Background: Transcription of the α7 nicotinic receptor gene (CHRNA7) is decreased in schizophrenia.
Results: Activating protein-2α (AP-2α) is a potent repressor of CHRNA7 transcription.
Conclusion: AP-2α may contribute to the decreased expression of the CHRNA7 gene in schizophrenia.
Significance: This is the first report of a transcriptional repressor in the human CHRNA7 gene.
Keywords: Nicotinic Acetylcholine Receptors, Promoters, Repressor Protein, Schizophrenia, siRNA, Transcription Factors, AP-2alpha, CHRNA7
Abstract
The CHRNA7 gene, which encodes the α7 nicotinic acetylcholine receptor (α7*nAChR), has been implicated as a candidate gene in schizophrenia. Expression of the α7*nAChR mRNA and protein are reduced in multiple regions of post-mortem brain from patients diagnosed with schizophrenia. Transcriptional regulation may therefore be an important mechanism for the regulation of this gene. A 230-bp proximal promoter fragment, necessary for transcription in cultured neuroblastoma cells, was used to study a putative AP-2α binding site. Mutation of the site indicates that AP-2α plays a negative role in regulating CHRNA7 transcription. This was confirmed through knockdown and overexpression of AP-2α. Electrophoretic mobility shift assays (EMSAs) identified positive DNA-protein interaction at this same site, and supershift assays indicate that the complex includes AP-2α. The interaction was confirmed in cells using chromatin immunoprecipitation (ChIP). DNA methylation was discovered as an anomalous mechanism for CHRNA7 regulation in one cell line. These studies suggest a role for AP-2α regulation of CHRNA7 mRNA expression in multiple tissues during development.
Introduction
Nicotinic acetylcholine receptors (nAChRs)2 are a family of ligand-gated ion channels expressed in both the brain and periphery (1, 2). Genes encoding α and β nAChR subunits assemble as heteromeric receptors, binding nicotine with low affinity. Receptors composed of α7 subunits are thought to assemble as homomers and bind nicotine with low affinity (3, 4). The gene for the α7 subunit (CHRNA7) and the chromosomal region containing the gene have both been implicated as candidate loci for schizophrenia in multiple genetic linkage studies (5–14).
A fragment of 2600 bp in the 5′-upstream regulatory region of the human CHRNA7 gene was cloned, and a proximal promoter of 230 bp upstream of the start codon was identified that is required for transcription (15, 16). As in most other nicotinic acetylcholine receptor genes, the CHRNA7 promoter region is GC-rich and has no canonical CCAAT- or TATA-box. Indirect evidence suggests that the polymorphisms in the proximal promoter of the gene are associated with a sensory processing deficit in schizophrenia and with the disease itself (16, 17). Some of these polymorphisms decrease transcription in an in vitro reporter gene assay and are associated with decreased expression of CHRNA7 mRNA in post-mortem brain (18). Further, a polymorphism in the 5′ upstream regulatory region is strongly associated with schizophrenia (19) and with a positive α7*nAChR agonist effect in the functional magnetic resonance imaging (fMRI) default network in schizophrenics (20).
Published studies of the rodent and bovine CHRNA7 genes showed a single promoter region directly upstream of the transcription start site, containing sequences for putative specificity protein (Sp) family members (Sp1, Sp3, and Sp4), activator protein 2α (AP-2α), and early growth response factor 1 (Egr-1) transcription factor binding (21–23). Whereas Sp1 and Egr-1 activate transcription of the bovine gene (24), Egr-1 represses the murine gene (25). AP-2α was shown to bind the human α3 neuronal nicotinic acetylcholine receptor gene promoter, but its effect on promoter activity was not studied (26–28).
The AP-2 family of transcription factors has, to date, five different proteins in humans and mice: AP-2α, AP-2β, AP-2γ, AP-2δ, and AP-2ϵ, which share a conserved basic helix-span-helix domain necessary and sufficient for dimerization and DNA binding, whereas the transactivation domains among family members are divergent, with only certain critical residues in common (29–38). The AP-2 proteins bind to the palindromic consensus sequence 5′-GCCN3GGC, and AP-2α can also bind the motifs 5′-GCCN4GGC and 5′-GCCN3/4GGG (39). AP-2α has roles in embryonic development, can be stimulated by retinoic acid, and is expressed in neural crest cell lineages in mice (40, 41). The expression pattern seen during murine development, with high expression in differentiating neuroblasts, suggests that AP-2α plays a role in the development of cells important for sensory perception (31). A screen for AP-2α target genes showed a role in repressing transcription of multiple genes involved in differentiation, such as KLF-4 (Krüppel-like factor 4), mEFEMP-1 (epidermal growth factor-containing fibulin-like extracellular matrix protein 1), Mtd (matador), and Stra13 (stimulated by retinoic acid 13), suggesting that AP-2α might be required for cell proliferation by suppressing genes that induce terminal differentiation and apoptosis (42). This is also seen in neuroblastomas, with poorly differentiated tumor cells expressing high levels of AP-2α compared with mature tumor cells (43). AP-2α is a repressor of FGFR1 (fibroblast growth factor receptor 1) transcription during skeletal myogenesis (44).
AP-2α also represses transcription of the human acetylcholinesterase gene (ACHE) and enhances transcription of the human choline acetyltransferase gene (ChAT), both of which are utilized by cholinergic neurons to modulate the effects of acetylcholine (45, 46). Down-regulation of AChE levels could potentially modulate neuronal nicotinic receptor function, because acetylcholine is one of the ligands of the α7*nAChR. Thus, AP-2α may function as a common regulatory element coordinating members of the cholinergic system.
In the current study, we identified an inverse correlation between the expression of CHRNA7 mRNA and the expression of AP-2α mRNA, suggesting that AP-2α may play a negative role in the regulation of the human CHRNA7 gene. We used electrophoretic mobility shift assays (EMSAs), chromatin immunoprecipitation (ChIP), and a luciferase reporter gene assay to identify an essential site of DNA/protein interaction ∼71 bp upstream of the translation start site that binds AP-2α both in vitro and in cells. Using RNA interference and AP-2α expression vectors, AP-2α was found to negatively regulate transcription of CHRNA7 in multiple cell lines.
EXPERIMENTAL PROCEDURES
Cell Culture
The human neuroblastoma cell lines SH-SY5Y, SK-N-BE, HepG2, HeLa, and SK-N-MC were obtained from American Type Culture Collection (ATCC, Manassas, VA). The cell line SH-EP1 was a generous gift from Dr. June Biedler (47). All were grown in 1:1 Ham F-12 Dulbecco's Eagle's medium supplemented with 15% fetal bovine serum, 100 μg/ml streptomycin, 100 units/ml penicillin, and 2 mm l-glutamine at 37 °C in a 5% CO2 incubator.
cDNA Preparation and Quantitative Real-time PCR
Total RNA was isolated from the cell lines SH-SY5Y, SK-N-BE, HeLa, HepG2, SK-N-MC, and SH-EP1 using the TRIzol protocol (Invitrogen). cDNA was prepared using the SuperScript® III First-Strand Synthesis SuperMix (Invitrogen), following the manufacturer's protocols. After preparation of cDNA, standards were produced by mixing a sample of each cDNA reaction together and performing a standard dilution. Quantitative PCR was performed using primers specific to AP-2α, GAPDH, CHRNA7, Sp1, Sp3, Sp4, or Egr-1 mRNA (Table 1) (Proligo, Sigma-Aldrich) as follows. 26-μl reactions containing 1× SYBR Green mix (Bio-Rad), 100 nm primers, and 10 ng of the cDNA were set up in 96-well plates, with standard curves from 2–50 ng of pooled cDNA run alongside the samples on a Bio-Rad iCycler with the MyIQ single color real-time PCR detection system. Values were calculated by subtracting the experimental Ct scores from the 10 ng standard Ct scores and controlling for the efficiency of the reaction and then dividing by the GAPDH scores, as outlined in Ref. 48.
