Background: Peroxiredoxin IV metabolizes endoplasmic reticulum-derived hydrogen peroxide.
Results: Peroxiredoxin IV structures reveal an unusually stable decamer and a sulfenylated intermediate in the enzymatic cycle.
Conclusion: The enzymatic cycle of peroxiredoxin IV involves destabilization of the active site followed by formation of a stable disulfide-bonded decamer.
Significance: Elucidating peroxiredoxin structures is required to understand how they function during oxidative stress.
Keywords: Endoplasmic Reticulum (ER), Oxidation-Reduction, Oxidative Stress, Peroxiredoxin, Protein Structure, Sulfhydryl, Hydrogen Peroxide
Abstract
Peroxiredoxin IV (PrxIV) is an endoplasmic reticulum-localized enzyme that metabolizes the hydrogen peroxide produced by endoplasmic reticulum oxidase 1 (Ero1). It has been shown to play a role in de novo disulfide formation, oxidizing members of the protein disulfide isomerase family of enzymes, and is a member of the typical 2-Cys peroxiredoxin family. We have determined the crystal structure of both reduced and disulfide-bonded, as well as a resolving cysteine mutant of human PrxIV. We show that PrxIV has a similar structure to other typical 2-Cys peroxiredoxins and undergoes a conformational change from a fully folded to a locally unfolded form following the formation of a disulfide between the peroxidatic and resolving cysteine residues. Unlike other mammalian typical 2-Cys peroxiredoxins, we show that human PrxIV forms a stable decameric structure even in its disulfide-bonded state. In addition, the structure of a resolving cysteine mutant reveals an intermediate in the reaction cycle that adopts the locally unfolded conformation. Interestingly the peroxidatic cysteine in the crystal structure is sulfenylated rather than sulfinylated or sulfonylated. In addition, the peroxidatic cysteine in the resolving cysteine mutant is resistant to hyper-oxidation following incubation with high concentrations of hydrogen peroxide. These results highlight some unique properties of PrxIV and suggest that the equilibrium between the fully folded and locally unfolded forms favors the locally unfolded conformation upon sulfenylation of the peroxidatic cysteine residue.
Introduction
The peroxiredoxins are a family of peroxidases important for anti-oxidant protection as well as intracellular signaling (1). PrxIV2 is a member of this family localized to the ER, where it forms (α2)5 decamers (2). Like other typical 2-Cys peroxiredoxins, it contains an active site cysteine residue that reacts with hydrogen peroxide to form a cysteine sulfenic acid (SOH) (3). The cysteine sulfenic acid is then resolved by reaction with a second cysteine residue in an adjacent polypeptide of the α2 dimer, resulting in the formation of an interchain disulfide bond (4). The disulfide formed as part of the reaction cycle is reduced by a member of the PDI family of oxidoreductases, resulting in their oxidation (5). The reaction cycle is depicted in Fig. 1. As long as there is a supply of hydrogen peroxide, PrxIV is able to form disulfides de novo and is, therefore, an important enzyme in the process of disulfide formation in proteins entering the secretory pathway (5, 6). Hydrogen peroxide can be generated within the ER by Ero1, which couples the formation of a disulfide with the reduction of oxygen to hydrogen peroxide (7). In addition, the acidity of ER-localized NADPH-oxidase 4 (Nox4) leads to the formation of hydrogen peroxide following oxidation of NADPH (8, 9). It is known that for other 2-Cys peroxiredoxins the sulfenic cysteine, rather than forming a disulfide, can react further with hydrogen peroxide to form a sulfinic (SO2H) and even a sulfonic acid (SO3H) (3). These hyper-oxidized forms of cysteine inactivate the enzyme; however, an additional enzyme called sulfiredoxin can reduce the sulfinic acid group back to sulfenic acid (10). This mechanism of inactivation and reactivation enables the peroxiredoxins to act as efficient peroxidases at low concentrations of peroxide, but once the concentration increases, the enzyme is inactivated, allowing the hydrogen peroxide to be stabilized and initiate signaling events (11). It is not known whether PrxIV is regulated in a similar manner, but as yet, there is no indication of the presence of a sulfiredoxin-like activity localized to the ER.
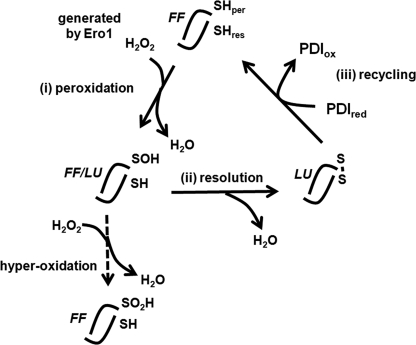
FIGURE 1.
Schematic depicting the various forms of PrxIV during the PrxIV reaction cycle. An individual dimer is depicted that is part of a decameric structure throughout the reaction cycle. Each dimer contains two active sites (only one is depicted here). (i) peroxidation, the peroxidatic cysteine (SHper) reacts with hydrogen peroxide generated by Ero1 or NADPH-oxidase 4 (Nox4) in the ER lumen. (ii) resolution, the resolving cysteine (SHres) reacts with the resulting sulfenylated cysteine to form an interchain disulfide bond. (iii) recycling, a reduced (PDIred) member of the PDI family is then oxidized (PDIox), resulting in the regeneration of the active site cysteine. Each stage of the reaction cycle is accompanied by a conformational change from the FF to the LU conformation as depicted. The sulfenylated intermediate exists in either the FF or the LU conformation and can be further oxidized to a sulfinic acid form if the hydrogen peroxide concentration is high.
