast tumor cell sensitivity to chemotherapy and to identify compounds that suppress the NFκB pathway before a molecular-targeted therapy can be effectively employed for breast cancer treatment.
In contrast to NFκB, the transcription factor p53 is a first-line tumor suppressor induced by stimuli endangering genome integrity (8). The exact regulation of p53-mediated cell cycle arrest or apoptosis is complex and depends on the cellular context and specific stress stimuli (8). Inactivation of the p53 pathway is observed in most human cancers, with mutations in p53 occurring in at least 50% of all tumors (9). Interestingly, in addition to the lack of tumor suppressive functions, p53 mutants gain oncogenic activities contributing to carcinogenesis and drug resistance (10). Considering the deregulation of NFκB and p53 pathways in numerous cancers, it is not surprising that an extensive cross-talk between these pathways exists at various levels. In fact, after chemotherapy-induced DNA damage, NFκB was shown to play a role in neoplastic transformation by inhibiting p53 gene expression (11). Also, NFκB attenuated p53 protein stability by inducing the E3 ubiquitin ligase MDM2 (12). Furthermore, the NFκB gene promoter is activated by p53 mutants, and p52 subunit of NFκB can modulate the promoter activity of p53 target genes (13). Moreover, NFκB and p53 compete for coactivators, for example, the histone acetyltransferases p300 and CBP (14). Interestingly this cross-talk is often biased toward NFκB proteins in drug-resistant tumors (15). An ideal therapeutic approach should, therefore, involve tailoring this cross-talk in favor of p53 to chemo-sensitize drug-resistant tumors. While talking about the competition between NFκB and p53 for “the survival of the fittest,” the possibility of SMAR1 in regulating the signaling cross-talk between NFκB and p53 cannot be ignored. SMAR1, a scaffold matrix-associated region-binding protein, is involved in chromatin-mediated gene regulation. Studies suggest that SMAR1, via p53, is involved in delaying tumor progression in vivo (16). SMAR1 stabilizes p53 by not allowing Mdm2 to bind and export p53 out of the nucleus for proteasome degradation (16). On the other hand, although SMAR1 facilitates nuclear translocation of anti-apoptotic transcription factor, p65NFκB, it inhibits NFκB-dependent transcription of a specific set of NFκB target genes by recruitment of a repressor complex like histone deacetylase (17). Interestingly, SMAR1 is also known to repress p53 target proteins Bax, PUMA, and Noxa while preventing apoptosis (18). Considering such diverse roles of SMAR1 in both inducing and inhibiting apoptosis, an ideal therapeutic approach should, therefore, involve tailoring SMAR1-signaling network against NFκB but essentially in favor of p53 to chemo-sensitize drug-resistant tumors.
Apart from drug resistance, tissue toxicity and immune dysfunctions as induced by doxorubicin most often amplify the problem. The major side effects include immune suppression, hepatotoxicity, neuropathy, alopecia, etc. (19–22). Doxorubicin also causes cardiac toxicity at high dose, and cardiomyopathy may even lead to irreversible congestive heart failure (19–22). A combinatorial therapy that not only shifts the cancer cells from resistance to apoptosis but also prevents systemic toxicity in the cancer patient will, therefore, be the ideal candidate for regressing drug-resistant cancers.
It has been well established that curcumin inhibits NFκB activation and expression of its target genes as induced by diverse agents and anticancer drugs (23, 24). Recently, Sreekanth and co-workers (25) have shown that curcumin could effectively down-regulate survival signals induced by paclitaxel, thereby sensitizing cancer cells toward that drug. According to Choi et al. (26) curcumin down-regulated the multidrug resistance mdr1b gene expression in multidrug-resistant L1210/Adr cells probably due to the suppression of P-glycoprotein expression inhibiting the PI3K/Akt/NFκB pathway. On the other hand, reports from our laboratory have established the involvement of p53 in curcumin-induced cancer cell death (27–30). On the basis of all these reports we hypothesized that by shifting of cellular microenvironment, curcumin may down-regulate the NFκB survival pathway and promote p53 apoptotic signal thereby sensitizing the drug resistance breast cancer cells to doxorubicin. Because curcumin also ameliorates immunosuppression and inhibits systemic toxicity in the tumor bearer (31–34), combinatorial application of this plant product with doxorubicin may also prevent systemic toxicity in the tumor bearer besides shifting the cancer cells from resistance to apoptosis.
To prove our hypothesis we utilized two experimental systems, (i) an in vitro mammary epithelial carcinoma cell model in which the molecular mechanisms can be verified and (ii) an in vivo mammary carcinoma-bearing mouse model that better reflects the molecular complexity of patient-derived tumor specimens. Here we report that curcumin sensitizes drug-resistant breast tumors to doxorubicin by inhibiting the NFκB-mediated defense pathway and activating p53 apoptotic signaling. Inhibition of p65NFκB by curcumin was both SMAR1-dependent and -independent. In fact, inactivation of the NFκB pathway by curcumin rescued p300 from p65NFκB and launched p53-p300 collaboration to induce p53-dependent Bax, PUMA, and Noxa transactivation and instigation of downstream mitochondria-dependent death cascade in drug-resistant breast cancer cells. Interestingly for induction of p53-dependent apoptosis, curcumin-mediated execution of PML-SMAR1 cross-talk was indispensable. A simultaneous decrease in drug-induced systemic toxicity might also have enhanced the efficacy of doxorubicin by improving the intrinsic defense machineries of the tumor bearer. Therefore, curcumin in combination with standard chemotherapeutics may serve as a double-edged sword in culminating both resistance and toxicity after chemotherapy.
EXPERIMENTAL PROCEDURES
Animal and Tumor Model
Swiss albino mice (NCLAS, Hyderabad, India) weighing 23–25 g were maintained in a temperature-controlled room with a light dark cycle. To determine the combinatorial therapeutic efficacy of curcumin and doxorubicin, 1 × 106 doxorubicin-resistant Ehrlich ascites carcinoma (EAC) cells (obtained from Choudhuri and Chatterjee (35)) were peritoneally injected in mice, and cancer cells were allowed to grow to 7 days. Then the mice were divided in to 7 groups (10 mice in each group) and treated as follows: 1) vehicle control, 2) LD50 doxorubicin (5 mg/kg body weight), 3) LD50 curcumin (50 mg/kg body weight), 4) ½LD50 doxorubicin (2.5 mg/kg body weight), 5) ½LD50 curcumin (25 mg/kg body weight), 6) LD50 doxorubicin + LD50 curcumin, and 7) ½LD50 doxorubicin + ½LD50 curcumin. A single dose of doxorubicin was given intraperitoneally on the seventh day, and starting from that day curcumin was fed orally every alternate day (up to 21 days). Then, after sacrificing the mice, cell count and cell cycle experiments were carried out. Initially, for the estimation of the LD50 dose of doxorubicin and curcumin, mice bearing sensitive tumors were treated with different doses of doxorubicin and curcumin, and the dose at which tumor burden was reduced to half that of untreated animals was considered to be the respective LD50 dose that in the present case was 5 mg/kg body weight and 50 mg/kg body weight for doxorubicin and curcumin respectively (supplemental Table 1). All animal experiments were performed following the principles of laboratory animal care (NIH publication No. 85-23, revised in 1985) as well as Indian laws on protection of animals under the prevision of authorized investigator.
Cell Culture and Treatments
EAC cells were collected from the peritoneal cavity of tumor-bearing mice and freed from adherent cells. More than 98% of the non-adherent population was found to be CD16-negative, among which >92% were characterized by Wright staining (34). The cells were routinely maintained in complete DMEM or RPMI 1640 at 37 °C in a humidified incubator containing 5% CO2 (36). Cells were allowed to reach confluence before use. Viable cell numbers were determined by a trypan blue exclusion test. Cells were treated with specified doses of doxorubicin and curcumin for definite time intervals. During chemosensitization with curcumin in vitro, cells were pretreated for 2 h with curcumin followed by doxorubicin treatment for 24 h. To inhibit mitochondrial pore formation, cells were treated with cyclosporine A 25 μm before 1 h of drug treatment. While determining the toxicity of doxorubicin, curcumin or the combinatorial doses, thymus, spleen, and femurs from drug-treated tumor-bearing animals were removed after 21 days of tumor inoculation, and a single cell suspension of thymocytes, lymphocytes, and bone marrow progenitor cells was made and suspended in RPMI 1640 medium (Sigma). Splenic lymphocytes were purified by Ficoll gradient (Sigma) centrifugation. Macrophages were allowed to adhere at 37 °C for 1 h. Viable cells were counted by a trypan blue dye exclusion test (31–34).
Plasmids, siRNA, and Transfections
The expression constructs (pcDNA3.0/HA-tagged IκBα-32A/36A (IκBα super-repressor (IκBα-SR), a kind gift from Dr. J. Didonato, The Cleveland Clinic Foundation), pcDNA3.1-p65NFκB/p53/Bcl-2 vectors (2 μg/million cells), or SMAR1-shRNA (300 pmol/million cells) were introduced into exponentially growing cancer cells using Lipofectamine 2000 (Invitrogen) according to the protocol provided by the manufacturer. Stably expressing clones were isolated by limiting dilution and selection with G418 sulfate (400 μg/ml; Sigma), and G418-resistant cells were cloned and screened by Western blotting and RT-PCR. For endogenous silencing of specific genes, cells were transfected with 300 pmol of control/p65NFκB-/p53-/Bax-siRNA (Santa Cruz) using Lipofectamine 2000 separately for 12 h (37). The mRNA and protein levels were determined by RT-PCR and Western blotting.