TABLE 1.
Primer sets and probes used in this study
For primers targeting the proximal promoter of CHRNA7, numbering is based on the translation initiation site.
Site-directed Mutagenesis
The promoter-luciferase construct Pr0.23B contains a CHRNA7 promoter fragment from −245 to −15 inserted upstream of the luciferase gene in the pGL3-Basic vector (16). Mutation of the AP-2 binding site was introduced by PCR with the QuikChange II XL site-directed mutagenesis kit, according to the manufacturer's protocols (Stratagene, Agilent Technologies, Santa Clara, CA) and using the mutagenesis primers AP-2mutF and AP-2mutR (Table 1) (Proligo). The mutant construct was confirmed by DNA sequencing and named Pr0.23B-AP-2mut.
Expression Vectors
Expression vectors for AP-2α were kind gifts from Dr. Trevor Williams (Craniofacial Biology, University of Colorado Denver). The expression vector SP(RSV)-AP2 expresses the full-length AP-2α protein under the control of the Rous sarcoma virus long terminal repeat (RSV LTR); SP(RSV)-AP2ΔN278 expresses AP-2α without DNA binding activity through deletion of 278 amino acid residues from the amino terminus, also under the RSV LTR; and SP(RSV)-NN contains the RSV LTR but no coding region (37).
Transient Transfection and Reporter Gene Assay
SH-SY5Y, SK-N-BE, HepG2, HeLa, SK-N-MC, and SH-EP1 cells grown to subconfluence were transfected with the ProFection mammalian transfection calcium phosphate system (Promega, Madison, WI). 1 × 105 cells/well (2 × 105 for SH-SY5Y) on 24-well plates were incubated with 1.25 μg of pGL3 luciferase reporter construct and 0.25 μg of pRL-0 vector as an internal control for the transfection efficiency plus 10 ng of expression vector or empty expression vector (SP (RSV)-AP2, SP(RSV)-AP2ΔN278, or SP(RSV)-NN). Cells were harvested after 48 h, and luciferase activity was determined with the dual luciferase reporter assay system according to the manufacturer's instructions (Promega). The pGL3-Control construct in which the luciferase gene is regulated by the SV40 promoter and enhancer was used as a positive control to measure the maximum reporter gene activity. The pGL3-Basic construct, which has no promoter, was used as a negative control. The pRL-0 construct used for transfection control has a constitutively active Renilla luciferase gene that is not affected by overexpression of transcription factors. All control vectors were obtained from Promega. Eight wells were used for each experiment, and experiments were replicated in triplicate.
Knockdown of AP-2α Using siRNA
RNA interference was performed using Silencer predesigned siRNA (Ambion, Austin, TX) that targets AP-2α (TFAP2A) at exon 5 (siRNA ID 4522) and random negative control siRNA 1, which has no significant sequence similarity to mouse, rat, or human transcript sequences and targets GAPDH at the 5′ medial region of the GAPDH human mRNA sequence. SH-SY5Y, SK-N-BE, HepG2, HeLa, SK-N-MC, and SH-EP1 cells were plated on 24-well plates in antibiotic-free 1:1 Ham F-12 Dulbecco's Eagle's medium supplemented with 15% fetal bovine serum at 1 × 105 cells/well in a total of 0.5 ml of medium. The following day, siRNA was transfected using Lipofectamine 2000 (Invitrogen). Multiple dilutions of the siRNAs were made (200 pmol, 50 pmol, 5 pmol, and 0.5 pmol) and transfected using the manufacturer's protocol. The cells were incubated for 36 h and then harvested using 0.5 ml of TRIzol reagent per well. RNA was extracted using the TRIzol protocol, cDNA was made, and quantitative PCR was performed.
Immunoblot
Cell lysates were prepared after RNAi using radioimmune precipitation buffer with 1× protease inhibitor mixture (Pierce), 1 mm DTT, and 1 mm PMSF. Lysates were run on NuPAGE Novex 4–12% BisTris gels (Invitrogen) using MOPS running buffer, transferred to PVDF, and probed using 1:1000 AP-2α antibody (C-18, sc-184X, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or 1:1000 GAPDH antibody (FL-335, sc-25778), with 1:5000 goat anti-rabbit IgG HRP-conjugated secondary antibody (sc-2004, Santa Cruz Biotechnology, Inc.). Quantification was performed using ImageJ with normalization to GAPDH levels. Experiments were performed in triplicate.
Preparation of Nuclear Extract
Nuclear extracts were prepared from SH-SY5Y, SK-N-BE, HepG2, HeLa, SK-N-MC, and SH-EP1 cell lines using a modified method of Dignam et al. (49). Briefly, cells grown to an 80% confluence (∼1 × 106/ml) were harvested and washed three times with cold PBS, and the pellet was resuspended in five volumes of a hypotonic buffer (10 mm HEPES (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 0.2 mm PMSF, 0.5 mm DTT) with protease inhibitor mixture added to 1× concentration (Sigma-Aldrich). Cells were centrifuged for 5 min in a JS-4.2 rotor at 3000 rpm, and the pellet was resuspended in three volumes of hypotonic buffer, left on ice to swell for 10 min, and then transferred to a Dounce homogenizer. Cell membranes were disrupted with 10 up and down strokes using a type B pestle. After centrifugation for 15 min at 4000 rpm, the pelleted nuclei were resuspended in a half volume of low salt buffer containing 20 mm HEPES (pH 7.9), 25% glycerol, 1.5 mm MgCl2, 200 mm KCl, 0.2 mm EDTA, 0.2 mm PMSF, 0.5 mm DTT, and protease inhibitor mixture (1×). This was followed by adding, in a dropwise fashion, a half volume of high salt buffer (the same as low salt buffer except that KCl was increased to 1.2 m). Nuclei were incubated for 30 min at 4 °C with gentle mixing. The extracted protein was centrifuged for 30 min in a refrigerated microcentrifuge at maximum speed. Supernatant (nuclear extract) was dialyzed at 4 °C against a buffer containing 20 mm HEPES (pH 7.9), 20% glycerol, 100 mm KCl, 0.2 mm EDTA, 0.2 mm PMSF, and 0.5 mm DTT. The extract was centrifuged for 20 min in a refrigerated microcentrifuge at maximum speed and then aliquoted, frozen in liquid nitrogen, and stored at −80 °C. Protein concentration was determined by a Bradford assay (Bio-Rad).