The reaction cycle occurs in three phases (Fig. 1): reaction with hydrogen peroxide (peroxidation), resolution of the cysteine sulfenic acid to form a disulfide (resolution), and finally reduction of the disulfide by a low molecular weight thiol or a member of the thioredoxin superfamily (recycling). Structural analysis of several different members of the peroxiredoxin family has demonstrated that transition between these phases is accompanied by a conformational change in the protein from an FF to a LU form (12). In the FF form, the active site or peroxidatic cysteine is present toward the end of an α-helix. The peroxidatic cysteine forms part of an active site pocket that both lowers the pKa of the cysteine as well as providing a substrate binding site that has been suggested to optimize the orientation of the substrate and to stabilize the transition state to allow extremely efficient catalysis (13). Indeed it has been shown that peroxiredoxins have apparent second order rate constants of ∼107 m−1 s−1 (1). The fast reaction kinetics coupled to the abundance of peroxiredoxins indicates that these are major cellular peroxidases. Once the peroxidatic cysteine is sulfenylated, the protein is thought to adopt the LU conformation, which involves unwinding of the α-helix containing the peroxidatic cysteine and positioning the sulfenylated cysteine close to the resolving cysteine residue to allow the formation of a disulfide. Additional changes to the structure involve the C-terminal α-helix unwinding, which repositions the resolving cysteine so that it is now well placed to react with the peroxidatic cysteine (14). In PrxIV, the resulting disordered C-terminal region may well act as a binding site for PDI, which is known to bind to unfolded regions of proteins and peptides via its b′ domain (15). Once the disulfide is reduced, the enzyme re-forms the FF conformation. For all the mammalian peroxiredoxins studied to date, this structural transition is also accompanied by changes in the quaternary structure (16). In the FF conformation, the enzyme typically exists as a decamer but may also form dodecamers. Upon transition to the LU form, the decamer dissociates to dimers ((α2)5–6 to α2). The dissociation and reassociation of dimers are thought to be required as part of the reaction cycle with the dimers being more easily reduced by thioredoxins and the oligomers being more active toward peroxides (4).
The instability of disulfide-bonded oligomers of the typical 2-Cys peroxiredoxins has meant that few structures of this form of the protein are available. Previous analysis of PrxIV suggests that the decamer is particularly stable (2). Here we have carried out crystallographic studies of human PrxIV and found that the disulfide-bonded LU form of the enzyme was indeed stable, allowing the structure to be determined at high resolution. Preventing the formation of the interchain disulfide between the peroxidatic and resolving cysteine by mutation of the resolving cysteine to alanine resulted in a mutant that was insensitive to hyper-oxidation of the peroxidatic cysteine. Structural analysis of this mutant shows that the enzyme can adopt the LU conformation when the peroxidatic cysteine is sulfenylated. This novel peroxiredoxin structure reveals a potential intermediate in the enzymatic cycling and supports the idea that sulfenylation of the peroxidatic cysteine causes the conformational shift from the FF to the LU form and prevents further oxidation occurring.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
All reagents were purchased from Sigma-Aldrich (Gillingham, Dorset, UK), and enzymes were from Promega (Southampton, Hampshire, UK) unless otherwise stated. Rabbit polyclonal anti-PrxIV and anti-Prx-SO2–3 were purchased from Ab Frontier (Seoul, Korea).
DNA Constructs
All human PrxIV constructs used were as described previously (5, 17) apart from the C-terminal truncation, which was constructed by introducing a stop codon after Pro-231.
Preparation of Purified Proteins
All proteins (minus secretory signal peptides) were expressed and purified using Escherichia coli BL21-DE3. The purification of His-tagged PrxIV and its mutants as well as ERp46 has been described previously (17). His-tagged PDI was similarly expressed and purified by Ni2+ column chromatography as described previously (18). Each protein was quantified using the relevant 280 nm absorption extinction coefficient.
Size Exclusion Chromatography
The oligomeric state of the recombinant proteins was analyzed at room temperature by size exclusion chromatography using a Superdex 200 PC 3.2/30 column (GE Healthcare). The buffer used was 50 mm HEPES, pH 7.0, containing 150 mm NaCl with or without 1 mm DTT. Proteins were loaded at 1 mg/ml in a volume of 25 μl, and the flow rate was 0.04 ml/min. Molecular weight calibration was performed with blue dextran, apoferritin, β-amylase, BSA, carbonic anhydrase, and cytochrome c as standards.
PrxIV Recycling Assay
GSH (10 mm) was mixed with ERp46 or PDI (0.6 μm) and then added to PrxIV (6 μm) and incubated at 37 °C in 50 mm NaH2PO4 buffer, pH 7.4, containing 150 mm NaCl, 1 mm EDTA. Reactions were quenched by the addition of 6 volumes of 10% w/v TCA and incubated on ice for 15 min. Precipitated proteins were isolated by centrifugation for 15 min at 15,000 × g at 4 °C. Following an acetone wash, pellets were air-dried for 15 min before being resuspended in SDS sample buffer (31.25 mm Tris buffer, pH 6.8, containing 2% w/v SDS, 5% v/v glycerol, 0.01% w/v bromphenol blue, 40 mm N-ethylmaleimide). Samples were heated immediately at 100 °C for 5 min before being loaded on the gel.
Hyper-oxidation Assay
Wild type or mutant PrxIV (5 μm) was mixed with 2.1 μm thioredoxin, 1.5 μm thioredoxin reductase, and 0.5 mm NADPH in 50 mm NaH2PO4 buffer, pH 7.4, containing 150 mm NaCl, 1 mm EDTA. Various concentrations of H2O2 were added, and samples were incubated for 10 min at room temperature. An equal amount of sample buffer containing 100 mm dithiothreitol (DTT) was added before boiling for 5 min. To denature PrxIV, 1% SDS was used in the buffer, and 50 mm DTT was used to reduce protein. Excess DTT was removed by size exclusion chromatography. The extent of hyper-oxidation of PrxIV was assayed by carrying out a Western blot using an antibody that recognizes the sulfinylated or sulfonylated form of PrxIV.