Flow Cytometry
For the determination of apoptosis, control and treated cells were stained with 7AAD and annexin V-PE (BD Pharmingen) and analyzed on flow cytometer (FACSCalibur, BD Biosciences). Electronic compensation of the instrument was done to exclude overlapping of the emission spectra. A total of 10,000 events were acquired for analysis using CellQuest software (BD Biosciences). annexin V/7AAD-positive cells were regarded as apoptotic cells (37, 38). Cell cycle phase distribution of nuclear DNA was determined using a fluorescence detector equipped with 488-nm argon laser light source and 623-nm band pass filter (linear scale) and CellQuest software. Ten thousand total events were acquired, and a histogram display of DNA content (x axis, propidium iodide fluorescence) versus counts (y axis) has been displayed. CellQuest software was employed to quantitate the data at different phases of the cell cycle. For determination of mitochondrial transmembrane potential, control and treated cells were harvested, washed twice with PBS, and incubated with 40 nm 3,3′-dihexyloxacarbocyanine iodide (DiOC6) in serum-free RPMI 1640 and incubated for 15 min at 37 °C in dark. The cells were analyzed flow cytometrically for 3,3′-dihexyloxacarbocyanine iodide fluorescence.
Immunoblotting and Co-immunoprecipitation
Cells were lysed in buffer (10 mm Hepes, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, and 0.5 mm DTT) and spun at 3300 × g to get cell lysates. The pellet was resuspended in buffer (20 mm Hepes, pH 7.9, 0.4 m NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, and 0.5 mm DTT) and spun down at 12,000 × g for 30 min to get nuclear fraction. For whole cell lysates, cells were resuspended and homogenized in buffer (100 mm Tris-Cl, pH 7.4, 300 mm NaCl, 1% Nonidet P-40, and 0.25% sodium deoxycholate). All the buffers were supplemented with protease and phosphatase inhibitor mixtures (39–41). For direct Western blot analysis, the cell lysates or the particular fractions were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with specific antibodies, e.g. anti-p53, -Ac-p53, -Bcl2, -Bax, -PUMA, -Noxa, -p65NFκB, -IκBα, -caspase-9 and -3, -cytochrome c, -PML, -p300, -SMAR1, produced from Santa Cruz; thereafter the immunoblots were visualized by chemiluminescence. Equal protein loading was confirmed with α-actin/histone-H1/manganese superoxide dismutase antibody (Santa Cruz). For the determination of direct interaction between two proteins, a co-immunoprecipitation technique was employed (36). p53-p300, p65NFκB-p300, and PML-SMAR1 interaction was determined by co-immunoprecipitation. Samples (300 μg of protein from the total lysate) were incubated at 4 °C overnight with anti-p53/-p65/-PML/-IgG antibody and then incubated for 2 h at 4 °C with protein A-Sepharose. Immunocomplexes were washed of unbound proteins with cold TBS with protease inhibitors, and pelleted beads were boiled for 5 min in SDS-PAGE sample buffer. The immunoprecipitated proteins were resolved on SDS-PAGE and analyzed by Western blotting for detection of associated proteins. Equal protein loading was confirmed using α-actin as an internal control.
Enzyme Assay
Serum glutamate pyruvate transaminase, glutamate oxaloacetate transaminase, and alkaline phosphatase were estimated according to standard protocol.
Reverse Transcriptase and Real-time Quantitative RT-PCR Analysis
Two μg of the total RNA each from doxorubicin/curcumin-treated doxorubicin-sensitive/-resistant cells was extracted by TRIzol (Invitrogen) and reverse-transcribed and then subjected to PCR with enzymes and reagents of the RTplusPCR system (Eppendorf, Hamburg, Germany) using GeneAmpPCR 2720 (Applied Biosystems Foster City, CA) (39, 40). The cDNAs were amplified with primers specific for Bax (5′-GGAATTCCAAGAAGCTGAGCGAGTGT-3′/5′-GGAATTCTTCTTCCAGATGGTGAGCGAG-3′), PUMA (5′-AGCTCCCATCCTGGCTCTGG-3′/5′-CAGGCAGTTGTCAGCTGGG-3′, Noxa (5′-ACTGTGGTTCTGGCGCAGAT-3′/5′-TGAGCACACTCGTCCTTCAAGT-3′), Bcl-2 (5′-CCTGTGCCACCATGTGTCCATC-3′/5′-GCTGAGAACAGGGTCTTCAGAGAC-3), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; internal control, 5′-TGATGACATCAAGAAGGTGGTGAAG-3′/5′-TCCTTGGAGGCCATGTAGGCCAT-3). For quantitative PCR, total RNA was prepared from 50 mg of heart ventricular tissue using Trizol (Invitrogen) isolation reagent. Different mRNA expression levels in mice ventricular tissue were measured by quantitative real-time PCR using Quanti TechTM SYBR Green real-time PCR kit and the iCycler real-time detection system and software according to manufacturer's instruction. To check B-type natriuretic peptide (BNP) (NCBI GenBankTM accession number M 25297) and GAPDH (NCBI GenBankTM accession number XM-216453), expression oligonucleotide primers are designed based on published information (42). Passive reference dye (ROX) was used to normalize the SYBR Green/double-stranded DNA complex signal during analysis to correct for well-to-well variation and sampling loading error. Amplification products using SYBR Green detection were checked using a melting curve with iCycler software (Version 3; Bio-Rad) and by 1.5% agarose gel electrophoresis to confirm the size of the DNA fragment and that single product was formed. Samples were compared using the relative (comparative) Ct method. The Ct value, which is inversely proportional to the initiate template copy number, is the calculated cycle number where the fluorescence signal emitted is significantly above background levels. Expression levels of the housekeeping gene, GAPDH, were used to normalize for variations in amount of RNA and RNA purity. The -fold induction or repression by real-time RT-PCR was calculated according to following formula: -fold change = 2−ΔΔCt, where ΔΔCt = ΔCt control − ΔCt treatment, and ΔCt = target gene Ct − GAPDH Ct. Oligonucleotide primer sequences used in real time quantitative RT-PCR were as follows: BNP sense (5′-TGGGAAGTCCTAGCCAGTCTC-3′) and antisense, (5′-GCCGATCCGGTCTATCTTCT-3′); GAPDH sense (5′-GCCATCAACGACCCCTTC-3′) and antisense (5′-AGCCC-CAGCCTTCTCCA-3′).
Chromatin Immunoprecipitation
ChIP assays were carried out using a ChIP assay kit (Millipore) according to a modification of the manufacturer's instructions. 2 × 106 cells were fixed with 1% formaldehyde for 10 min at 37 °C to cross-link the protein-DNA complexes. Then they were harvested and washed twice with ice-cold PBS containing protease inhibitors. Cells were added to SDS lysis buffer and incubated on ice for 10 min. Cell lysates were sonicated to shear the DNA to lengths between 200 and 1000 base pairs and then centrifuged at 13,000 rpm for 10 min at 4 °C. The sonicated cell supernatants were diluted 10-fold in ChIP dilution buffer and precleared with protein A-agarose/salmon sperm DNA for 30 min at 4 °C with agitation. The supernatant was recovered after pelleting the agarose by centrifugation and then incubated with specific antibody against p53/p65NFκB overnight at 4 °C. The antibody-protein-DNA complexes were collected by adding protein A-agarose/salmon sperm DNA for 1h at 4 °C with rotation. Immunoprecipitated antibody-protein-DNA complexes were washed according to described protocol. Chromatin complexes were eluted with freshly prepared extraction buffer (1% SDS, 0.1 m NaHCO3). To reverse cross-links, 5 m NaCl was added to each eluate, and then the solution was heated to 65 °C for 5 h. Proteins were digested with 10 mg/ml proteinase K for 1 h at 45 °C, and DNA was recovered by phenol/chloroform extraction and ethanol precipitation. DNA fragments were amplified by PCR. The sequences of promoter-specific primers are as follows: Bax forward primer 5′-TCAGCACAGATTAGTTTCTG-3′, Bax reverse primer 5′-GGGATTACAGGCATGAGCTA-3′; PUMA forward primer 5′-GATTACAGGCATGCGCCACA-3′, PUMA reverse primer 5′-ACCCACACTGATGATCACA C-3′; Noxa forward primer 5′-TTTTCTGGGCTTGTTTAC CC-3′, Noxa reverse primer 5′-TACAAAACGAGGTGG GAGGA-3′.
Histological Studies
Histological studies of liver and heart tissue were carried out after curcumin and doxorubicin administration. At the third week of treatment, the liver and heart were removed from each animal and fixed in 10% buffered formalin. The tissues were then dehydrated in graded alcohol and embedded in paraffin. Sections were obtained, and standard staining techniques using hematoxylin and eosin were used to detect morphological changes.
Statistical Analyses
Values are shown as mean ± S.E. except where otherwise indicated. Comparison of multiple experimental groups was performed by 2-way analysis of variance followed by a post hoc Bonferroni multiple comparison test. Data were analyzed, and when appropriate significance of the differences between mean values was determined by Student's t test. Results were considered significant at p < 0.05.