Electrophoretic Mobility Shift Assay
Nuclear extracts (10 μg of protein) were incubated with biotin-labeled double-stranded DNA probes, 3 μg of poly(dI·dC), 3 μg of sonicated salmon sperm DNA, 8% glycerol, 5 μg of bovine serum albumin, and 10× binding buffer (Thermo-Pierce); electrophoresed on a polyacrylamide gel; transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare); UV-cross-linked; and developed using a chemiluminescent reagent. Supershift was performed using 2 μg of an AP-2α antibody (C-18, sc-184X, Santa Cruz Biotechnology, Inc.). The probes used are AP-2 sense annealed to AP-2 antisense or AP-2mut sense annealed to AP-2mut antisense (Table 1).
ChIP and Real-time PCR
ChIP was performed as described (50) using an antibody to human AP-2α (C-18) raised against a peptide at the carboxyl terminus of human AP-2α (Santa Cruz Biotechnology, Inc.) and lysates from SH-SY5Y, SK-N-BE, HeLa, SK-N-MC, and SH-EP1 cells. DNA was purified from the ChIP and used as template in real-time PCRs. Primers against multiple regions of the CHRNA7 promoter were used to generate PCR products ranging in size from 50 to 100 bp using the primers found in Table 1. Twenty-six-microliter reactions containing 1× SYBR Green mix (Bio-Rad), 100 nm primers, and 1:50 dilutions of the ChIP DNA were set up in 96-well plates, with standard curves from 1.25–40 ng of sonicated genomic DNA run alongside the ChIP samples on a Bio-Rad iCycler with the MyIQ single color real-time PCR detection system. Antibody-free ChIP reactions and input DNA values were used to normalize the values from experimental ChIP samples. Values are based on the fold change from beads incubated with no antibody.
DNA Isolation for Methylation Analysis
Cells in TRIzol were bead-homogenized using 1.0-mm high density zirconium oxide beads (Glen Mills). RNA was phenol/chloroform-extracted following the manufacturer's directions. DNA was extracted from the interphase/organic phase left over after RNA isolation by using the back extraction buffer method (TRI Reagent Protocol, Ambion). The protocol was modified to include a more extensive ethanol wash. DNA was quantified by spectrophotometry and stored at 4 °C.
Sodium Bisulfite Conversion, Cloning, and Sequencing
Sample DNA (500 ng) was bisulfite-modified using an EZ DNA methylation kit (Zymo Research, Orange, CA). CpGenome universal methylated and unmethylated DNA (500 ng; Millipore) were used to control for conversion efficiency. After this treatment, >98% of unmethylated C residues were converted to T. Bisulfite-converted DNA (4 μl) was used in a nested PCR with the following primer pairs: outer forward and reverse and nested forward and reverse (Table 1). Both rounds of amplification were performed using a Roche GC-Rich kit (Roche Applied Science). Primers for primary and secondary PCRs are summarized in Table 1. Amplicons were gel-isolated using a Promega Wizard SV gel and PCR clean-up system (Promega). The PCR product was then poly(A)-tailed and cloned using a TOPO TA cloning kit (Invitrogen). Plasmids were extracted using a S.N.A.P. Miniprep kit (Invitrogen). Sequencing was performed in an Applied Biosystems ABI 3100-Avant Genetic Analyzer (Applied Biosystems, Carlsbad, CA) using the Applied Biosystems BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). All procedures followed the manufacturer's directions with the addition of 2.5 μl of a 5 m betaine solution (Sigma-Aldrich) to the sequencing reaction.
Statistical Analysis
All data are presented as means ± S.E. Statistical calculations were performed using GraphPad Prism 4.0 analysis tools. Differences between individual groups were analyzed by paired t test.
RESULTS
AP-2α and CHRNA7 mRNA Expression Are Inversely Correlated
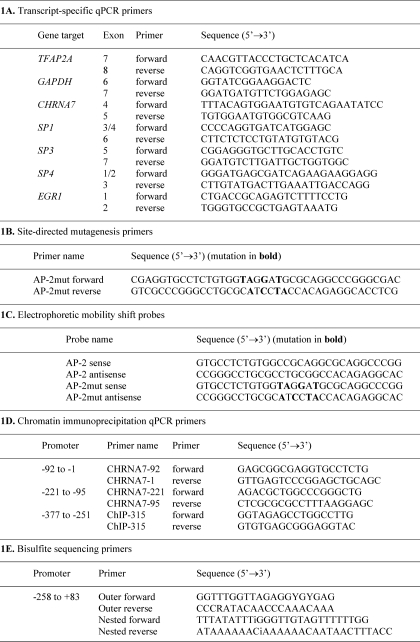
We used in silico analysis to identify a putative transcription factor binding site in the proximal CHRNA7 promoter (−245 to −1), including multiple Sp factor and Egr-1 binding sites, and a single AP-2α binding site. Using quantitative PCR, we measured the levels of mRNA for transcription factors with predicted binding sites in the CHRNA7 promoter region (Fig. 1A; Sp1, Sp3, Sp4, AP-2α, and Egr-1), in multiple human cell lines derived from different tissues. GAPDH was utilized as a control gene. Although levels of Sp1, Sp3, Sp4, and Egr-1 mRNA showed no correlation with levels of CHRNA7 mRNA, we discovered a significant inverse correlation between the levels of AP-2α and the levels of CHRNA7 mRNA. As seen in Fig. 1B, the neuroblastoma cell lines SH-SY5Y and SK-N-BE and the liver carcinoma HepG2 show high levels of CHRNA7 mRNA expression and low levels of AP-2α. No AP-2α mRNA was detected in HepG2 cells, consistent with previous findings (30). The neuroblastoma cell line SK-N-MC and the cervical carcinoma line HeLa instead show high levels of AP-2α and very low to no CHRNA7 mRNA, suggesting possible involvement of AP-2α in the regulation of CHRNA7 transcription.
FIGURE 1.
Inverse correlation between AP-2α and CHRNA7 mRNA expression. A, sequence of the proximal promoter of the CHRNA7, with putative Sp and Egr-1 sites indicated by black and gray bars, respectively, and the AP-2α binding site indicated by the red bar. Mutation of the AP-2α site used in this study is shown in red. B, using quantitative real-time PCR, RNA isolated from SH-SY5Y, SK-N-BE, HepG2, HeLa, SK-N-MC, and SH-EP1 cells grown in complete medium was reverse transcribed to cDNA and analyzed for the expression of Sp1, Sp3, Sp4, Egr-1, AP-2α, and CHRNA7 mRNA, which was normalized to GAPDH mRNA levels. Triplicates were used for each experiment, and experiments were replicated in triplicate. Data are presented as mean ± S.E. (error bars). Ø, undetectable cDNA level after 40 PCR cycles.