Electrophoresis, Western Blotting, and Densitometry
SDS-PAGE was carried out under reducing conditions in the presence of DTT (50 mm) or non-reducing conditions where DTT was omitted. Separated proteins were visualized by staining overnight using colloidal Coomassie Blue (10% w/v phosphoric acid, 10% w/v ammonium sulfate, 0.12% Coomassie Brilliant Blue G250, 20% v/v methanol) followed by destaining in distilled water. For Western blotting, proteins were transferred to nitrocellulose membrane (LI-COR Biosciences, Cambridge, UK). Nonspecific binding was blocked using 5% (w/v) nonfat dried skimmed milk in Tween/Tris-buffered saline (50 mm Tris buffer, pH 7.5, containing 150 mm NaCl and 0.05% (v/v) Tween 20). Primary antibody was diluted 1:2000, and incubations were performed for 1 h at room temperature in Tween/Tris-buffered saline. Secondary antibody was diluted 1:2500 in Tween/Tris-buffered saline, and incubations were performed in a light-shielded box for 1 h at room temperature. Specific proteins were visualized using an Odyssey SA imaging system (LI-COR Biosciences). For quantification of fluorescent Western blots, scans were performed at minimum intensities required to see all relevant proteins. Densitometry was then performed upon unmodified output images using ImageJ (National Institutes of Health). Where required, bound antibodies were stripped from nitrocellulose membranes by 5-min washes in 0.2 m NaOH followed by 5 min in distilled water. Primary antibodies were then reapplied without the need for further blocking.
X-ray Crystallography
All crystallizations were performed by sitting drop vapor diffusion at 16 °C. For wild type PrxIV or oxidized C51A mutant, the crystallization conditions were initially identified using the Crystal Screen from Hampton Research. Conditions were subsequently optimized to 0.1 m HEPES buffer, pH 7.3, containing 11% (w/v) polyethylene glycol (PEG) 8000, 8% (v/v) ethylene glycol with 16 mg/ml protein for wild type or 0.1 m Tris buffer, pH 7.0, containing 48% (w/v) PEG 200 with 15 mg/ml protein for the C51A mutant. Crystals were grown using a 0.5-ml reservoir solution and a 2-μl drop containing a 1:1 ratio of protein to reservoir solution. Single crystals were flash-cooled in liquid N2 using 40% (v/v) ethylene glycol in the cryo solution. For the reduced C51A mutant, the crystallization conditions were initially identified using the Cryo Screen from Emerald Biosystems. Conditions were optimized to 0.1 m Tris buffer, pH 8.5, containing 1.55 m (NH4)2SO4, 12% (v/v) glycerol. Protein was concentrated to 15 mg/ml in 10 mm DTT, and crystals were grown in a 0.5-ml reservoir solution and a 2-μl drop containing a 1:1 ratio of protein to reservoir solution. Single crystals were flashed-cooled in liquid N2. For the oxidized C245A mutant, protein was treated with 1 mm H2O2 for 5 min at room temperature before being applied to a size exclusion column. Protein was then concentrated to 11.8 mg/ml. Crystals were obtained directly from a Crystal Screen from Hampton Research, the optimal conditions being 0.1 m HEPES buffer, pH 7.5, 10% (w/v) PEG 8000, 8% (v/v) ethylene glycol. For the reduced C245A mutant, protein was concentrated to 15 mg/ml in 10 mm DTT. Crystals were obtained directly from a Joint Center for Structural Genomics-plus Screen from Molecular Dimensions, the optimal conditions being 0.1 m HEPES buffer, pH 7.0, containing 1.0 m succinic acid, 1% (w/v) PEG 2000 monomethyl ether. Both crystallizations were set up in 96-well plates using a 0.1-ml reservoir solution and a 0.5-μl drop containing a 1:1 ratio of protein to reservoir solution. Single crystals were flash-cooled in liquid N2 using 40% (v/v) ethylene glycol in the cryo solution.
Data were collected at the Diamond Light Source (Didcot, Oxfordshire, UK) stations I02, I03, and I04. All crystals belong to space group C2 with five monomers in the asymmetric unit. The data of oxidized C245A were integrated and scaled using d*TREK (19), and the remaining data were integrated and scaled using XDS (20). The initial phases were obtained by molecular replacement with the program PHASER (21) by using a half-decamer of truncated human PrxIV (Protein Data Bank (PDB) code 2PN8) as a search model. Structural modeling and refinement were performed with COOT (22) and REFMAC (23) in the CCP4 suite (24). The TLS thermal mode was used, which allows a separation of the overall lattice vibrations before the standard restrained refinement of atomic coordinates and the individual atom isotropic B-factors (25). For all the PrxIV structures reported here, five monomers in the asymmetric unit were treated as separate TLS groups. Molprobity (26) was used to monitor the model geometry. The data collection and refinement statistics are presented in Table 1. The figures presented in this study were generated using PyMOL (37).
TABLE 1.