RESULTS
Curcumin Effectively Sensitizes Doxorubicin-resistant Ascites Carcinoma Cells to Apoptosis in Vitro
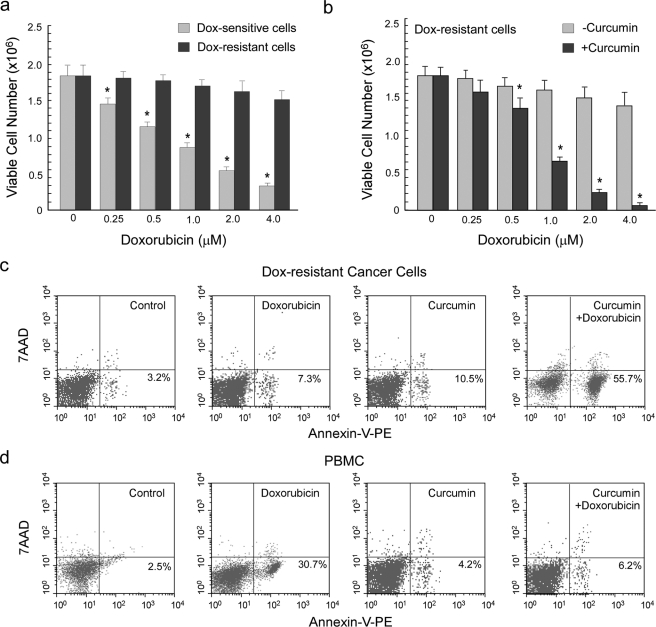
Results of Fig. 1a depict that in comparison to sensitive EAC cells, increasing concentrations of doxorubicin (0–4 μm) had minimal effect on viability of doxorubicin-resistant carcinoma cells, as observed by trypan blue dye exclusion assay. Therefore, to chemosensitize doxorubicin-resistant cells, they were pretreated with LD50 (10 μm) curcumin for 2 h followed by exposure to doxorubicin (0–4 μm) and then subjected to determination of cell viability. Interestingly, it was observed that, in contrast to doxorubicin treatment alone, combination of doxorubicin and curcumin at different dose ratios was highly effective in significantly reducing cell viability in drug-resistant cells (Fig. 1b). Indeed treatment with a combinatorial regimen, i.e. 1 μm doxorubicin and 10 μm curcumin, which are the LD50 values for sensitive EAC cells, demonstrated significant (50–60%) annexin V positivity of drug-resistant cells, thereby confirming apoptosis to be the mode of cell death underlying curcumin-mediated chemo-sensitization (Fig. 1c). On the other hand, LD50 doxorubicin or curcumin alone failed to induce any significant apoptosis (Fig. 1c). To establish the non-toxic nature of the combinatorial regimen toward normal cells of the host, we treated peripheral blood mononuclear cells collected from normal mice with different combinations of doxorubicin and curcumin. Our annexin V-PE/7AAD double-labeling assay revealed that the combination of LD50 curcumin and LD50 doxorubicin, although significantly apoptotic for resistant cells, furnished minimal toxic effect to peripheral blood mononuclear cells (Fig. 1d). We therefore, have used this combinatorial dose for further studies.
FIGURE 1.
Treatment of drug-resistant ascites carcinoma cells with curcumin restored their sensitivity toward doxorubicin. a, 1 million doxorubicin-sensitive and 1 million doxorubicin-resistant ascites carcinoma cells were treated with a dose range (0–4 μm) of doxorubicin for a time interval of 24 h and subjected to determination of cell viability test by trypan blue dye exclusion assay. b, to assay the in vitro chemosensitizing potential of curcumin, doxorubicin-resistant cells were pretreated with 10 μm curcumin for a period of 2 h followed by 24 h of doxorubicin treatment. Doxorubicin-resistant cells (c) and peripheral blood mononuclear cells (PBMC; d) were treated with doxorubicin (1 μm) and curcumin (10 μm) separately or in combination and subjected to flow cytometric determination of percentage apoptosis by annexin V-PE/7AAD binding. Values are the mean ± S.E. of five independent experiments or are representative of a typical experiment in the case of flow cytometric analysis. *. p < 0.05 when compared with respective control sets.
These results not only highlight the role of curcumin in sensitizing doxorubicin-resistant carcinoma cells but also point toward the effectiveness of the combinatorial therapy in rejuvenating the “forgotten” apoptotic program in drug-resistant cancer cells along with minimizing the side effects of drug-induced toxicity.
Curcumin Chemosensitizes Drug-resistant Cancer Cells by Inhibiting Doxorubicin-induced Nuclear Translocation of NFκB via SMAR1-dependent and -independent Mechanisms
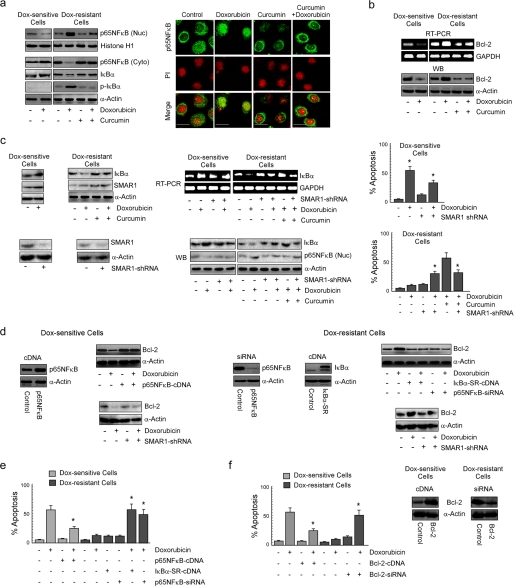
Because p65NFκB has been reported to be globally involved in tumor drug resistance whereas curcumin is known to inhibit NFκB activation, we examined whether this plant flavonoid suppresses the NFκB pathway to combat NFκB-mediated chemoresistance, thereby sensitizing drug-resistant breast cancer cells. Our search revealed that in contrast to sensitive cells, doxorubicin was found to enhance nuclear translocation of p65NFκB at early time point (1 h) in resistant carcinoma cells (Fig. 2a). Interestingly, curcumin pretreatment efficiently blocked such translocation (Fig. 2a) as observed by both Western blot and confocal imaging experiments. In addition, the mRNA and protein levels of the NFκB-target gene, Bcl-2, were found to be up-regulated by doxorubicin alone in drug-resistant cells, which could be efficiently blocked by the combinatorial regimen (Fig. 2b). However, in sensitive cells doxorubicin alone inhibited Bcl-2 expression (Fig. 2b). A search for underlying mechanisms revealed that curcumin prevented drug-induced phosphorylation and degradation of IκBα, as observed after 1 h of drug treatment (Fig. 2a).
FIGURE 2.
Curcumin inhibits drug-induced NFκB activation and Bcl-2 up-regulation maneuvering SMAR1 to induce apoptosis of doxorubicin-resistant cancer cells. a, doxorubicin-treated drug-sensitive cells and doxorubicin/curcumin alone or in combination-treated drug-resistant cells were subjected to isolation of nuclear and cytosolic fractions for Western blot (WB) analysis or to confocal microscopy and evaluated for nuclear localization of p65NFκB after 1 h of doxorubicin treatment. At the same time cell lysates from the same experimental set up was verified for IκBα and phospho-IκBα levels. The concentration of doxorubicin and curcumin were 1 and 10 μm, respectively. Magnification bars in confocal microscopy images indicate 20 μm. b, RT-PCR and Western blot images depict expression of Bcl-2 at 24 h in the same set of experiments. c, at the same time the lysates from the previous experimental sets were screened for IκBα and SMAR1 levels. To study the role of SMAR1 in regulation of IκBα expression, SMAR1-shRNA-transfected drug-sensitive or drug-resistant cells were treated with doxorubicin or in combination with curcumin to follow the expression of IκBα by RT-PCR and Western blot analysis. In parallel, a portion of the same set of cells was subjected to Western blot analysis to determine nuclear translocation of p65NFκB, and the remaining portion was screened for percentage apoptosis. d, p65NFκB-cDNA-/SMAR1-shRNA-transfected drug-sensitive and IκBα-SR-cDNA-/p65NFκB-siRNA-/SMAR1-shRNA-transfected drug-resistant cells were treated with doxorubicin for 24 h and explored for Bcl-2 expression by Western blot analysis. e, flow cytometric determination of doxorubicin-induced apoptosis in p65NFκB-cDNA-transfected drug-sensitive and IκBα-SR-cDNA-/p65NFκB-siRNA-transfected drug-resistant cells is shown. f, similarly, Bcl-2-cDNA-transfected drug-sensitive cells and Bcl-2-siRNA-transfected drug-resistant cells were evaluated for doxorubicin sensitivity by measuring annexin V-PE/7AAD positivity. GAPDH and α-actin/histone H1 were used as internal loading controls for Western blot and RT-PCR. Values are the mean ± S.E. of five independent experiments in each case or are representative of a typical experiment in the case of RT-PCR analysis and Western blots. Confocal images shown are representative of >10 images taken in different fields from two independent experiments. *, p < 0.05 when compared with respective untransfected/control sets.
Apart from IκBα degradation preceding NFκB activation, earlier reports by Singh et al. (17) demonstrated that SMAR1 also induces the nuclear accumulation of p65NFκB. They showed that SMAR1 in association with p65NFκB could inhibit transcription of IκBα as well as other NFκB target genes. However, in our system, because NFκB activation coincided with Bcl-2 induction, it may be relevant to hypothesize that SMAR1-dependent repression of p65NFκB-target genes was not applicable for Bcl-2. This can be further supported by the fact that bclxl and xiap promoters lack the presence of Matrix Attachments Regions (as predicted by the MARWIZ software (17). On the other hand, although doxorubicin reduced IκBα levels by inducing IκBα phosphorylation (during early time period; 1 h), the possibility of SMAR1-mediated IκBα repression during a later time period (24 h) and hence sustained p65NFκB activation cannot be ruled out. Therefore, we next investigated the role of SMAR1 in IκBα repression. We indeed observed that doxorubicin induced SMAR1 in sensitive and resistant cells; however, -fold induction in sensitive cells was significantly higher than in resistant cells, but curcumin pretreatment of resistant cells augmented doxorubicin-mediated SMAR1 induction (Fig. 2c), suggesting that networks regulating SMAR1 are differentially regulated in apoptosis versus resistance responses. Interestingly, we observed that in comparison to sensitive cells, doxorubicin reduced IκBα expression (24 h) in resistant cells, the effect of which was significantly nullified in cells transfected with SMAR1-shRNA (Fig. 2c). However, in curcumin-pretreated resistant cells, doxorubicin failed to reduce IκBα expression, as was the case with doxorubicin-treated sensitive cells (Fig. 2c). In fact in these cells SMAR1 knockdown reduced IκBα levels, suggesting differential function of SMAR1 in resistant and sensitive cells. In light of these findings it may not be out of context to state that the differential SMAR1 expression status in sensitive/resistant cells undergoing genotoxic damage could probably be one of the reasons or effects underlying such discrepancies of SMAR1 signaling. Consistently SMAR1 knock-out-resistant cells were partially sensitive to doxorubicin-mediated apoptosis but resistant to combination treatment, and sensitive cells in absence of SMAR-1 became partially resistant to doxorubicin (Fig. 2c). However, it is to be noted that SMAR1 repression alone induced IκBα expression irrespective of the type of cell (Fig. 2c), which can be justified by the fact that SMAR1, independent of NFκB, can also repress IκBα (17).