Mutation of an AP-2α Binding Site Results in Increased Reporter Gene Activity
To expand on the inverse correlation between AP-2α and CHRNA7 mRNA expression, the putative AP-2α binding site ∼71 bases upstream of the translation start site indicated in Fig. 1A was mutated as shown. This was performed in a 230-bp promoter fragment driving a luciferase reporter gene, using site-directed mutagenesis. As seen in Fig. 2, a significant increase in luciferase expression was seen upon mutation of the putative AP-2α site in all but the SH-SY5Y cell line. This may be due to the low levels of AP-2α expression because the increase in reporter gene transcription correlates with the original levels of AP-2α mRNA. These data, combined with the inverse correlation of mRNA levels, suggest that AP-2α may play a repressive role in CHRNA7 transcription.
FIGURE 2.
Mutation analysis of a putative AP-2α binding site using a reporter gene assay. A 230-bp CHRNA7 proximal promoter fragment was isolated and inserted upstream of a luciferase reporter gene in the vector pGL3-Basic and named Pr0.23B, with or without the AP-2α site mutated by site-directed mutagenesis. Transient transfection was performed using the Pr0.23B vector, AP-2mut vector, or pGL3-Basic with the Renilla luciferase vector pRL-TK as a control for transfection efficiency in multiple cell lines. Activity was determined by taking the ratio of luciferase to Renilla activity, and Pr0.23B activity was set to 100% for each cell line. The empty vector pGL3-Basic shows low expression of the luciferase reporter gene. Eight wells were used for each experiment, and experiments were replicated in triplicate. Data are presented as mean ± S.E. (error bars). *, p < 0.005. MCS, most common sequence.
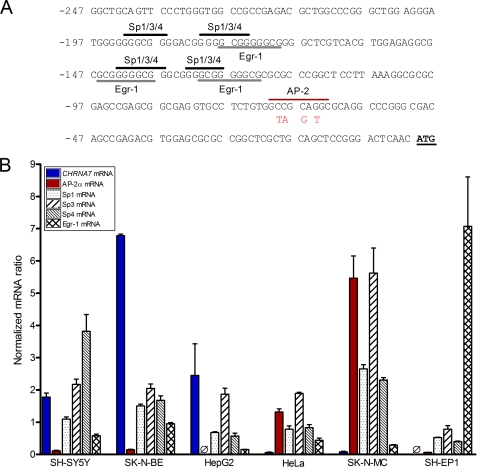
AP-2α Exhibits DNA Binding-dependent Repression of Reporter Gene Activity
Because the previous experiment suggested that the levels of AP-2α might affect the significance of changes in reporter gene expression, we performed overexpression studies using multiple AP-2α expression vectors. These overexpression vectors include a full-length AP-2α expression vector (SP(RSV)-AP2) as well as a vector with truncated AP-2α protein that has dimerization activity but no DNA binding activity (SP(RSV)-AP2ΔN278) (37). SP(RSV)-NN was used as a control for no protein overexpression. Fig. 3A shows that AP-2α overexpression in multiple cell lines causes a significant decrease in expression of a luciferase reporter gene driven by the CHRNA7 proximal promoter, and this decrease no longer occurs when using an AP-2α expression vector that expresses AP-2α without DNA binding activity. SK-N-MC cells and SK-N-BE cells, however, showed no significant response to overexpression of AP-2α. It is interesting to note that the cell lines that show the most robust response in this luciferase reporter assay were both of non-neuronal derivation (HepG2 and HeLa).
FIGURE 3.
AP-2α overexpression and effects on dual luciferase reporter gene assay and endogenous CHRNA7 mRNA expression. A, transient transfection of the Pr0.23B vector with either the empty expression vector SP(RSV)-NN, full-length AP-2α expression vector SP(RSV)-AP2, or the truncated AP-2α expression vector that lacks DNA binding (SP(RSV)-AP2ΔN278), along with the Renilla luciferase vector pRL-0 as a control for transfection efficiency. Activity was determined by taking the ratio of luciferase to Renilla activity, and activity of Pr0.23B with the empty vector was set to 100% for each cell line. Eight wells were used for each experiment, and experiments were replicated in triplicate. Data are presented as means ± S.E. (error bars). *, p < 0.005. B, transient transfection of the empty expression vector SP(RSV)-NN, full-length AP-2α expression vector SP(RSV)-AP2, or the truncated AP-2α expression vector that lacks DNA binding (SP(RSV)-AP2ΔN278) in multiple cell lines. RNA was extracted, cDNA was made, and levels of CHRNA7 mRNA were measured using quantitative PCR and normalized to GAPDH mRNA levels. The level of CHRNA7 mRNA from cells transfected with the empty expression vector was set to 100%. Triplicates were used for each experiment, and experiments were replicated in triplicate. Data are presented as mean ± S.E. *, p < 0.0005.
We also observed a significant reduction of CHRNA7 mRNA in the non-neuronal cell line HepG2 with overexpression of AP-2α, based on quantitative real-time PCR (Fig. 3B), with no significant effect on endogenous CHRNA7 mRNA expression in either neuronal cell line that normally expresses high levels of CHRNA7 mRNA (SH-SY5Y and SK-N-BE). This may indicate either tighter control over CHRNA7 expression in neuron-derived cells or the presence of a potent enhancer of CHRNA7 transcription in the two neuronal cell lines that compensates for the effects of AP-2α overexpression. HeLa cells also showed a significant reduction of CHRNA7 mRNA, although this reduction did not appear to be DNA binding-dependent. Taken together with the previous data, AP-2α shows a significant repressive effect on CHRNA7 transcription.
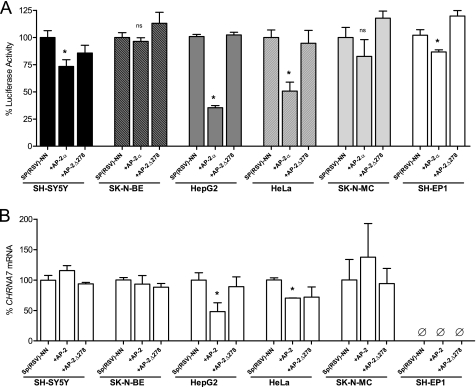
Knockdown of AP-2α Increases CHRNA7 mRNA Levels
RNA interference was performed, using either randomized siRNAs or siRNA directly targeting AP-2α, to determine if the reduction of AP-2α has a direct effect on CHRNA7 mRNA levels. As seen in Fig. 4A, siRNA directly targeting AP-2α reduced AP-2α protein expression in multiple cell lines, excluding HepG2 cells, which do not express AP-2α. GAPDH, used as a loading control, was unaffected by AP-2α knockdown. The protein levels were quantified in Fig. 4B, along with mRNA levels of both AP-2α and CHRNA7, and showed significant decreases of AP-2α protein in all cell lines tested. There was also a significant decrease of AP-2α mRNA in all cell lines except SK-N-BE, although the level trends low as well, with corresponding increases of endogenous CHRNA7 mRNA levels. This confirms that AP-2α expression has a significant negative effect on CHRNA7 mRNA expression. HepG2 cells, which do not express AP-2α, showed no change in CHRNA7 mRNA levels, as expected. SH-EP1 cells did not show an increase of CHRNA7 mRNA levels when AP-2α was reduced; this suggests that regulation of CHRNA7 in this line is through alternate means.
FIGURE 4.