Data collection and refinement statistics
| Oxidized WT (PDB ID 3TJB) | Reduced C51A (PDB ID 3TJF) | Oxidized C51A (PDB ID 3TJG) | Reduced C245A (PDB ID 3TJK) | Sulfenylated C245A (PDB ID 3TJJ) | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2 | C2 | C2 | C2 | C2 |
| Unit cell (Å) | a = 108.0, b = 139.4, c = 95.9 | a = 108.3, b = 138.8, c = 96.24 | a = 107.2, b = 130.1, c = 95.1 | a = 108.2, b = 138.4, c = 96.0 | a = 108.7, b = 140.0, c = 96.28 |
| β (°) | 102.81 | 103.97 | 102.29 | 103.80 | 103.18 |
| Resolution (Å)a | 69.71-2.37 (2.44-2.37) | 28.77-2.04 (2.09-2.04) | 28.96-2.24 (2.30-2.24) | 83.71-2.09 (2.15-2.09) | 42.72-1.91 (1.98-1.91) |
| No. of unique observationsa | 55,077 (4093) | 87,136 (6233) | 64,808 (4733) | 80,529 (5992) | 103,152 (10,808) |
| Multiplicitya | 3.7 (3.8) | 4.3 (4.0) | 5.0 (5.0) | 4.7 (4.7) | 2.1 (2.08) |
| Completeness(%)a | 98.8 (99.0) | 99.4 (96.4) | 99.8 (99.8) | 99.6 (99.8) | 95.2 (95.8) |
| Mean I/σa | 9.6 (1.7) | 16.3 (2.2) | 16.5 (2.6) | 14.3 (2.4) | 11.7 (1.6) |
| Rmerge(%)a | 6.8 (73.0) | 5.1 (70.8) | 4.9 (58.4) | 5.3 (68.3) | 3.4 (38.7) |
| Refinement | |||||
| Rwork/Rfree (%) | 16.3/22.1 | 16.0/19.7 | 15.8/20.1 | 16.0/20.1 | 15.6/18.9 |
| r.m.s.d.b for bond lengths (Å) | 0.017 | 0.016 | 0.018 | 0.017 | 0.017 |
| r.m.s.d.b for bond angles (°) | 1.623 | 1.458 | 1.618 | 1.598 | 1.47 |
| Mean/Wilson plot B factor (Å2) | 53.1/55.1 | 41.1/33.8 | 55.9/45.8 | 45.0/35.4 | 41.8/34.9 |
| Ramachandran plot (%)c | |||||
| Favored | 96.99 | 97.00 | 96.52 | 97.31 | 98.05 |
| Allowed | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
a Values in parentheses are for the highest-resolution shell.
b r.m.s.d., root mean square deviation.
c Percentages of residues in favored/allowed regions calculated by the program Molprobity (25).
RESULTS
Structural Characterization of Human PrxIV
We have recently shown that PrxIV is an ER-localized protein synthesized as a preprotein with an amino-terminal signal sequence that is cleaved following translocation into the ER (2). The mature protein differs from other peroxiredoxins in that it contains an additional amino-terminal extension of 40 amino acids. This N-terminal extension contains a single cysteine residue that forms an interchain disulfide bond between individual dimers. Mutating this residue to alanine (C51A) has no effect on enzymatic activity or on the ability of the protein to form decamers (2, 5). To determine the structure of various redox states of PrxIV, we expressed and purified the full-length mature protein (residues 37–271 of the preprotein). We have previously demonstrated that the purified recombinant protein is disulfide-linked between the peroxidatic and resolving cysteine residues (17), so we surmised that crystals formed would be of the oxidized disulfide-bonded form and that crystals formed in the presence of DTT would be of the reduced form. We were able to obtain diffractable crystals for oxidized wild type protein as well as oxidized and reduced protein containing the C51A mutation. There was no difference in the overall folds of the wild type and C51A mutant PrxIV, and as the C51A mutant gave slightly higher resolution, we describe its structure here. Data collection and refinement statistics can be found in Table 1 for all atomic models described.
As with other mammalian typical 2-Cys peroxiredoxins (12), the reduced form adopts a (α2)5 decamer consisting of five dimers (Fig. 2A) forming a ring donut structure with the N termini pointing toward the inside and the C termini pointing toward the outside of the ring. The N-terminal region was not visible in either the wild type (not shown) or the C51A structure but probably extends into the center of the ring. We were able to model residues 73–269 for the reduced protein at 2.05 Å resolution. Unlike other mammalian peroxiredoxins, the oxidized protein also formed a stable decamer with a very similar arrangement of dimers (Fig. 2A). Here the last 25 amino acids were not visible in the structure, and hence, we were only able to model residues 76–246, but still at high resolution (2.30 Å). The individual dimers of the reduced and oxidized protein had similar core globular structures with a thioredoxin-like fold (Fig. 2B, reduced protein). A central twisted β-sheet is surrounded by α-helices with the β1- and β8-strands interacting between the two monomers in the dimer. In addition, the α6-helix in the reduced protein extends from one monomer to interact and form a cap over the α2-helix of the partner subunit. The α2-helix contains the peroxidatic cysteine, which sits within a pocket surrounded by the β4- and β5-strands as well as the α3 and α5-helices. The α6-helix is at the C terminus and is the helix that becomes disordered in the oxidized protein (Fig. 2C). The dimers are, therefore, stabilized by the close interaction of the β-sheets for both the reduced and oxidized forms.
FIGURE 2.
Structures of reduced and oxidized form of PrxIV. A, diagram of reduced and oxidized PrxIV (C51A) decamer structures. The N and C termini of the each chain are labeled as N and C, respectively. B, diagram of reduced PrxIV dimer structure. The peroxidatic cysteine sulfur atom is shown as a yellow ball. The secondary structure of one monomer is labeled. C, expanded representation of the structure between Cys-124 and Cys-245 from different chains colored blue and red. The cysteine side chains are shown as yellow sticks, and sulfur atoms are shown as yellow balls.
In addition to the disordering of the α6-helix, there is also an unwinding of the α2-helix when comparing the reduced and oxidized forms (Fig. 2C). In the reduced form, the peroxidatic and resolving cysteines are separated by ∼13 Å. The disulfide between the peroxidatic (Cys-124) and the resolving (Cys-245) cysteine is clearly visible in the oxidized structure. To form this disulfide, a combination of the unwinding of the α2-helix containing the peroxidatic cysteine and the repositioning of the resolving cysteine is required. The transition from the FF to the LU conformation is characteristic of all typical 2-Cys peroxiredoxins (12) and is usually accompanied by the dissolution of the decameric structure to form individual dimers (Fig. 3A). The ability to form stable decamers containing a disulfide between the peroxidatic and resolving cysteine residue is, therefore, a unique feature of PrxIV.