Because SMAR1-dependent or -independent activation of NFκB appeared to be the major chemoresistance pathway, we attempted to explore the consequences of pathway modulation on the expression of chemo-resistance factor Bcl-2. To this end we observed that transfecting resistant cells with super repressor IκBα-SR-cDNA or p65NFκB-siRNA decreased Bcl-2 followed by significant apoptosis in response to doxorubicin (Fig. 2, d and e). On the other hand, sensitive cells expressing p65NFκB-cDNA manifested enhanced Bcl-2 with significant resistance upon doxorubicin exposure (Fig. 2, d and e). Interestingly, SMAR1 silencing in resistant cells reduced doxorubicin-induced Bcl-2 up-regulation, in harmony with reduced nuclear expression of p65NFκB, whereas its absence augmented Bcl-2 levels in sensitive or resistant cells treated with doxorubicin or a combination of curcumin and doxorubicin, respectively (Fig. 2, c and d). The anti-apoptotic role of NFκB-dependent Bcl-2 up-regulation in drug resistance was confirmed by evaluating drug sensitivity of Bcl-2-engineered cells. We observed that transfection of resistant cells with Bcl-2-siRNA efficiently reverted drug resistance, whereas overexpression of Bcl-2 in sensitive cells bestowed them with doxorubicin resistance (Fig. 2f). Collectively, these results confirmed that drug-induced p65NFκB activation and Bcl-2 up-regulation were primarily involved in chemoresistance, which upon inhibition by curcumin were sensitized to drug-induced apoptosis.
Inhibition of p65NFκB and Induction of PML-SMAR1 Cross-talk by Curcumin Trigger p53-mediated Apoptosis in Drug-resistant Cells
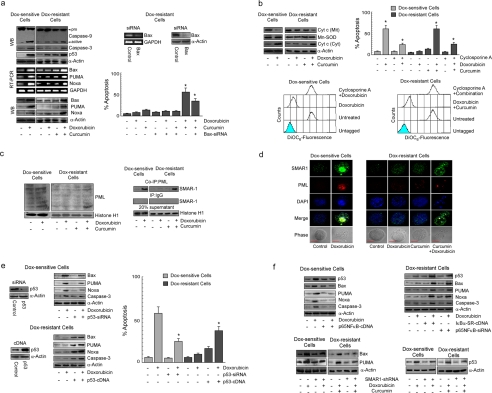
Because inhibition of p65NFκB activity in resistant cells induced a powerful apoptotic response, we predicted the involvement of the cellular apoptotic proteins during curcumin-mediated chemosensitization. At this end we evaluated the status of apoptotic proteases, i.e. caspase-9 and caspase-3, in response to curcumin-doxorubicin combinatorial treatment. It was noted that in comparison to curcumin or doxorubicin treatment alone, resistant cells undergoing combinatorial therapy manifested significantly up-regulated levels of cleaved caspase-9 and caspase-3 (Fig. 3a). In contrast, doxorubicin treatment alone could significantly increase both caspase-9 and caspase-3 in sensitive cells (Fig. 3a). The above results tempted us to compare the p53 activation status upon doxorubicin exposure in both the sensitive and resistant cells. Results of Fig. 3a revealed that doxorubicin induced p53 in both sensitive and resistant cells; however, p53 expression in resistant cells was slightly less when compared with sensitive cells (Fig. 3a). Interestingly, although doxorubicin alone could not induce Bax expression in resistant cells when compared with sensitive cells (Fig. 3a), combinatorial therapy, which elevated p53 expression only slightly more than doxorubicin treatment, showed significantly elevated Bax levels, silencing, which reversed the chemosensitizing ability of curcumin (Fig. 3a). Other apoptotic targets of p53 like PUMA and Noxa also revealed similar expression patterns (Fig. 3a). Elevated levels of these proteins were consistent with an increase in cytosolic cytochrome c in sensitive- and resistant cells treated with doxorubicin or combination treatment, respectively. Consistently, like doxorubicin treatment in sensitive cells, curcumin pretreatment in resistant cells enabled doxorubicin-induced loss of mitochondrial transmembrane potential, whereas cyclosporine A treatment partly ablated such mitochondrial transmembrane potential loss, suggesting the existence of mitochondria-independent apoptotic pathways as well (Fig. 3b).
FIGURE 3.
Curcumin-mediated NFκB inhibition triggers p53-dependent Bax activation in doxorubicin-resistant cancer cells. a, sensitive/resistant cells were treated with doxorubicin alone or in combination with curcumin for 24 h and Western-blotted (WB) for the evaluation of (pro)active-caspase-9 and caspase-3. In parallel, p53 levels in the nuclear fractions was also determined. Expression levels of Bax, PUMA, and Noxa in the same experimental set were determined by RT-PCR and Western blot analysis. Control/Bax-siRNA-transfected resistant cells were treated with doxorubicin alone or in combination with curcumin, and percent apoptosis was determined by annexin V-PE/7AAD positivity. b, sensitive/resistant cells were treated with doxorubicin alone or in combination with curcumin and were subjected to isolation of cytosolic and mitochondrial fractions and Western-blotted for cytochrome c (Cyt c). In parallel, flow cytometry was used to determine mitochondrial transmembrane potential and percentage apoptosis in doxorubicin/combination dose-treated sensitive/resistant cells in the presence and absence of cyclosporine A. Mn-SOD, manganese superoxide dismutase antibody. c, lysates of sensitive/resistant cells treated with doxorubicin alone or in combination with curcumin for 24 h were Western-blotted with anti-PML antibody or immunoprecipitated with PML/IgG, and the immunoprecipitates (co-IP) were Western-blotted with SMAR1. d, confocal images depict co-localization of SMAR1 with PML in sensitive/resistant cells treated with doxorubicin alone or in combination with curcumin, respectively. Bar length in confocal microscopy images indicate 10 μm (e) p53-siRNA-transfected sensitive cells, and p53-cDNA-transfected resistant cells were treated with doxorubicin and evaluated for Bax, PUMA, Noxa, and caspase-3 levels at 24 h by Western blot and percent apoptosis by flow cytometry. f, IκBα-SR-cDNA-/p65NFκB-siRNA-transfected resistant cells and p65NFκB-overexpressed sensitive cells were evaluated for change in p53, Bax, PUMA, Noxa, and caspase-3 levels at 24 h. SMAR1-shRNA-transfected sensitive/resistant cells were treated with doxorubicin alone or in combination with curcumin, respectively, and the expression of p53 at 8 h or Bax and PUMA at 24 h was assayed by Western blot analysis. GAPDH and α-actin were used as internal loading controls for RT-PCR and Western blot. Values are the mean ± S.E. of five independent experiments in each case or is representative of a typical experiment in the case of RT-PCR and Western blot analysis. Confocal images shown are representative of >10 images taken in different fields from two independent experiments. *, p < 0.05 when compared with respective untransfected/control sets.
Although our earlier results suggested the role of SMAR1 in NFκB-mediated drug resistance (Fig. 2b), our parallel findings of increased expression of SMAR1 in cells undergoing apoptosis (doxorubicin-treated sensitive cells or combination-treated resistant cells) when compared with cells resisting apoptosis (doxorubicin-treated resistant cells) (Fig. 2b) questioned the role of SMAR1 in apoptosis as well. Based on the findings of Sinha et al. (18), who elaborated SMAR-1-mediated repression of Bax or PUMA, it may not be erroneous to postulate that SMAR1 knockdown may amplify the already existing apoptotic response. However, in our study we observed increased expression of Bax or PUMA despite the expression of SMAR1, which suggested other mechanisms to be involved in surpassing SMAR1-mediated repression of apoptosis. Consistently, we observed that sensitive cells treated with doxorubicin or resistant cells treated with a combination dose exhibited nuclear accumulation of PML, whereas in doxorubicin-treated resistant cells PML expression was not significantly visual (Fig. 3c). Immunoprecipitation studies further showed that SMAR1 was co-localized in PML bodies of sensitive cells treated with doxorubicin or resistant cells treated with a combination dose. This suggested that the possibility of SMAR1-mediated repression of Bax and PUMA was ameliorated by PML that sequestered SMAR1 to facilitate transcription of p53 apoptotic targets (Fig. 3c).