Knockdown of AP-2α and effects on endogenous CHRNA7 mRNA levels. Multiple cell lines were transfected with 50 pmol of siRNA (random or AP-2α-specific) for 48 h. A, total cell lysates were used in Western blot analysis to determine AP-2α protein levels using specific antibody. GAPDH served as a loading control. B, RNA was extracted, and cDNA was made. Levels of AP-2α and CHRNA7 mRNA were measured using quantitative PCR and normalized to GAPDH levels. Triplicates were used for each experiment, and experiments were replicated in triplicate. Protein levels from Fig. 4A were quantified using ImageJ (National Institutes of Health) and normalized to GAPDH protein levels. Data are presented as mean ± S.E. *, p < 0.05; **, p < 0.0005.
AP-2α Specifically Binds to the CHRNA7 Promoter AP-2 Binding Site
The previous experiments assume that the site of AP-2α binding is the one found at 71 bases upstream of the translation start site. To confirm this, we performed electrophoretic mobility shift assays using a double-stranded DNA oligonucleotide to assay binding of AP-2α. Fig. 5 shows these in vitro protein-binding assays. Cell line nuclear extracts, prepared from SH-SY5Y, SK-N-BE, and SK-N-MC, contained DNA binding proteins. Binding to the AP-2 site (Fig. 5, lane 2) was specifically competed out with an excess of unlabeled probe (Fig. 5, lane 3) but is not competed out with an excess of mutant probe (Fig. 5, lane 4). Nonspecific bands (ns) remain regardless of competitor, as indicated, with multiple bands (asterisks) suggesting the presence of ternary complexes that may or may not include AP-2α. These ternary bands have yet to be investigated for potential function. Labeled mutant probe alone (Fig. 5, lane 5) shows differential and nonspecific binding, indicating that the mutant cannot bind AP-2α. Supershift analysis by the addition of the AP-2α antibody indicates that the site is specifically bound by AP-2α (Fig. 5, lane 6). Although the supershift in SK-N-BE cells overlaps a nonspecific band, the band is significantly darker only in the lane with the antibody added and was consistently replicated as such.
FIGURE 5.
EMSA of binding to the CHRNA7 promoter AP-2 binding site at −71. Double-stranded oligonucleotides containing the normal (wt) or the mutated (mt) AP-2 site and the flanking sequences as found in the human most common sequence CHRNA7 promoter were 3′-end-labeled with biotin and incubated with nuclear extracts prepared from SH-SY5Y, SK-N-BE, SK-N-MC, HeLa (not shown), HepG2 (not shown), or SH-EP1 (not shown) cell lines as indicated. Extract was present in lanes 2–6 of each gel, wild type labeled probe was used in lanes 2–4 and 6, and mutated labeled probe was used in lane 5. Unlabeled competitor wild type probe was present in 100-fold excess in lane 3, and unlabeled competitor mutated probe was present in 100-fold excess in lane 4. Anti-AP-2α antibody was used for supershifting in lane 6. Protein-DNA complexes are indicated by the arrows labeled AP-2α, and the antibody supershifted complexes are indicated by the arrows labeled SS. Protein complexes indicated by the arrows labeled with asterisks are unknown ternary complexes. EMSAs were replicated two times for each cell extract, and three separately prepared extracts were used for each cell line.
As expected, the AP-2α site probe did not bind protein in HepG2 cells, which express no AP-2α protein; nor did we see specific binding in SH-EP1 cells, which have relatively low levels of AP-2α expression. Interestingly, we found no protein binding in HeLa nuclear extracts, which may indicate that this specific site is not able to bind protein in non-neuronal cells. EMSAs that showed no DNA-protein interaction consistently replicated as such and are not shown.
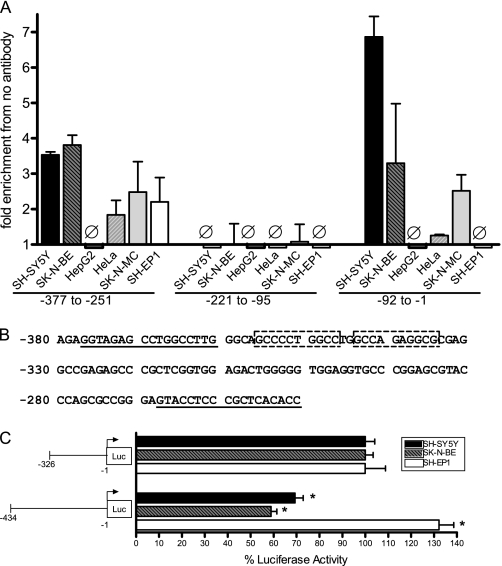
AP-2α Specifically Binds to the CHRNA7 Promoter Region Containing the AP-2 Binding Site
To determine if the protein binding seen in vitro is also present in cells, we used ChIP to isolate DNA fragments bound by AP-2α, using the anti- AP-2α antibody, followed by quantitative PCR to identify the DNA regions isolated for the CHRNA7 promoter. As a negative control, we performed the ChIP with no antibody. In Fig. 6A, we show that AP-2α is indeed bound at the −71 region of the CHRNA7 promoter in multiple cell lines. Interestingly, AP-2α is not bound at −71 in SH-EP1 cells, which suggests an alternate mechanism of CHRNA7 regulation, possibly through DNA methylation (see below), which could block AP-2α binding to the proximal promoter. AP-2α is also not bound at −71 in HeLa cells, which is consistent with the in vitro binding results and suggests that this site may not be bound by AP-2α in this line, either through polymorphisms in the genomic DNA or through regulation by other factors.
FIGURE 6.
Chromatin immunoprecipitation identification of AP-2α binding sites in the CHRNA7 promoter. A, SH-SY5Y, SK-N-BE, SK-N-MC, HeLa, HepG2, or SH-EP1 cells were formaldehyde-cross-linked and lysed, and lysates were sonicated to obtain DNA fragments 200–300 bp in length. Immunoprecipitation was performed using Sepharose beads and anti-AP-2α antibody or no antibody plus lysate. DNA fragments isolated by immunoprecipitation were used for quantitative PCR to amplify relevant CHRNA7 promoter sequences as indicated, using the primers shown in Table 1. Fold enrichment compared with no antibody control is indicated. Ø, no change from controls. Triplicates were used for each experiment, and experiments were replicated in triplicate. Data are presented as mean ± S.E. (error bars). B, a second region containing AP-2α binding was identified using chromatin immunoprecipitation. Two putative AP-2α consensus binding sites are indicated by boxes within the CHRNA7 promoter region. The positions of primers ChIP-315F and ChIP-315R are underlined. C, transient transfection of SH-SY5Y, SK-N-BE, and SH-EP1 cells with luciferase reporter vectors containing 5′ deletions of the CHRNA7 promoter at the second putative AP-2α binding region, normalized to Renilla expression from the pRL-TK vector. Eight wells were used for each experiment, and experiments were replicated in triplicate. −326 was set to 100% for each cell line. Data are presented as mean ± S.E. *, p < 0.0005.