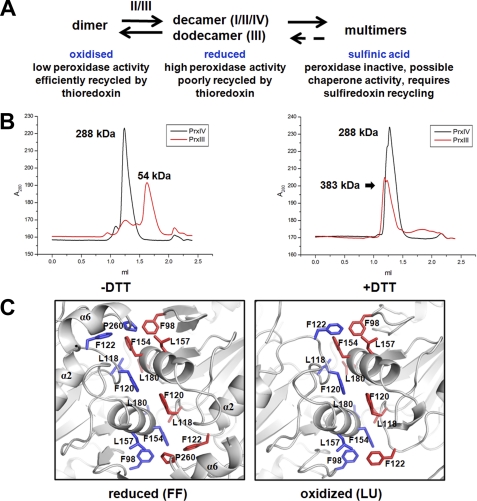
FIGURE 3.
PrxIV forms stable decamer in both reduced and oxidized conditions. A, schematic representation of the dynamic changes between oligomeric states of peroxiredoxins in different redox conditions and their possible functions. The roman numbers represent different peroxiredoxins. B, size exclusion chromatograms of PrxIII and PrxIV carried out under normal or reducing conditions on a Superdex 200 column. The column was pre-equilibrated with buffer with or without DTT as indicated. C, detailed representation of the dimer-dimer interface in PrxIV. The hydrophobic side chains are highlighted. Blue and red represent different chains of PrxIV.
Stabilization of the Decamer in PrxIV
For typical 2-Cys peroxiredoxins, a transition from a decamer to a dimer is thought to be required to complete the enzymatic cycle as the decamer is poorly recycled by thioredoxin, whereas the dimer has only low peroxidase activity (27) (Fig. 3A). Our previous work has shown that the recombinant protein used in our crystallization studies is enzymatically active and is efficiently recycled by a thioredoxin domain containing proteins such as PDI and ERp46 (5). To investigate whether the oligomeric state of PrxIV in solution was altered during reduction, we carried out size exclusion chromatography in the presence or absence of DTT (Fig. 3B). As a control, we also carried out a similar experiment with PrxIII, which is known to undergo a dodecamer-dimer transition in solution upon reduction (28). Although the PrxIII eluted as a dimer in the absence of reduction and a higher sized oligomer in the presence of DTT, the elution profile of PrxIV was similar regardless of DTT treatment. These results demonstrate that the stable oxidized decamer seen in the crystal structure can also be identified by size exclusion chromatography.
To try to explain the unusual stability of the PrxIV, we compared the dimer-dimer interface of the FF and LU structures (Fig. 3C). The reduced structure reveals the presence of several hydrophobic interactions between amino acid side chains that are likely to account for the stability of the decamer. When the FF and LU structures are compared, it can clearly be seen that the α6-helix becomes disordered, resulting in the displacement of Pro-260 from the interface. In addition, the consequence of the α2-helix unwinding is that Phe-122 now relocates to the space previously occupied by Pro-260. The reorganization of these two residues during the FF to LU conformations may well account, at least in part, for the maintenance of the decameric structure. Interestingly it has been speculated that the conformational changes leading to dissociate the decamer to dimer are initiated by the unwinding of the α2-helix in other 2-Cys peroxiredoxins (27). It would appear that the equivalent transition in PrxIV does not cause the same dissociation as the interface remains stabilized due to the repositioning of Phe-122.
Structural Characterization of a Resolving Cysteine Mutant of PrxIV
To gain greater insights into the mechanism of action of PrxIV, we prepared a mutant of PrxIV in which the resolving cysteine (Cys-245) was mutated to alanine. Such a mutant would not be able to complete its enzymatic cycle and should be blocked following oxidation of the peroxidatic cysteine. We solved the structure of the C245A mutant following incubation of the protein in either hydrogen peroxide or DTT (Fig. 4A). Under both conditions, the protein formed a stable decamer. As with the wild type protein, in the presence of DTT, the protein adopts an FF conformation with an ordered α6-helix with the peroxidatic cysteine in the active site pocket. The C245A mutant treated with hydrogen peroxide adopted the LU conformation, indicating that the FF form becomes destabilized upon oxidation of the peroxidatic cysteine. The C-terminal region of the protein, including Ala-245, became disordered and was not visible in the structure. We had predicted that preventing the resolution of the peroxidatic cysteine may lead to hyper-oxidation of this residue. However, the peroxidatic cysteine in the crystal structure was only oxidized to a sulfenic acid and not further modified to a sulfinic or sulfonic form (Fig. 4A, inset). These results suggest that for further modification of the peroxidatic cysteine to occur, it must be present in the active site pocket.
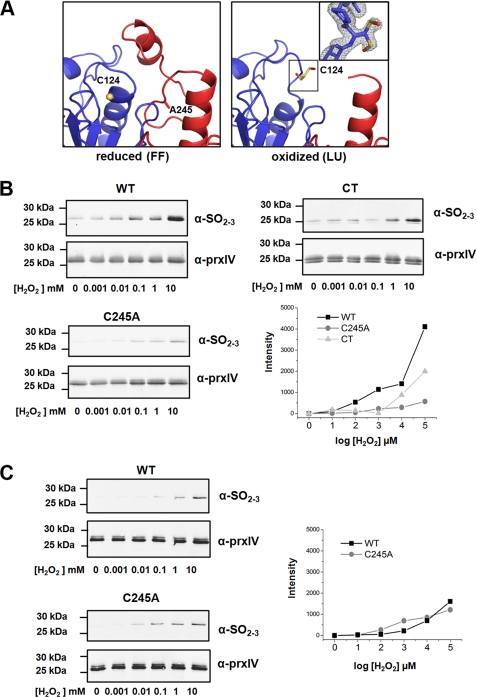
FIGURE 4.