In a parallel experiment, transfecting resistant cells with p53-cDNA increased Bax, PUMA, and Noxa expression and caspase-3 activation upon doxorubicin treatment, subsequently enabling doxorubicin-induced apoptosis, whereas transiently silencing p53 in sensitive cells made them significantly resistant to doxorubicin (Fig. 3d). Because doxorubicin failed to induce p53 transcriptional function in resistant cells, which otherwise could be maneuvered by deliberately increasing p53 levels, it seemed logical to hypothesize that increasing p53 in resistant cells restored p53 transcriptional functions and, therefore, drug sensitivity. However, this assumption appeared to be incompletely true, as in resistant cells undergoing combinatorial therapy, p53 expression was only slightly more than in doxorubicin-treated cells, Bax expression in these cells unexpectedly surpassed that of cells treated with doxorubicin. This raised the possibility of the involvement of the p53 transcriptional inhibitor(s) in drug-treated resistant cells that somehow opposed p53-dependent transcription of apoptotic genes. Curcumin, on the other hand, by restraining this “inhibitor” might have activated the p53-transcriptional program. Because our previous results have demonstrated curcumin-induced inhibition of NFκB activation, we hypothesized that doxorubicin-activated NFκB might block p53-dependent apoptotic program in resistant cells. To confirm this hypothesis we utilized IκBα-SR-cDNA/p65NFκB-siRNA-transfected resistant cells and checked p53-dependent execution of apoptosis in these cells on doxorubicin treatment. Indeed these transfectants not only manifested NFκB inhibition, as mentioned earlier (Fig. 2c), but also displayed robust p53 induction along with up-regulation of Bax, PUMA, and Noxa (Fig. 3e). Activation of caspase-3 in these cells (Fig. 3e) finally confirmed that drug-induced NFκB intervened the functioning of p53-dependent apoptotic program. Consistently, in sensitive cells overexpression of p65NFκB resisted up-regulation of p53 followed by inhibition of apoptotic targets upon doxorubicin treatment (Fig. 3e). In addition to NFκB, SMAR1 also appeared to be an inhibitor of p53 as SMAR1-mediated repression of p53 target genes is well documented (18), but its inhibitory function must have been negated by PML-SMAR1 cross-talk as observed in our experiments. This suggests that silencing SMAR-1 might not significantly affect apoptosis in sensitive cells treated with doxorubicin or resistant cells treated with a combination dose. However, contrasting our hypothesis, we observed that SMAR1-silenced sensitive or resistant cells manifested partially reduced levels of Bax and PUMA when treated with doxorubicin or combinatorial therapy, respectively, which also explains why these cells became partially resistant under these conditions (Fig. 2c). This indicated that instead of resisting apoptosis, SMAR1 under these conditions was indeed involved in triggering apoptosis. A more detailed study revealed that in sensitive cells treated with doxorubicin or resistant cells treated with a combination dose, SMAR1 silencing partly reduced p53 levels during the early hours of drug treatment (8 h) when compared with untransfected cells (Fig. 3f). Briefly, this suggests that in cells marked to undergo apoptosis, SMAR1 during the initial hours is required for p53 activation and during later hours is sequestered by PML to nullify its apoptosis inhibiting properties. All these results together signified that curcumin, by modulating the SMAR1 signaling network, inhibited NFκB and skewed the cellular microenvironment in favor of p53-transcriptional activation to result in doxorubicin-induced apoptosis.
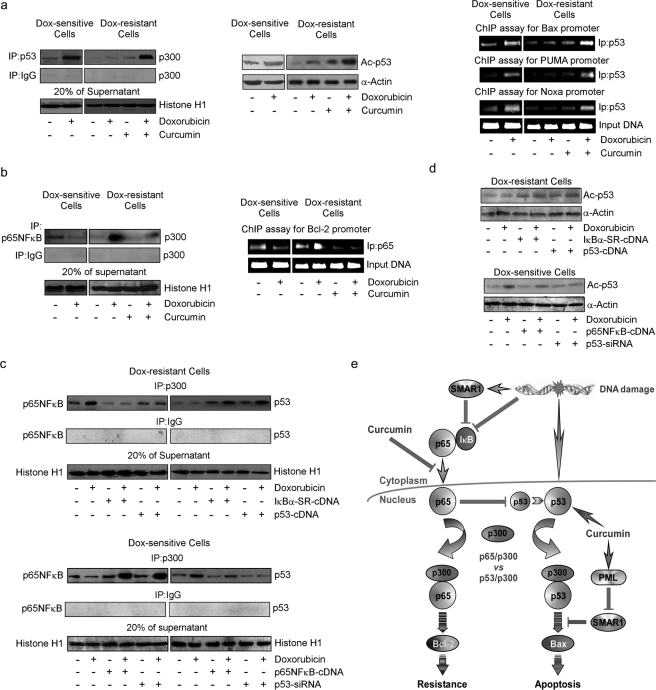
Curcumin Rescues p300 from p65NFκB to Establish p53-p300 Collaboration in Drug-resistant Cells
We next attempted to unveil the mechanisms underlying NFκB-mediated inhibition of p53 transcription functions. Recent studies indicate that the transcriptional activity of p53 in response to genotoxic stress is regulated by its interaction with transcriptional co-activator, p300 (39). To verify the effects of curcumin on p53-p300 cross-talk, if any, we immunoprecipitated nuclear p53 in resistant cells treated with doxorubicin, curcumin, or both and verified its interaction with p300 by Western blotting. It was observed that in contrast to sensitive cells, doxorubicin failed to induce p53-p300 interaction in resistant cells, but curcumin pretreatment restored this interaction (Fig. 4a). Consistently, curcumin also restored drug-induced p53 acetylation (lysine 373) and p53-dependent transcription of Bax, PUMA, and Noxa in resistant cells (Fig. 4a) as observed by CHIP analysis. This subsequently enabled p53-dependent apoptosis as observed earlier (Fig. 3a). Because doxorubicin triggered p53-p300 interaction in sensitive cells where p65NFκB activation did not take place, we proposed that nuclear translocation of p65NFκB in drug-treated resistant cells might have sequestered p300 and thereby abridged p53-p300 cross-talk. As anticipated, doxorubicin treatment manifested significant p65NFκB-bound p300 in nuclear lysates of resistant cells (Fig. 4b). Interestingly, curcumin treatment inhibited p65NFκB-p300 cross-talk and prevented p65NFκB-mediated transcription of Bcl-2 (Fig. 4b). However, in IκBα-SR-overexpressing or curcumin-pretreated cells where p65NFκB activation was inhibited, doxorubicin induced p53-p300 interaction and p53 acetylation thereby confirming the role of p65NFκB in inhibiting p53-p300 cross-talk (Fig. 4, c and d). Similarly, in doxorubicin-treated resistant cells where p300 was sequestered by p65NFκB, increasing p53 levels by p53-cDNA overwhelmingly increased p53 concentration, intervened-p65NFκB-p300 cross-talk, and enabled p53-p300 interaction and p53 acetylation (Fig. 4, c and d). The observation was further strengthened when in p65NFκB-cDNA- or p53-siRNA-transfected sensitive cells, doxorubicin favored p65NFκB-p300 interaction over p53-p300 association (Fig. 4c). These results indicate a competition between NFκB and p53 for availing p300, and depending on the relative availability the winner, and the fate of the cell are decided.
FIGURE 4.
Curcumin induced p53-p300 interaction by inhibiting drug-induced NFκB activation in doxorubicin-resistant cells. a, left panel; b, left panel, p53/p65NFκB-associated p300 was immunopurified with anti-p53/p65NFκB antibodies from nuclear lysates of sensitive/resistant cells treated with doxorubicin alone or in combination with curcumin for 24 h and were Western-blotted (WB) with p300 antibody. A portion of nuclear lysates from the same set were used for Western blot analysis of acetylated p53 at lysine 373 (a, middle panel), and remaining cells from the same experimental set were subjected to ChIP assay for the determination of p53 and p65NFκB activity on Bax/PUMA/Noxa and Bcl-2 promoter, respectively (a, right panel; b, right panel). p53-cDNA/IκBα-SR-cDNA-transfected doxorubicin-resistant cells and p53-siRNA/p65NFκB-cDNA-transfected doxorubicin-sensitive cells were treated with doxorubicin for 24 h, and the nuclear lysates obtained were either subjected to immunoprecipitation (IP) with p300/IgG antibody and the immunoprecipitates were Western-blotted with anti-p65NFκB/-p53 antibodies (c) or were subjected to Western blot analysis of acetylated p53 at lysine 373 (d). To verify comparable protein input during immunoprecipitation, 20% of supernatant from the nuclear lysates was blotted with histone H1 antibody. Values are the mean ± S.E. of five independent experiments in each case or representative of typical experiment in case of ChIP assay and Western blots. *, p < 0.05 when compared with respective untransfected/control sets. e, shown is a schematic illustration depicting differential regulation of anti- and pro-apoptotic network by curcumin in drug-resistant cells.
In summary, the above results conclude that in resistant cells, doxorubicin activated p65NFκB by SMAR1-dependent and independent mechanisms, which by competing with p53 for the transcriptional co-activator p300 inhibited the apoptotic program and up-regulated the resistant machinery of the cell (Fig. 4e). On the other hand, curcumin, by inhibiting p65NFκB and inducing PML-SMAR1 cross-talk, censored the resistance pathway, thereby making p300 available for p53 interaction upon doxorubicin treatment. resulting in transcription of pro-apoptotic protein Bax that effectively sensitized drug-resistant cancer cells (Fig. 4e).