We also identified a second site further upstream that shows significant binding of AP-2α, which in silico analysis suggests may be found at −347, indicated in Fig. 6B. This site has yet to be studied using mutagenesis and in vitro protein binding assays. However, deletion of the region containing this putative site does increase reporter gene expression in SH-SY5Y and SK-N-BE cell lines (Fig. 6C). The presence of AP-2α binding in cell lines that have little AP-2α and very high levels of CHRNA7 mRNA suggests that the presence and binding of AP-2α alone is not enough to repress transcription and may indicate that transcription in these cell lines is driven by a potent neuronal enhancer.
SH-EP1 Cells Have Robust DNA Methylation in the CHRNA7 Proximal Promoter Region
After multiple experiments showed that SH-EP1 cells have no CHRNA7 mRNA yet no presence of AP-2α binding, we performed analysis of DNA methylation in the CHRNA7 proximal promoter region in multiple cell lines. As seen in Fig. 7, the proximal promoter region of SH-EP1 is highly methylated, whereas the other cell lines studied showed very little methylation. It is interesting to note that the SH-EP1 methylation is contained in the region that normally binds AP-2α, and previous studies suggest that AP-2α cannot bind methylated DNA (51, 52). This provides a possible explanation for the lack of AP-2α binding as well as suggesting an alternative mechanism for repression of CHRNA7 mRNA levels.
FIGURE 7.
DNA methylation in the proximal promoter of CHRNA7. DNA isolated from SH-SY5Y, SK-N-BE, HeLa, and SH-EP1 cells was used for sodium bisulfite conversion to identify methylated CpG islands within the CHRNA7 promoter region. The proximal promoter region is indicated by the location of the CpG islands and the percentage of samples showing methylation. SH-EP1 cells show heavy methylation over the region where the original AP-2α binding site is located (−71).
DISCUSSION
The prevalence of smoking in the common mental illness schizophrenia is much higher than in the general population, implicating nicotinic receptors in the disorder (53, 54). Expression of the α7 neuronal nicotinic acetylcholine receptor, α7*nAChR, is decreased in post-mortem hippocampus, cortex, and reticular nucleus of the thalamus in schizophrenia (55–58), suggesting that smoking may be a form of self-medication (59, 60). The α7*nAChR is a major pharmacological target in the development of treatments for schizophrenia and other neurological disorders (61).
Multiple polymorphisms have been isolated in the 5′ regulatory region of the CHRNA7 gene, some of which are associated with schizophrenia and sensory processing or fMRI responses in the disorder (16, 19, 20). Although the function of specific transcription factors that bind in the bovine and rat genes has been reported (21, 22), transcription factor function in the human CHRNA7 promoter has not been well defined. The current study describes a potent transcriptional repressor of this gene in humans, which may have long reaching effects on the production of α7 subunits and, subsequently, α7*nAChRs.
The AP-2α gene maps near the marker D6S470 on chromosome 6p24, which has previously been linked to schizophrenia (62–65). Linkage analysis performed on schizophrenics and controls showed that three promoter polymorphisms found in AP-2α are not significantly associated with schizophrenia, although one polymorphism did show significant genotype distribution differences in patients with an episodic course versus controls, suggesting that a larger cohort of schizophrenia patients might be required for significance (66).
AP-2α is a well studied transcription factor that is involved in multiple signaling pathways, both as an activator and as a repressor, with roles in regulating differentiation, proliferation, and apoptosis during development and in specific tissues. Specifically, it represses genes that lead to differentiation and apoptosis while activating genes involved in cell proliferation and is abundantly expressed in neural tube progenitors during embryogenesis (42, 44, 67).
For example, AP-2α acts as a repressor of peroxisome proliferator-activated receptor β/δ (PPARβ/δ). In lung carcinoma, nicotine increases the expression of PPARβ/δ through activation of α7*nAChRs. The α7*nAChRs mediate activation of phosphatidylinositol 3-kinase (PI3K) and mammalian target of rapamycin (mTOR), which inhibits AP-2α protein expression to increase levels of PPARβ/δ. Nicotine itself reduces AP-2α mRNA levels while increasing the levels of α7*nAChRs (67–69). This suggests a potential feedback mechanism in which active α7*nAChRs can further increase expression of CHRNA7 mRNA in response to nicotine binding by reducing the levels of AP-2α expression.
Mood stabilizers used to treat bipolar disorder affect both AP-2α expression and DNA binding. In the rat brain, some of the treatments for mania (lithium, carbamazepine, valproate) or depression (lamotrigine) in bipolar disorder reduce levels of AP-2α, which is involved in expression of cytosolic phospholipase A(2) (70, 71). Activation of AP-2α requires phosphorylation by PKA or PKCϵ (72). Chronic lithium treatment decreased PKAα and PKCϵ protein levels as well as arachadonic acid-dependent PKC activity in rat brain, decreasing phosphorylation and activity of AP-2α (73). These drugs are sometimes used in combination with antipsychotics in the treatment of schizophrenia (74, 75) and could affect both α7*nAChR receptor function and CHRNA7 transcription.
In summary, repression of CHRNA7 mRNA by AP-2α offers an extra layer of control over CHRNA7 expression and might be evaluated as a potential modulatory factor in the development of pharmacological targets to treat the symptoms of schizophrenia and other mood disorders.
Acknowledgments
We thank Trevor Williams for the AP-2α expression vectors and David Bentley, Soojin Kim, Benjamin Erickson, and Ralph Berger for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DA009457 and MH081177 and a Veterans Affairs Medical Research Service Merit Review (to S. L.).