The effect of protein conformation on PrxIV hyper-oxidation. A, expanded view of the active sites of PrxIV C245A mutant in both reduced and oxidized form. Peroxidatic cysteines are shown as yellow sticks, and the oxygen atom is shown as red. The inset is the 2Fo − Fc electron density map around Cys-124 contoured at 1.0 σ. Dual conformation of sulfenylated cysteine is modeled. B, dose-dependent increase in PrxIV wild type, C245A, and C-terminally truncated (CT) mutant hyper-oxidation after exposure to hydrogen peroxide at the indicated concentrations. PrxIV hyper-oxidation was monitored by Western blotting using an antibody that recognizes the sulfinic or sulfonic forms of PrxIV (α-SO2–3). The assay was carried out in the presence of a recycling system consisting of thioredoxin, thioredoxin reductase, and NADPH. Western blots were used for quantification of hyper-oxidized PrxIV by fluorescent intensity. Intensity of hyper-oxidized PrxIV was adjusted for loading and subtracted by lane 1 (no H2O2). Data presented are representative experiments performed three times with similar results. C, wild type or C245A mutant PrxIV was reduced with DTT before excess DTT was removed. Protein was exposed to the indicated concentrations of hydrogen peroxide without a recycling system. Western blots were developed and quantified as in B.
To further investigate the consequence of the C245A mutation on the ability of PrxIV peroxidatic cysteine to become modified to sulfinic or sulfonic derivatives, we determined the relative sensitivity of the wild type or mutant proteins to hydrogen peroxide. Hyper-oxidation was assayed by the reactivity of the protein toward an antibody that specifically recognizes the sulfinic or sulfonic forms of the peroxidatic cysteine (Fig. 4B). The assay was carried out in the presence of a recycling system consisting of thioredoxin, thioredoxin reductase, and NADPH. Note that purified recombinant PrxIV often migrates as a doublet under reducing conditions, probably due to modification of the histidine tag (29). Under these conditions, the C245A mutant is relatively insensitive to hyper-oxidation when compared with the wild type protein even at quite high concentrations of hydrogen peroxide. As expected, the wild type protein is insensitive to hyper-oxidation if denatured prior to the addition of hydrogen peroxide (data not shown), confirming that the active site pocket needs to be retained for further oxidation to occur. Hence, the lack of resolution of the peroxidatic cysteine in the C245A mutant leads to most of the protein adopting the LU conformation, thereby preventing further oxidation, whereas the recycling of the wild type enzyme ensures that a proportion of the molecules is always in the FF conformation and is, therefore, subject to hyper-oxidation. In addition, the inability of the recycling system to reduce the sulfenylated peroxidatic cysteine would suggest that thioredoxin cannot access this residue even when PrxIV is in the LU conformation.
To demonstrate that the hyper-oxidation was due to recycling of wild type PrxIV, we carried out a similar experiment, but this time, we did not include the recycling system. We reduced the protein with DTT to start with to ensure that all the molecules were in the FF form, removed excess DTT, and then assessed the sensitivity of the reduced protein to hydrogen peroxide (Fig. 4C). We now found that the peroxidatic cysteine in the wild type protein was as insensitive to hyper-oxidation as the C245A mutant and much less sensitive than in the presence of the recycling system. Hence, our ability to crystallize a stable sulfenylated decamer in the LU conformation indicates that the initial oxidation of the peroxidatic cysteine triggers the unwinding of the α2-helix and that, once unwound, it does not re-form the FF conformation until the peroxidatic cysteine is reduced. This reduction step normally occurs following the formation of a disulfide with the resolving cysteine and is catalyzed by a PDI family member.
Consequence of Removing the C-terminal Helix on Peroxidatic Cysteine Oxidation and Recycling
Previous work has demonstrated that the C-terminal helix containing a conserved “YF motif” is required to stabilize the FF conformation and in the process allow hyper-oxidation of the peroxidatic cysteine (30). Indeed removing this region from peroxiredoxins containing a structured C terminus makes the peroxidatic cysteine more resistant to hyper-oxidation (31). We also found that the peroxidatic cysteine within PrxIV lacking a C-terminal helix was more resistant to hyper-oxidation than the wild type protein (Fig. 4B). The C-terminally truncated PrxIV was more sensitive to the effects of oxidation than the C245A mutant, confirming the role of the recycling step for hyper-oxidation of the peroxidatic cysteine (Fig. 4B). The presence of the C-terminal α6-helix and the conserved YF motif in PrxIV would suggest that this enzyme is regulated by hyper-oxidation.
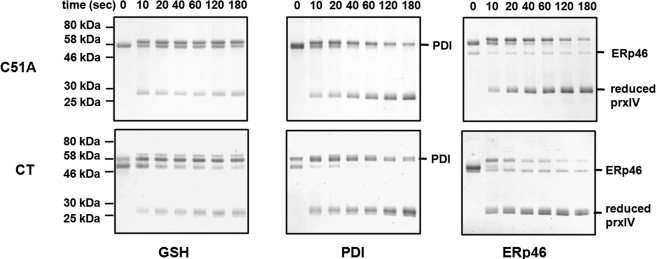
The disordering of the α6-helix upon the formation of the LU conformation could provide a binding site for the PDI family members to allow recycling and oxidation of these oxidoreductases. To test the possibility that the disordered α6-helix forms a binding site for PDI, we assessed the ability of either PDI or ERp46 to reduce the C-terminal truncated PrxIV. Both enzymes accelerated the reduction of PrxIV in the presence of glutathione when compared with glutathione alone (Fig. 5). There was no difference between the efficiency of reduction of PrxIV between wild type and the C-terminal truncation, indicating that the α-6-helix does not provide a binding site for these enzymes.