Validation of the Efficiency of the Combinatorial Therapy in Mice Bearing Doxorubicin-resistant Carcinoma Cells
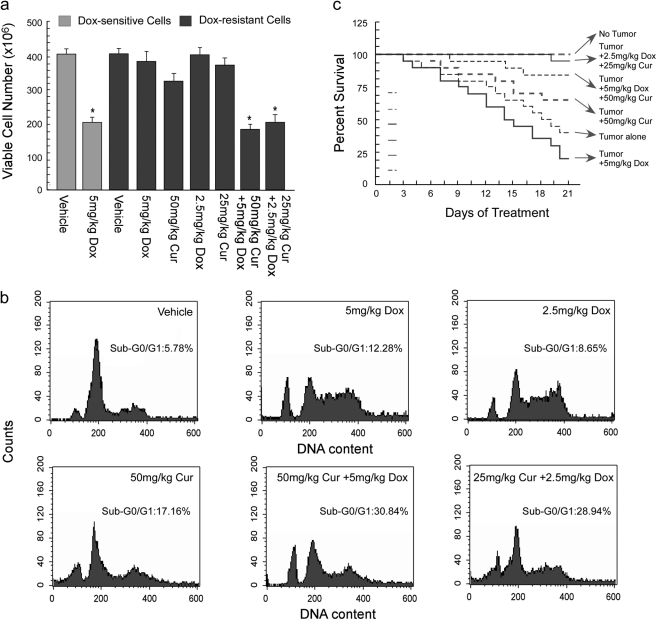
After delineating the mechanism underneath curcumin-induced sensitization of doxorubicin-resistant cancer cells toward apoptosis, we next attempted to validate these results in doxorubicin-resistant tumor-bearing mice. As furnished in Fig. 5a, the effective dose of doxorubicin that reduced 50% of sensitive-cell number (LD50, 5 mg/kg body weight) failed to significantly reduce the resistant tumor load alone. Interestingly, doxorubicin, when treated in combination with curcumin (LD50, 50 mg/kg body weight) at the LD50 dose of each or at ½LD50 dose of each, resistant tumor load was reduced from ∼390 × 106 to ∼190 × 106 and ∼210 × 106, respectively (Fig. 5a). Flow cytometric analysis further demonstrated 12% hypoploidy of tumor cells in LD50 doxorubicin-treated tumor-bearing mice when compared with untreated cells (Fig. 5b). However, percent hypoploidy (sub-G0/G1 cells) in drug-resistant tumor cells increased up to 31 and 28% at the LD50 and ½LD50 combined doses of both the drugs, respectively (Fig. 5b). These results clearly validated the candidature of curcumin in imparting sensitivity or reversing doxorubicin resistance not only in in vitro conditions but also in in vivo condition. Interestingly, in contrast to doxorubicin alone that severely reduced the longevity of the doxorubicin-resistant tumor-bearing animals, curcumin along with the drug provided a robust survival advantage to these mice (Fig. 5c). Among the dose combinations used, although ½LD50 of the drugs in combination killed a slightly lesser number of tumor cells than did the LD50 of combinatorial drugs, it gave a better survival advantage than the latter. These findings not only led us to the conclusion that even a low dose of curcumin could lower the effective dose of doxorubicin but also prompted us to hypothesize that at this low dose, doxorubicin might have imparted lesser systemic toxicity than the LD50 dose, and ½LD50 curcumin could completely eradicate that toxicity including immune toxicity. Such a decrease in drug-induced systemic toxicity might also have enhanced the efficacy of doxorubicin by improving the intrinsic defense machineries, thereby extending superior survival in tumor bearer.
FIGURE 5.
Curcumin reverted drug resistance and provided survival advantage of doxorubicin-treated tumor-bearing mice. a, doxorubicin-sensitive/resistant tumor-bearing mice were treated with doxorubicin alone or in combination with curcumin at different doses, and the total number of viable tumor cells in the peritoneal cavity was assayed by trypan blue dye-exclusion test. b, doxorubicin-resistant tumor-bearing mice were treated with doxorubicin alone or in combination with curcumin, and the tumor cells were subjected to flow cytometric determination of percentage hypoploidy (sub-G0/G1 phase cells). c, the survival rates of doxorubicin-resistant tumor-bearing animals treated with doxorubicin alone or in combination with curcumin were calculated by counting the number of live animals at a fixed time interval and were represented as Kaplan-Meier curve. Values are the mean ± S.E. of five independent experiments in each case. *, p < 0.05 when compared with respective control sets.
Curcumin Protects Doxorubicin-resistant Tumor-bearing Mice from Systemic Toxicity to Provide Survival Advantage
It is known that during the treatment of drug-resistant tumor cells, the concentration of drug employed for regressing tumor often exceeds that of the tolerable threshold, complicating the situation further with additive problems of systemic toxicity (19–22). On the other hand, reports have validated curcumin with immuno-, hepato-, and cardio-protective effects (31–34). Keeping this information in mind, we intended to verify our hypothesis that curcumin could eliminate drug-induced systemic toxicity in doxorubicin-resistant tumor-bearing mice.
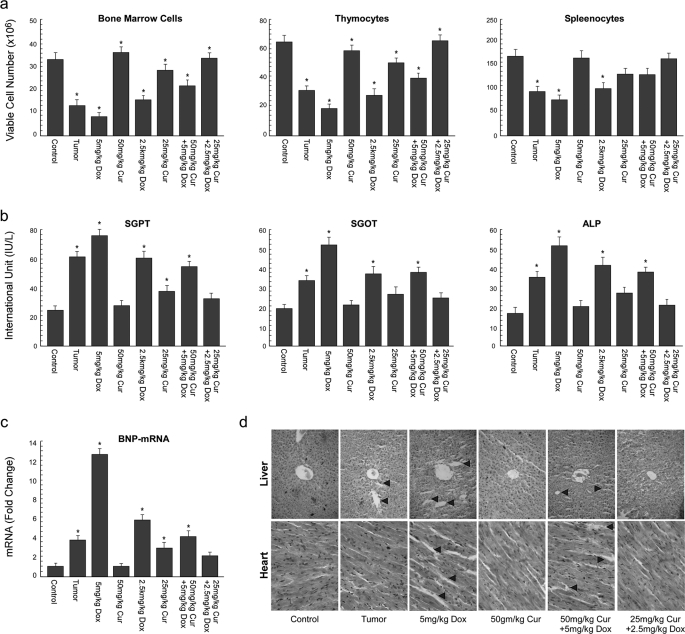
An attempt to examine the effect of curcumin/doxorubicin combinatorial treatment on the immune system of the tumor bearer revealed that tumor burden itself reduced the viable cell numbers in thymus, bone marrow, and spleen (Fig. 6a). Doxorubicin at LD50 further lowered these functional immune cell numbers, whereas curcumin at the same dose provided partial protection to the tumor bearer from such immune suppression when applied in combination (Fig. 6a). Importantly, in comparison to LD50, ½LD50 doxorubicin imparted less immune toxicity, which could be significantly eliminated when applied with ½LD50 curcumin (Fig. 6a). Further studies to monitor doxorubicin-induced hepatic toxicity and its amelioration, if any, by curcumin exposed significant elevation in serum glutamate pyruvate transaminase, serum glutamate oxaloacetate transaminase, and alkaline phosphatase levels in the serum of mice due to tumor burden, which was further augmented by LD50 doxorubicin treatment (Fig. 6b). In this case too, although LD50 curcumin endowed with a partial safeguard, combination of ½LD50 of both the drugs provided complete protection to the liver (Fig. 6b). Our hypothesis was further strengthened by the next set of experiments which disclosed that in the ventricle of the hearts of the tumor bearer, mRNA levels of BNP, a marker for cardio toxicity whose expression correlates positively with heart failure and diastolic dysfunction (42), was brought down toward normal values by the combinatorial administration of curcumin with doxorubicin, where again the combination of ½LD50 of both the drugs furnished better protection (Fig. 6c). Histopathological data produced further support to these observations (Fig. 6d).
FIGURE 6.
Curcumin ameliorated doxorubicin-induced systemic toxicity in drug-resistant tumor-bearing mice. a, total number of viable cells in the thymus, bone marrow, and spleen from normal and doxorubicin/curcumin (Cur) alone or in combination-treated doxorubicin-resistant tumor-bearing mice were determined by trypan blue dye-exclusion assay. b, serum glutamate pyruvate transaminase (SGPT), glutamate oxaloacetate transaminase (SGOT), and alkaline phosphatase (ALP) from the above set of mice were assayed as described under “Experimental Procedures” and plotted graphically. c, real-time PCR experiments were carried out to compare -fold change in BNP-mRNA levels from the same set of mice and were plotted graphically (left panels). Histological sections of liver and heart from the animals were stained with hematoxylin and counter-stained with eosin and microscopically analyzed for histopathological examinations of tissue toxicity like cellular damage and vacuolization. Values are the mean ± S.E. of five independent experiments in each case. *, p < 0.05 when compared with respective control sets or representative of typical experiment in case histological analysis.
These results together confirm our hypothesis that curcumin, at a low dose like ½LD50, not only brought down the effective dose of doxorubicin that itself lowered the drug-induced systemic toxicity in the tumor bearer but also eradicated that residual toxicity, thereby rejuvenating the intrinsic defense system of the host. As a cumulative effect of all these consequences, the tumor-bearing mice were extended with better survival advantage at this dose combination.
DISCUSSION
Doxorubicin is one of the most widely used therapeutic modalities for breast cancer. Unfortunately, many breast tumor cells are inherently resistant to doxorubicin or can acquire chemoresistance shortly after therapy, which inevitably leads to treatment failure and relapse of the disease (2, 3). An accumulating body of evidence suggests that constitutive activation of the NFκB pathway can contribute to cancer development, progression, and resistance to cancer therapy (4, 5), whereas activation of this pathway by doxorubicin can also render tumor cells more resistant to chemotherapy (6). Therefore, inhibition of the NFκB defense pathway has the potential to increase the therapeutic index of doxorubicin. In this regard NFκB inhibitors may emerge as the most promising anti-tumor agents and novel tumor sensitizers for doxorubicin.
Using a rationally targeted approach, we have demonstrated the unique role of curcumin in sensitizing doxorubicin-resistant cancer cells. Curcumin-mediated sensitization to cancer therapy relies on its ability to suppress the NFκB pathway through reduced p65NFκB translocation to nucleus via both SMAR1-dependent and -independent manners. SMAR1-independent mechanisms included inhibition of phosphorylation-mediated degradation of IκBα, the canonical NFκB inhibitory pathway, whereas SMAR1-dependent pathways involved reversal of SMAR1-mediated transcription repression of IκBα. Such effects of curcumin rescued p300 from p65NFκB, thereby setting off p53-p300 cross-talk that resulted in Bax, PUMA, and Noxa transactivation, mitochondrial transmembrane potential loss, and activation of downstream caspase cascade. Importantly curcumin-mediated induction of PML-SMAR1 cross-talk was also indispensable for p53 transcriptional functions and chemosensitization of resistant cells. In contrast to doxorubicin-treated sensitive cells or combination dose-treated resistant cells, SMAR1 knockdown in resistant cells ameliorated doxorubicin-mediated IκBα repression, suggesting differential function of SMAR1 in modulating IκBα-dependent NFκB responses in resistant and sensitive cells; more because the probability of SMAR1-mediated IκBα repression, if any, during apoptotic condition was probably inhibited due to sequestration of SMAR1 by PML. This is in accordance with studies by Sinha et al. (18), where SMAR1-mediated repression of apoptosis is quenched by PML induction in severe DNA damaging conditions. Importantly, mild genotoxic stress fails to achieve PML-SMAR1 cross-talk. Based on these studies, we can further argue that curcumin pretreatment in resistant cells might amplify doxorubicin-induced genotoxic insult, thereby mimicking conditions of severe DNA damage. In other words, due to high DNA damage repair capacity typical of resistant cells (42), doxorubicin alone fails to achieve DNA damage response in favor of apoptosis.