- nAChR
- nicotinic acetylcholine receptor
- RSV
- Rous sarcoma virus
- PPARβ/δ
- peroxisome proliferator-activated receptor β/δ
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W. (2009) Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leonard S., Bertrand D. (2001) Nicotine Tob. Res. 3, 203–223 [DOI] [PubMed] [Google Scholar]
- 3. Gerzanich V., Anand R., Lindstrom J. (1994) Mol. Pharmacol. 45, 212–220 [PubMed] [Google Scholar]
- 4. Drisdel R. C., Green W. N. (2000) J. Neurosci. 20, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freedman R., Coon H., Myles-Worsley M., Orr-Urtreger A., Olincy A., Davis A., Polymeropoulos M., Holik J., Hopkins J., Hoff M., Rosenthal J., Waldo M. C., Reimherr F., Wender P., Yaw J., Young D. A., Breese C. R., Adams C., Patterson D., Adler L. E., Kruglyak L., Leonard S., Byerley W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaufmann C. A., Suarez B., Malaspina D., Pepple J., Svrakic D., Markel P. D., Meyer J., Zambuto C. T., Schmitt K., Matise T. C., Harkavy Friedman J. M., Hampe C., Lee H., Shore D., Wynne D., Faraone S. V., Tsuang M. T., Cloninger C. R. (1998) Am. J. Med. Genet. 81, 282–289 [PubMed] [Google Scholar]
- 7. Stöber G., Saar K., Rüschendorf F., Meyer J., Nürnberg G., Jatzke S., Franzek E., Reis A., Lesch K. P., Wienker T. F., Beckmann H. (2000) Am. J. Hum. Genet. 67, 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu C. M., Hwu H. G., Lin M. W., Ou-Yang W. C., Lee S. F., Fann C. S., Wong S. H., Hsieh S. H. (2001) Am. J. Med. Genet. 105, 658–661 [DOI] [PubMed] [Google Scholar]
- 9. Tsuang D. W., Skol A. D., Faraone S. V., Bingham S., Young K. A., Prabhudesai S., Haverstock S. L., Mena F., Menon A. S., Bisset D., Pepple J., Sauter F., Baldwin C., Weiss D., Collins J., Boehnke M., Schellenberg G. D., Tsuang M. T. (2001) Am. J. Med. Genet. 105, 662–668 [PubMed] [Google Scholar]
- 10. Xu J., Pato M. T., Torre C. D., Medeiros H., Carvalho C., Basile V. S., Bauer A., Dourado A., Valente J., Soares M. J., Macedo A. A., Coelho I., Ferreira C. P., Azevedo M. H., Macciardi F., Kennedy J. L., Pato C. N. (2001) Am. J. Med. Genet. 105, 669–674 [DOI] [PubMed] [Google Scholar]
- 11. Harrison P. J., Weinberger D. R. (2005) Mol. Psychiatry 10, 40–68 [DOI] [PubMed] [Google Scholar]
- 12. International Schizophrenia Consortium (2008) Nature 455, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stefansson H., Rujescu D., Cichon S., Pietiläinen O. P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J. E., Hansen T., Jakobsen K. D., Muglia P., Francks C., Matthews P. M., Gylfason A., Halldorsson B. V., Gudbjartsson D., Thorgeirsson T. E., Sigurdsson A., Jonasdottir A., Jonasdottir A., Bjornsson A., Mattiasdottir S., Blondal T., Haraldsson M., Magnusdottir B. B., Giegling I., Möller H. J., Hartmann A., Shianna K. V., Ge D., Need A. C., Crombie C., Fraser G., Walker N., Lonnqvist J., Suvisaari J., Tuulio-Henriksson A., Paunio T., Toulopoulou T., Bramon E., Di Forti M., Murray R., Ruggeri M., Vassos E., Tosato S., Walshe M., Li T., Vasilescu C., Mühleisen T. W., Wang A. G., Ullum H., Djurovic S., Melle I., Olesen J., Kiemeney L. A., Franke B., Sabatti C., Freimer N. B., Gulcher J. R., Thorsteinsdottir U., Kong A., Andreassen O. A., Ophoff R. A., Georgi A., Rietschel M., Werge T., Petursson H., Goldstein D. B., Nöthen M. M., Peltonen L., Collier D. A., St Clair D., Stefansson K. (2008) Nature 455, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shinawi M., Schaaf C. P., Bhatt S. S., Xia Z., Patel A., Cheung S. W., Lanpher B., Nagl S., Herding H. S., Nevinny-Stickel C., Immken L. L., Patel G. S., German J. R., Beaudet A. L., Stankiewicz P. (2009) Nat. Genet. 41, 1269–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gault J., Robinson M., Berger R., Drebing C., Logel J., Hopkins J., Moore T., Jacobs S., Meriwether J., Choi M. J., Kim E. J., Walton K., Buiting K., Davis A., Breese C., Freedman R., Leonard S. (1998) Genomics 52, 173–185 [DOI] [PubMed] [Google Scholar]
- 16. Leonard S., Gault J., Hopkins J., Logel J., Vianzon R., Short M., Drebing C., Berger R., Venn D., Sirota P., Zerbe G., Olincy A., Ross R. G., Adler L. E., Freedman R. (2002) Arch. Gen. Psychiatry 59, 1085–1096 [DOI] [PubMed] [Google Scholar]
- 17. Houy E., Raux G., Thibaut F., Belmont A., Demily C., Allio G., Haouzir S., Fouldrin G., Petit M., Frebourg T., Campion D. (2004) Mol. Psychiatry 9, 320–322 [DOI] [PubMed] [Google Scholar]
- 18. Mexal S., Berger R., Logel J., Ross R. G., Freedman R., Leonard S. (2010) J. Mol. Neurosci. 40, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stephens S. H., Logel J., Barton A., Franks A., Schultz J., Short M., Dickenson J., James B., Fingerlin T. E., Wagner B., Hodgkinson C., Graw S., Ross R. G., Freedman R., Leonard S. (2009) Schizophr. Res. 109, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tregellas J. R., Tanabe J., Rojas D. C., Shatti S., Olincy A., Johnson L., Martin L. F., Soti F., Kem W. R., Leonard S., Freedman R. (2011) Biol. Psychiatry 69, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagavarapu U., Danthi S., Boyd R. T. (2001) J. Biol. Chem. 276, 16749–16757 [DOI] [PubMed] [Google Scholar]
- 22. Carrasco-Serrano C., Campos-Caro A., Viniegra S., Ballesta J. J., Criado M. (1998) J. Biol. Chem. 273, 20021–20028 [DOI] [PubMed] [Google Scholar]
- 23. Carrasco-Serrano C., Viniegra S., Ballesta J. J., Criado M. (2000) J. Neurochem. 74, 932–939 [DOI] [PubMed] [Google Scholar]
- 24. Criado M., Domínguez del Toro E., Carrasco-Serrano C., Smillie F. I., Juíz J. M., Viniegra S., Ballesta J. J. (1997) J. Neurosci 17, 6554–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reynolds P. R., Hoidal J. R. (2005) J. Biol. Chem. 280, 32548–32554 [DOI] [PubMed] [Google Scholar]
- 26. Yang X., Fyodorov D., Deneris E. S. (1995) J. Biol. Chem. 270, 8514–8520 [DOI] [PubMed] [Google Scholar]
- 27. Fornasari D., Battaglioli E., Flora A., Terzano S., Clementi F. (1997) Mol. Pharmacol. 51, 250–261 [DOI] [PubMed] [Google Scholar]
- 28. Terzano S., Flora A., Clementi F., Fornasari D. (2000) J. Biol. Chem. 275, 41495–41503 [DOI] [PubMed] [Google Scholar]