FIGURE 5.
The effect of truncating the C-terminal helix of PrxIV. A Coomassie Blue stain of a non-reducing gel showing reduction of oxidized PrxIV by PDI or ERp46 in the presence of glutathione or by glutathione alone is shown. Purified C51A or C-terminally truncated mutant PrxIV (6 μm) was incubated with 0.6 μm ERp46 or PDI and 10 mm glutathione or with 10 mm glutathione alone at 37 °C for the indicated times.
DISCUSSION
PrxIV is an important ER-localized peroxiredoxin as it is able to not only metabolize the hydrogen peroxide produced by Ero1 but can also introduce disulfide bonds into members of the PDI family of oxidoreductases (5, 6). The peroxiredoxin family of enzymes is well characterized in terms of their structure, mechanism of action, and regulation (12). Here we have shown that although PrxIV shares many of the characteristics of the other members of the typical 2-Cys peroxiredoxins, it has some unique features that were revealed by the crystal structures of different redox states. In particular, the enzyme forms a decamer that is stable even in its oxidized form. These data confirm previous studies that show that the oxidized or reduced recombinant protein behaves as a decamer as judged by size exclusion chromatography (32) and that the endogenous cellular protein is also a decamer as judged by sucrose gradient analysis (2). We also reveal that when the resolving cysteine residue is mutated to alanine, the enzyme still adopts a decameric structure even in the LU form and that the peroxidatic cysteine is modified to a sulfenic acid. Such a structure has not been seen previously and is likely to be an intermediate in the reaction cycle.
The most striking difference between PrxIV and other peroxiredoxins is the presence of an N-terminal extension in PrxIV. To date there is no indication as to what role this extension plays in the structure or function of the enzyme. The fact that it is not visible in the crystal structure precludes any conclusions to be made concerning its structural organization. The extension does contain a cysteine residue that forms a disulfide between individual dimers. Mutating the cysteine to alanine has no effect on the stability of the decamer (17) or enzymatic activity (5), ruling out the presence of the cysteine as a reason for the N-terminal extension. A structure of PrxIV has been deposited in the PDB (2PN8), which was formed from a recombinant protein lacking the N-terminal extension. There are no obvious differences between this structure and the structure of the reduced full-length protein reported here, indicating that the extension has no effect on the ordered structure. In addition, its removal does not alter the stability of the decamer but does seem to prevent the formation of higher order structures (32) associated with a potential chaperone activity in other typical 2-Cys peroxiredoxins (33). The sequence is rich in acidic amino acid residues, so it could play a role in calcium binding in a similar way to other ER resident proteins (34). Alternatively it may play a role in the retention of PrxIV in the ER because although the enzyme is known to be resident in this organelle, it contains none of the classic ER retrieval sequences (35).
A partial explanation for the stability of the decamer in PrxIV comes from an analysis of the changes to the dimer-dimer interface during the transition from the FF to the LU conformation. As most of the stability of the interface in peroxiredoxins is provided by hydrophobic interactions (36), any changes to this hydrophobic interface are likely to affect the stability of the decamer. We clearly see a repositioning of hydrophobic residues to compensate for the removal of hydrophobic contacts upon the unwinding of the α6-helix. Such a repositioning of hydrophobic residues at the dimer-dimer interface does not occur in other 2-Cys peroxiredoxins (27). The repositioning of residues in PrxIV may well restrict the mobility of the residues lining the dimer-dimer interface, thereby stabilizing the decamer. In other peroxiredoxins, the decamer-to-dimer transition is thought to be crucial for the recycling step with thioredoxin (36). We see no such effect with PrxIV as the oxidized decamer is efficiently reduced by members of the PDI family (5). In contrast, PrxIV is poorly reduced by glutathione (5). Such specificity for the PDI family members would optimize their oxidation in the presence of relatively high glutathione concentrations, ensuring the passage of disulfides from PrxIV to PDI rather than glutathione and subsequently to proteins folding in the ER.
The presence of a C-terminal helix containing a conserved YF motif in PrxIV ensures that the enzyme will be inactivated at high concentrations of hydrogen peroxide. The physiological necessity for this type of regulation of activity still needs to be established. PrxIV can become hyper-oxidized in vivo following the addition of the reducing agent DTT, presumably due to an increase in hydrogen peroxide generated by Ero1 (17); however, there are currently no studies that show PrxIV hyper-oxidation under normal physiological conditions. We show here that the presence of the C-terminal helix clearly renders the enzyme more sensitive to hyper-oxidation, so the fact that this region is conserved in PrxIV would suggest a functional role for this mechanism of inactivation. The presence of the C-terminal helix has no consequence for the recycling step with the PDI family members, arguing against any role in electron transfer between these two proteins.
The structure of the LU conformation of the resolving cysteine mutant reveals that sulfenylation of the peroxidatic cysteine locks the enzyme in this conformation. The inability to hyper-oxidize the peroxidatic cysteine in this mutant demonstrates that there is little or no conversion back to the FF form. In the wild type protein, hyper-oxidation occurs due to the ability to rapidly recycle the peroxidatic cysteine back to its free thiol form once the disulfide between the resolving and peroxidatic cysteine is reduced. An efficient recycling step would ensure that at high concentrations of hydrogen peroxide, there are sufficient molecules in the FF form to allow more than a single hydrogen peroxide to react with the peroxidatic cysteine prior to conversion to the LU conformation. Hence, the efficiency of recycling is an important aspect of hyper-oxidation; inefficient recycling would prevent hyper-oxidation. These observations add detail to our understanding of the mechanism of action of PrxIV. In addition, they may go some way to explaining the specificity of the reduction step for thioredoxin-like proteins rather than the more abundant low molecular weight thiol glutathione. If peroxiredoxins were reduced efficiently by glutathione, they would be predominantly in the reduced FF form and, therefore, more sensitive to inactivation by hydrogen peroxide. It is interesting to note that in cells, despite the high concentration of glutathione, the peroxidatic cysteine of PrxIV is predominantly in a disulfide-bonded form, which would obviously protect it from hyper-oxidation (17). Although the focus of previous studies has been on the ability of these enzymes to become inactivated by hyper-oxidation, it is equally important to recognize that the conformational changes, as well as the recycling step, combine to affect inactivation during the reaction cycle of this important enzyme family.