The regulatory contribution of NFκB and p53 to cancer development and progression is well documented where inactivation of p53 and hyperactivation of NFκB are the common occurrences (43). In agreement with such complex regulation of NFκB and p53 at several steps, these transcription factors can functionally antagonize, cooperate, or exhibit independence (44–46). Likewise in our study, we observed that NFκB, being induced in drug-treated resistant cells, interfered with p53 functions by p300 sequestration. Inhibition of NFκB by curcumin or IκBα super repressor or SMAR1-shRNA rescued p300 from NFκB-clutch to restrain the resistance pathway. Consistently, there are studies reporting that NFκB has a high affinity for p300 that may lead to its sequestration, thereby making it unavailable to other transcription factors (47). In line with these studies we observed that upon combinatorial treatment of curcumin and doxorubicin, inhibition of NFκB rescued p300, making it available to another transcription factor(s) like p53 in the present case, thereby allowing p53-dependent transactivation of apoptotic proteins.
Interestingly, we observed that in cells transfected with p53-cDNA, NFκB-p300 cross-talk was intervened, consequently inducing p53-p300 interaction. Our findings were consistent with those of Webster and Perkins (48) who first reported that the RelA (p65) subunit of NFκB antagonized p53 transactivation through sequestration of the p300 and CBP co-activators. It is acknowledged that p300 and CBP participate at various stages of the p53 response, functioning as essential co-activators in p53-dependent transactivation of target genes (49). They promote transcription of specific p53 targets by two mechanisms. First, p300 and CBP are recruited by p53 to target gene promoters where they acetylate histones (49). Second, p53 acetylation secondary to DNA damage stabilizes the p53-DNA complex at target gene promoters (49). Similarly, acetylation of NFκB is important for NFκB-DNA binding activity, and p300 activation is known to enhance p65NFκB acetylation (50). The N- and C-terminal domains of both CBP/p300 functionally interact with a region of p65NFκB containing the transcriptional activation domain and thereby promote the transactivating functions of NFκB transcription factors (50). Therefore, our results along with others suggest that NFκB and p53 compete for transcriptional co-activator p300, and depending upon whether NFκB or p53 hires p300, execution of downstream effector pathways oscillates between chemoresistance and chemosensitivity responses.
Our observation of curcumin-sensitizing doxorubicin-resistant ascites carcinoma cells, whose origin is mammary epithelial carcinoma (51), emphasizes that curcumin in combination with doxorubicin can be used as an effective treatment strategy to reverse breast cancer drug resistance. Consistently, curcumin has been reported to increase the efficacy of doxorubicin by modulating the function of the multidrug resistance-linked ATP-binding cassette transporter ABCG2 (52) and to sensitize glioma cells in a p53- and caspase-independent manner by inhibition of AP-1 and NFκB signaling pathways (53). Another finding in the same line of studies demonstrated that curcumin sensitized non-small-cell lung cancer cells to cisplatin-induced apoptosis by superoxide-mediated Bcl-2 degradation (54). A study conducted by another group of researchers highlighted the chemosensitizing efficacy of curcumin in TRAIL (TNF-related apoptosis-inducing ligand Transwells)-resistant LNCaP xenografts (55). Reversal of P-glycoprotein-mediated doxorubicin resistance in human sarcoma MES-SA/Dx-5 cells by curcumin has also been reported (56). In support to these studies, our study provides another mechanism where curcumin, by antagonizing SMAR1-mediated apoptosis resistance, inhibits NFκB-p300 cross-talk for successful execution of p53-p300 interaction. Although there have been multiple in vitro studies indicating the apoptotic and chemosensitizing properties of curcumin, this is the first report demonstrating the in vivo chemosensitizing properties of curcumin where it not only reverses drug resistance in doxorubicin-resistant tumor-bearing mice but also ameliorates drug-induced systemic toxicity.
In fact, tumor itself as well as doxorubicin induced severe immunosuppression, increased liver toxicity, and caused cardiovascular injury. Although previous reports from our laboratory already established several mechanisms of tumor-induced immune toxicity and its inhibition by curcumin (31–34), this is the first report describing that this phytochemical can protect the host immune system from the toxicity rendered by the anti-cancer drug in tumor bearer. Our results showing hepatoprotective effect of curcumin were supported by the study of Chuang et al. (57) who showed that curcumin-containing diet inhibits murine hepatocarcinogenesis. Curcumin has also been shown to prevent alcohol-induced liver disease in rats by inhibiting the expression of NFκB-dependent genes (58). At the same time curcumin inhibited endotoxin-mediated activation of NFκB and suppressed the expression of cytokines, chemokines, COX-2, and inducible nitric-oxide synthase in Kupffer cells (59). Apart from immunotoxicity and liver damage, in our study curcumin also protected cardiac tissue from doxorubicin-induced toxicity. Consistent with the reports depicting the cardioprotective effects of curcumin (60), complete inhibition of BNP levels, while reversing doxorubicin-induced myocardial toxicity, was observed upon combinatorial application of curcumin. Interestingly, among the dose combinations used, although combination of ½LD50 of each drug killed a slightly lesser number of tumor cells than the combination of LD50, the former bestowed better survival advantage to the tumor-bearing mice than the latter. Our attempt to explore the cause underlying such “contradictory” results revealed that curcumin, even at low dose, was effective in lowering the effective dose of doxorubicin, thereby lessening the drug-induced systemic toxicity. The remaining toxicity was then taken care of by this low dose of curcumin that in combination with doxorubicin also sensitized resistant cancer cells of the tumor bearer. These results, therefore, could identify the optimal combination of these drugs that not only took advantage of the genetic aberrations that drive drug resistance and targeted the same but also provided a modern approach to treatment strategies by curbing the toxicity. This observation is in agreement with phase I clinical data showing that curcumin is well tolerated and suggested that curcumin could be a potential therapeutic agent for combination chemotherapy with DNA-damaging agents (61). Such differential activities of curcumin strongly support its candidature as a potential chemosensitizing agent, suggesting that a combinatorial regimen of curcumin and doxorubicin can be framed and tested for reversal of anthracycline resistance in future human clinical trials.
Supplementary Material
Acknowledgments
We thank R. Sarkar for editing the manuscript. We also thank U. Ghosh and R. Dutta for technical help and Dr. S. Choudhuri of Chittaranjan National Cancer Institute, Kolkata, India for providing drug sensitive and resistant EACs.
This work was supported by the grants from Department of Science and Technology, Government of India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- NFκB
- nuclear factor κB
- Bax
- Bcl-2-associated X protein
- Bcl-2
- B cell lymphoma 2
- BNP
- B-type natriuretic peptide
- EAC
- Ehrlich ascites carcinoma
- LD50
- lethal dose 50
- scaffold
- small matrix-associated region-binding protein
- PML
- promyelocyte leukemia
- CBP
- cAMP-response element-binding protein (CREB)-binding protein
- 7AAD
- 7-aminoactinomycin D
- PE
- phosphatidylethanolamine
- Dox
- doxorubicin
- DiOC6
- 3,3'-dihexyloxacarbocyanine iodide.