- 29. Bosher J. M., Williams T., Hurst H. C. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williams T., Admon A., Lüscher B., Tjian R. (1988) Genes Dev. 2, 1557–1569 [DOI] [PubMed] [Google Scholar]
- 31. Moser M., Imhof A., Pscherer A., Bauer R., Amselgruber W., Sinowatz F., Hofstädter F., Schüle R., Buettner R. (1995) Development 121, 2779–2788 [DOI] [PubMed] [Google Scholar]
- 32. Cheng C., Ying K., Xu M., Zhao W., Zhou Z., Huang Y., Wang W., Xu J., Zeng L., Xie Y., Mao Y. (2002) Int. J. Biochem. Cell Biol. 34, 78–86 [DOI] [PubMed] [Google Scholar]
- 33. Oulad-Abdelghani M., Bouillet P., Chazaud C., Dollé P., Chambon P. (1996) Exp. Cell Res. 225, 338–347 [DOI] [PubMed] [Google Scholar]
- 34. Zhao F., Satoda M., Licht J. D., Hayashizaki Y., Gelb B. D. (2001) J. Biol. Chem. 276, 40755–40760 [DOI] [PubMed] [Google Scholar]
- 35. García M. A., Campillos M., Ogueta S., Valdivieso F., Vázquez J. (2000) J. Mol. Biol. 301, 807–816 [DOI] [PubMed] [Google Scholar]
- 36. Wankhade S., Yu Y., Weinberg J., Tainsky M. A., Kannan P. (2000) J. Biol. Chem. 275, 29701–29708 [DOI] [PubMed] [Google Scholar]
- 37. Williams T., Tjian R. (1991) Genes Dev. 5, 670–682 [DOI] [PubMed] [Google Scholar]
- 38. Williams T., Tjian R. (1991) Science 251, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 39. Mohibullah N., Donner A., Ippolito J. A., Williams T. (1999) Nucleic Acids Res. 27, 2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitchell P. J., Timmons P. M., Hébert J. M., Rigby P. W., Tjian R. (1991) Genes Dev. 5, 105–119 [DOI] [PubMed] [Google Scholar]
- 41. Lüscher B., Mitchell P. J., Williams T., Tjian R. (1989) Genes Dev. 3, 1507–1517 [DOI] [PubMed] [Google Scholar]
- 42. Pfisterer P., Ehlermann J., Hegen M., Schorle H. (2002) J. Biol. Chem. 277, 6637–6644 [DOI] [PubMed] [Google Scholar]
- 43. Schulte J. H., Kirfel J., Lim S., Schramm A., Friedrichs N., Deubzer H. E., Witt O., Eggert A., Buettner R. (2008) Cancer Lett. 271, 56–63 [DOI] [PubMed] [Google Scholar]
- 44. Mitchell D. L., DiMario J. X. (2010) Exp. Cell Res. 316, 194–202 [DOI] [PubMed] [Google Scholar]
- 45. Getman D. K., Mutero A., Inoue K., Taylor P. (1995) J. Biol. Chem. 270, 23511–23519 [DOI] [PubMed] [Google Scholar]
- 46. Baskin F., Li Y. P., Hersh L. B., Davis R. M., Rosenberg R. N. (1997) Neuroscience 76, 821–827 [DOI] [PubMed] [Google Scholar]
- 47. Biedler J. L., Roffler-Tarlov S., Schachner M., Freedman L. S. (1978) Cancer Res. 38, 3751–3757 [PubMed] [Google Scholar]
- 48. Wong M. L., Medrano J. F. (2005) BioTechniques 39, 75–85 [DOI] [PubMed] [Google Scholar]
- 49. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gomes N. P., Bjerke G., Llorente B., Szostek S. A., Emerson B. M., Espinosa J. M. (2006) Genes Dev. 20, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hermann R., Doerfler W. (1991) FEBS Lett. 281, 191–195 [DOI] [PubMed] [Google Scholar]
- 52. Comb M., Goodman H. M. (1990) Nucleic Acids Res. 18, 3975–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Leon J., Diaz F. J. (2005) Schizophr. Res. 76, 135–157 [DOI] [PubMed] [Google Scholar]
- 54. Leonard S., Adler L. E., Benhammou K., Berger R., Breese C. R., Drebing C., Gault J., Lee M. J., Logel J., Olincy A., Ross R. G., Stevens K., Sullivan B., Vianzon R., Virnich D. E., Waldo M., Walton K., Freedman R. (2001) Pharmacol. Biochem. Behav. 70, 561–570 [DOI] [PubMed] [Google Scholar]
- 55. Freedman R., Hall M., Adler L. E., Leonard S. (1995) Biol. Psychiatry 38, 22–33 [DOI] [PubMed] [Google Scholar]
- 56. Guan Z. Z., Zhang X., Blennow K., Nordberg A. (1999) Neuroreport 10, 1779–1782 [DOI] [PubMed] [Google Scholar]
- 57. Court J., Spurden D., Lloyd S., McKeith I., Ballard C., Cairns N., Kerwin R., Perry R., Perry E. (1999) J. Neurochem. 73, 1590–1597 [DOI] [PubMed] [Google Scholar]
- 58. Marutle A., Zhang X., Court J., Piggott M., Johnson M., Perry R., Perry E., Nordberg A. (2001) J. Chem. Neuroanat. 22, 115–126 [DOI] [PubMed] [Google Scholar]
- 59. Kumari V., Postma P. (2005) Neurosci. Biobehav. Rev. 29, 1021–1034 [DOI] [PubMed] [Google Scholar]
- 60. Leonard S., Mexal S., Freedman R. (2007) J. Dual Diagn. 3, 43–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sharma G., Vijayaraghavan S. (2008) Curr. Med. Chem. 15, 2921–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Davies A. F., Stephens R. J., Olavesen M. G., Heather L., Dixon M. J., Magee A., Flinter F., Ragoussis J. (1995) Hum. Mol. Genet. 4, 121–128 [DOI] [PubMed] [Google Scholar]
- 63. Olavesen M. G., Bentley E., Mason R. V., Stephens R. J., Ragoussis J. (1997) Genomics 46, 303–306 [DOI] [PubMed] [Google Scholar]
- 64. Schwab S. G., Albus M., Hallmayer J., Hönig S., Borrmann M., Lichtermann D., Ebstein R. P., Ackenheil M., Lerer B., Risch N., Maier W., Wildenauer D. B. (1995) Nat. Genet. 11, 325–327 [DOI] [PubMed] [Google Scholar]
- 65. Straub R. E., MacLean C. J., O'Neill F. A., Burke J., Murphy B., Duke F., Shinkwin R., Webb B. T., Zhang J., Walsh D. (1995) Nat. Genet. 11, 287–293 [DOI] [PubMed] [Google Scholar]
- 66. Kawanishi Y., Harada S., Tachikawa H., Okubo T., Shiraishi H. (2000) J. Hum. Genet. 45, 24–30 [DOI] [PubMed] [Google Scholar]
- 67. Sun X., Ritzenthaler J. D., Zhong X., Zheng Y., Roman J., Han S. (2009) Cancer Res. 69, 6445–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zheng Y., Ritzenthaler J. D., Roman J., Han S. (2007) Am. J. Respir. Cell Mol. Biol. 37, 681–690 [DOI] [PubMed] [Google Scholar]
- 69. Fu X. W., Lindstrom J., Spindel E. R. (2009) Am. J. Respir. Cell Mol. Biol. 41, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rao J. S., Rapoport S. I., Bosetti F. (2005) Neuropsychopharmacology 30, 2006–2013 [DOI] [PubMed] [Google Scholar]
- 71. Rao J. S., Rapoport S. I. (2009) Curr. Mol. Pharmacol. 2, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Imagawa M., Chiu R., Karin M. (1987) Cell 51, 251–260 [DOI] [PubMed] [Google Scholar]
- 73. Bazinet R. P., Rao J. S., Chang L., Rapoport S. I., Lee H. J. (2005) Psychopharmacology 182, 180–185 [DOI] [PubMed] [Google Scholar]
- 74. Zink M., Englisch S., Meyer-Lindenberg A. (2010) Curr. Opin. Psychiatry 23, 103–111 [DOI] [PubMed] [Google Scholar]
- 75. Citrome L. (2009) J. Clin. Psychiatry 70, 932–933 [DOI] [PubMed] [Google Scholar]