Acknowledgments
We thank Gordon Lindsay for critical reading of the manuscript. We acknowledge the support of the Diamond Light Source for provision of beam time at stations I02, I03, and I04 and thank Katherine McAuley, James Nicholson, and Thomas Sorensen for their assistance. We also thank Mads Gabrielsen for collecting data for the sulfenylated C245A mutant.
The work was supported by grants from the Wellcome Trust (Grant 88053) and the Scottish University Life Science Alliance (to N. J. B.).
- PrxIV
- peroxiredoxin IV
- ER
- endoplasmic reticulum
- Ero1
- ER oxidase 1
- PDI
- protein disulfide isomerase
- FF
- fully folded
- LU
- locally unfolded.
REFERENCES
- 1. Winterbourn C. C. (2008) Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 2. Tavender T. J., Sheppard A. M., Bulleid N. J. (2008) Biochem. J. 411, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner E., Luche S., Penna L., Chevallet M., Van Dorsselaer A., Leize-Wagner E., Rabilloud T. (2002) Biochem. J. 366, 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 5. Tavender T. J., Springate J. J., Bulleid N. J. (2010) EMBO J. 29, 4185–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zito E., Melo E. P., Yang Y., Wahlander Å., Neubert T. A., Ron D. (2010) Mol. Cell 40, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tavender T. J., Bulleid N. J. (2010) Antioxid. Redox Signal. 13, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 8. El-Benna J., Dang P. M., Gougerot-Pocidalo M. A., Elbim C. (2005) Arch. Immunol. Ther. Exp. (Warsz.) 53, 199–206 [PubMed] [Google Scholar]
- 9. Wu R. F., Ma Z., Liu Z., Terada L. S. (2010) Mol. Cell. Biol. 30, 3553–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeong W., Park S. J., Chang T. S., Lee D. Y., Rhee S. G. (2006) J. Biol. Chem. 281, 14400–14407 [DOI] [PubMed] [Google Scholar]
- 11. Phalen T. J., Weirather K., Deming P. B., Anathy V., Howe A. K., van der Vliet A., Jönsson T. J., Poole L. B., Heintz N. H. (2006) J. Cell Biol. 175, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall A., Nelson K., Poole L. B., Karplus P. A. (2011) Antioxid. Redox Signal. 15, 795–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hall A., Parsonage D., Poole L. B., Karplus P. A. (2010) J. Mol. Biol. 402, 194–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hall A., Karplus P. A., Poole L. B. (2009) FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 16. Barranco-Medina S., Lázaro J. J., Dietz K. J. (2009) FEBS Lett. 583, 1809–1816 [DOI] [PubMed] [Google Scholar]
- 17. Tavender T. J., Bulleid N. J. (2010) J. Cell Sci. 123, 2672–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baker K. M., Chakravarthi S., Langton K. P., Sheppard A. M., Lu H., Bulleid N. J. (2008) EMBO J. 27, 2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pflugrath J. W. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Comparative Computational Project, Number Four (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 26. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 27. Wood Z. A., Poole L. B., Hantgan R. R., Karplus P. A. (2002) Biochemistry 41, 5493–5504 [DOI] [PubMed] [Google Scholar]
- 28. Cao Z., Bhella D., Lindsay J. G. (2007) J. Mol. Biol. 372, 1022–1033 [DOI] [PubMed] [Google Scholar]
- 29. Geoghegan K. F., Dixon H. B., Rosner P. J., Hoth L. R., Lanzetti A. J., Borzilleri K. A., Marr E. S., Pezzullo L. H., Martin L. B., LeMotte P. K., McColl A. S., Kamath A. V., Stroh J. G. (1999) Anal. Biochem. 267, 169–184 [DOI] [PubMed] [Google Scholar]
- 30. Koo K. H., Lee S., Jeong S. Y., Kim E. T., Kim H. J., Kim K., Song K., Chae H. Z. (2002) Arch. Biochem. Biophys. 397, 312–318 [DOI] [PubMed] [Google Scholar]
- 31. Sayed A. A., Williams D. L. (2004) J. Biol. Chem. 279, 26159–26166 [DOI] [PubMed] [Google Scholar]
- 32. Ikeda Y., Ito R., Ihara H., Okada T., Fujii J. (2010) Protein Expr. Purif. 72, 1–7 [DOI] [PubMed] [Google Scholar]
- 33. Jang H. H., Lee K. O., Chi Y. H., Jung B. G., Park S. K., Park J. H., Lee J. R., Lee S. S., Moon J. C., Yun J. W., Choi Y. O., Kim W. Y., Kang J. S., Cheong G. W., Yun D. J., Rhee S. G., Cho M. J., Lee S. Y. (2004) Cell 117, 625–635 [DOI] [PubMed] [Google Scholar]
- 34. Macer D. R., Koch G. L. (1988) J. Cell Sci. 91, 61–70 [DOI] [PubMed] [Google Scholar]
- 35. Trombetta E. S., Parodi A. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 649–676 [DOI] [PubMed] [Google Scholar]
- 36. Matsumura T., Okamoto K., Iwahara S., Hori H., Takahashi Y., Nishino T., Abe Y. (2008) J. Biol. Chem. 283, 284–293 [DOI] [PubMed] [Google Scholar]
- 37. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.4, Schrödinger, LLC, New York [Google Scholar]