REFERENCES
- 1. Chintamani, Tandon M., Mishra A., Agarwal U., Saxena S. (2011) World J. Surg. Oncol 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campbell K. J., O'Shea J. M., Perkins N. D. (2006) BMC Cancer 6, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arafa el-S. A., Zhu Q., Shah Z. I., Wani G., Barakat B. M., Racoma I., El-Mahdy M. A., Wani A. A. (2011) Mutat. Res. 706, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu X., Wang B., Ma X., Guo Y. (2009) Jpn. J. Clin. Oncol. 39, 418–424 [DOI] [PubMed] [Google Scholar]
- 5. Gionet N., Jansson D., Mader S., Pratt M. A. (2009) J. Cell. Biochem. 107, 448–459 [DOI] [PubMed] [Google Scholar]
- 6. Montagut C., Tusquets I., Ferrer B., Corominas J. M., Bellosillo B., Campas C., Suarez M., Fabregat X., Campo E., Gascon P., Serrano S., Fernandez P. L., Rovira A., Albanell J. (2006) Endocr. Relat. Cancer 13, 607–616 [DOI] [PubMed] [Google Scholar]
- 7. Baud V., Karin M. (2009) Nat. Rev. Drug Discov. 8, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryan K. M. (2011) Eur. J. Cancer 47, 44–50 [DOI] [PubMed] [Google Scholar]
- 9. Saeki H., Kitao H., Yoshinaga K., Nakanoko T, Kubo N., Kakeji Y., Morita M., Maehara Y. (2011) Clin. Cancer Res. 17, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 10. Rohwer N., Dame C., Haugstetter A., Wiedenmann B., Detjen K., Schmitt C. A., Cramer T. (2010) PLoS One 5, e12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim D. S., Park S. S., Nam B. H., Kim I. H., Kim S. Y. (2006) Cancer Res. 66, 10936–10943 [DOI] [PubMed] [Google Scholar]
- 12. Lin Y., Bai L., Chen W., Xu S. (2010) Expert Opin. Ther. Targets 14, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aylon Y., Oren M. (2007) Cell 130, 597–600 [DOI] [PubMed] [Google Scholar]
- 14. Schneider G., Henrich A., Greiner G., Wolf V., Lovas A., Wieczorek M., Wagner T., Reichardt S., von Werder A., Schmid R. M., Weih F., Heinzel T., Saur D., Krämer O. H. (2010) Oncogene 29, 2795–2806 [DOI] [PubMed] [Google Scholar]
- 15. Bourguignon L. Y., Xia W., Wong G. (2009) J. Biol. Chem. 284, 2657–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaul R., Mukherjee S., Ahmed F., Bhat M. K., Chhipa R., Galande S., Chattopadhyay S. (2003) Int. J. Cancer. 103, 606–615 [DOI] [PubMed] [Google Scholar]
- 17. Singh K., Sinha S., Malonia S. K., Bist P., Tergaonkar V., Chattopadhyay S. (2009) J. Biol. Chem. 284, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 18. Sinha S., Malonia S. K., Mittal S. P., Singh K., Kadreppa S., Kamat R., Mukhopadhyaya R., Pal J. K., Chattopadhyay S. (2010) EMBO J. 29, 830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tao Z., Jones E., Goodisman J., Souid A. K. (2008) Anal. Biochem. 381, 43–52 [DOI] [PubMed] [Google Scholar]
- 20. Patel N., Joseph C., Corcoran G. B., Ray S. D. (2010) Toxicol. Appl. Pharmacol. 245, 143–152 [DOI] [PubMed] [Google Scholar]
- 21. Constantinidou A., Jones R. L., Scurr M., Al-Muderis O., Judson I. (2011) Acta Oncol. 50, 455–461 [DOI] [PubMed] [Google Scholar]
- 22. Auvinen P. K., Mähönen U. A., Soininen K. M., Paananen P. K., Ranta-Koponen P. H., Saavalainen I. E., Johansson R. T. (2010) Tumori 96, 271–275 [DOI] [PubMed] [Google Scholar]
- 23. Kunnumakkara A. B., Anand P., Aggarwal B. B. (2008) Cancer Lett. 269, 199–225 [DOI] [PubMed] [Google Scholar]
- 24. Singh S., Aggarwal B. B. (1995) J. Biol. Chem. 270, 24995–25000 [DOI] [PubMed] [Google Scholar]
- 25. Bava S. V., Sreekanth C. N., Thulasidasan A. K., Anto N. P., Cheriyan V. T., Puliyappadamba V. T., Menon S. G., Ravichandran S. D., Anto R. J. (2011) Int. J. Biochem. Cell Biol. 43, 331–341 [DOI] [PubMed] [Google Scholar]
- 26. Choi B. H., Kim C. G., Lim Y., Shin S. Y., Lee Y. H. (2008) Cancer Lett. 259, 111–118 [DOI] [PubMed] [Google Scholar]
- 27. Choudhuri T., Pal S., Das T., Sa G. (2005) J. Biol. Chem. 280, 20059–20068 [DOI] [PubMed] [Google Scholar]
- 28. Choudhuri T., Pal S., Agwarwal M. L., Das T., Sa G. (2002) FEBS Lett. 512, 334–340 [DOI] [PubMed] [Google Scholar]
- 29. Pal S., Choudhuri T., Chattopadhyay S., Bhattacharya A., Datta G. K., Das T., Sa G. (2001) Biochem. Biophys. Res. Commun. 288, 658–665 [DOI] [PubMed] [Google Scholar]
- 30. Chakraborty J., Banerjee S., Ray P., Hossain D. M., Bhattacharyya S., Adhikary A., Chattopadhyay S., Das T., Sa G. (2010) J. Biol. Chem. 285, 33104–33112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhattacharyya S., Md, Sakib, Hossain D., Mohanty S., Sankar, Sen G., Chattopadhyay S., Banerjee S., Chakraborty J., Das K., Sarkar D., Das T., Sa G. (2010) Cell. Mol. Immunol. 7, 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhattacharyya S., Mandal D., Saha B., Sen G. S., Das T., Sa G. (2007) J. Biol. Chem. 282, 15954–15964 [DOI] [PubMed] [Google Scholar]
- 33. Bhattacharyya S., Mandal D., Sen G. S., Pal S., Banerjee S., Lahiry L., Finke J. H., Tannenbaum C. S., Das T., Sa G. (2007) Cancer Res. 67, 362–370 [DOI] [PubMed] [Google Scholar]
- 34. Pal S., Bhattacharyya S., Choudhuri T., Datta G. K., Das T., Sa G. (2005) Cancer Detect. Prev. 29, 470–478 [DOI] [PubMed] [Google Scholar]
- 35. Choudhuri S. K., Chatterjee A. (1998) Anticancer Drugs 9, 825–832 [DOI] [PubMed] [Google Scholar]
- 36. Lahiry L., Saha B., Chakraborty J., Adhikary A., Mohanty S., Hossain D. M., Banerjee S., Das K., Sa G., Das T. (2010) Carcinogenesis 31, 259–268 [DOI] [PubMed] [Google Scholar]
- 37. Chattopadhyay S., Bhattacharyya S., Saha B., Chakraborty J., Mohanty S., Sakib, Hossain D. M, Banerjee S., Das K., Sa G., Das T. (2009) PLoS One 4, e7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sa G., Das T., Moon C., Hilston C. M., Rayman P. A., Rini B. I., Tannenbaum C. S., Finke J. H. (2009) Cancer Res. 69, 3095–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lahiry L., Saha B., Chakraborty J., Bhattacharyya S., Chattopadhyay S., Banerjee S., Choudhuri T., Mandal D., Bhattacharyya A., Sa G., Das T. (2008) Apoptosis 13, 771–781 [DOI] [PubMed] [Google Scholar]
- 40. Das T., Sa G., Paszkiewicz-Kozik E., Hilston C., Molto L., Rayman P., Kudo D., Biswas K., Bukowski R. M., Finke J. H., Tannenbaum C. S. (2008) J. Immunol. 180, 4687–4696 [DOI] [PubMed] [Google Scholar]
- 41. Das T., Sa G., Hilston C., Kudo D., Rayman P., Biswas K., Molto L., Bukowski R., Rini B., Finke J. H., Tannenbaum C. (2008) Cancer Res. 68, 2014–2023 [DOI] [PubMed] [Google Scholar]
- 42. Ghose, Roy S., Mishra S., Ghosh G., Bandyopadhyay A. (2007) Matrix Biol. 26, 269–279 [DOI] [PubMed] [Google Scholar]
- 43. Wang Q. E., Milum K., Han C., Huang Y. W., Wani G., Thomale J., Wani A. A. (2011) Mol. Cancer 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 45. Scian M. J., Stagliano K. E., Anderson M. A., Hassan S., Bowman M., Miles M. F., Deb S. P., Deb S. (2005) Mol. Cell. Biol. 25, 10097–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schumm K., Rocha S., Caamano J., Perkins N. D. (2006) EMBO J. 25, 4820–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Furia B., Deng L., Wu K., Baylor S., Kehn K., Li H., Donnelly R., Coleman T., Kashanchi F. (2002) J. Biol. Chem. 277, 4973–4980 [DOI] [PubMed] [Google Scholar]
- 48. Webster G. A., Perkins N. D. (1999) Mol. Cell. Biol. 19, 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iyer N. G., Chin S. F., Ozdag H., Daigo Y., Hu D. E., Cariati M., Brindle K., Aparicio S., Caldas C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7386–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong H., Voll R. E., Ghosh S. (1998) Mol. Cell 1, 661–671 [DOI] [PubMed] [Google Scholar]
- 51. Lehmann W., Graetz H., Samtleben R., Schütt M., Langen P. (1980) Acta Biol. Med. Ger. 39, 93–105 [PubMed] [Google Scholar]
- 52. Chearwae W., Shukla S., Limtrakul P., Ambudkar S. V. (2006) Mol. Cancer Ther. 5, 1995–2006 [DOI] [PubMed] [Google Scholar]
- 53. Dhandapani K. M., Mahesh V. B., Brann D. W. (2007) J. Neurochem. 102, 522–538 [DOI] [PubMed] [Google Scholar]
- 54. Chanvorachote P., Pongrakhananon V., Wannachaiyasit S., Luanpitpong S., Rojanasakul Y., Nimmannit U. (2009) Cancer Invest. 27, 624–635 [DOI] [PubMed] [Google Scholar]
- 55. Shankar S., Ganapathy S., Chen Q., Srivastava R. K. (2008) Mol. Cancer 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Angelini A., Iezzi M., Di Febbo C., Di Ilio C., Cuccurullo F., Porreca E. (2008) Oncol. Rep. 20, 731–735 [PubMed] [Google Scholar]
- 57. Chuang S. E., Kuo M. L., Hsu C. H., Chen C. R., Lin J. K., Lai G. M., Hsieh C. Y., Cheng A. L. (2000) Carcinogenesis 21, 331–335 [DOI] [PubMed] [Google Scholar]
- 58. Nanji A. A., Jokelainen K., Tipoe G. L., Rahemtulla A., Thomas P., Dannenberg A. J. (2003) Am. J. Physiol. Gastrointest Liver Physiol. 284, G321–G327 [DOI] [PubMed] [Google Scholar]
- 59. Matsuda H., Ninomiya K., Morikawa T., Yoshikawa M. (1998) Bioorg. Med. Chem. Lett. 8, 339–344 [DOI] [PubMed] [Google Scholar]
- 60. Venkatesan N. (1998) Br. J. Pharmacol. 124, 425–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng A. L., Hsu C. H., Lin J. K., Hsu M. M., Ho Y. F., Shen T. S., Ko J. Y., Lin J. T., Lin B. R., Ming-Shiang W., Yu H. S., Jee S. H., Chen G. S., Chen T. M., Chen C. A., Lai M. K., Pu Y. S., Pan M. H., Wang Y. J., Tsai C. C., Hsieh C. Y. (2001) Anticancer Res. 21, 2895–2900 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.