Background: Plants have a unique form of cap-binding complex.
Results: Correct and mixed complexes show differential translation, and mixed complex subunits have lower binding affinity than correct complex subunits.
Conclusion: The subunits of the cap-binding complexes show specificity for complex formation, and the translational efficiency is determined by the large subunit.
Significance: The results suggest the potential for differential translation by the two plant cap-binding complexes.
Keywords: Eif4e, mRNA, Protein Complexes, Protein Synthesis, Translation Initiation Factors, Cap-binding Complex, eIF4G
Abstract
The initiation of translation in eukaryotes requires a suite of eIFs that include the cap-binding complex, eIF4F. eIF4F is comprised of the subunits eIF4G and eIF4E and often the helicase, eIF4A. The eIF4G subunit serves as an assembly point for other initiation factors, whereas eIF4E binds to the 7-methyl guanosine cap of mRNA. Plants have an isozyme form of eIF4F (eIFiso4F) with comparable subunits, eIFiso4E and eIFiso4G. Plant eIF4A is very loosely associated with the plant cap-binding complexes. The specificity of interaction of the individual subunits of the two complexes was previously unknown. To address this issue, mixed complexes (eIF4E-eIFiso4G or eIFiso4E-eIF4G) were expressed and purified from Escherichia coli for biochemical analysis. The activity of the mixed complexes in in vitro translation assays correlated with the large subunit of the respective correct complex. These results suggest that the eIF4G or eIFiso4G subunits influence translational efficiency more than the cap-binding subunits. The translation assays also showed varying responses of the mRNA templates to eIF4F or eIFiso4F, suggesting that some level of mRNA discrimination is possible. The dissociation constants for the correct complexes have KD values in the subnanomolar range, whereas the mixed complexes were found to have KD values in the ∼10 nm range. Displacement assays showed that the correct binding partner readily displaces the incorrect binding partner in a manner consistent with the difference in KD values. These results show molecular specificity for the formation of plant eIF4F and eIFiso4F complexes and suggest a role in mRNA discrimination during initiation of translation.
Introduction
Initiation of protein synthesis requires the concerted effort of a large number of proteins and protein complexes, as well as multiple methods of control and control elements (1–5). Among the protein complexes required is eIF4F, the cap-binding complex, which binds to the m7G2 cap found at the 5′ end of most eukaryotic cellular mRNAs. Mammalian eIF4F consists of three subunits: eIF4G, eIF4E, and eIF4A. eIF4G is a large, multidomain protein of ∼180 kDa. eIF4G is largely responsible for ribosome attachment and enhancing efficiency of mRNA translation through multiple protein and/or RNA interactions (6). eIF4E is a small protein of ∼24 kDa that directly binds the m7G cap and facilitates initiation events. Mammalian eIF4F complexes contain eIF4A, the prototype of the DEAD box helicase family (7, 8); however, plant eIF4A is loosely associated and is easily removed during purification (9).
eIF4G is a multifunctional protein that is an important structural platform for the assembly or nucleation of several initiation factors (eIF4A, eIF3, eIF4B, eIF5, and poly(A)-binding protein) and interaction with the 40 S ribosome during the initiation process (10, 11). Furthermore, in mammals there is also interaction of eIF4G with MNK kinase that then phosphorylates associated eIF4E (12). Mammalian eIF4G consists of several domains for interaction with these factors. The eIF4E and poly(A) binding protein binding sites are located in the N-terminal region, whereas the C-terminal region contains three HEAT domains, MIF4G (eIF4A and eIF3), MA3 (eIF4A), and W2 (MNK) (1, 13–18). However, yeast eIF4G has only retained the MIF4G domain and lost the MA3 and W2 HEAT domains. Plants have retained the MIF4G and MA3 HEAT domains but have also lost the W2 HEAT domain (see Fig. 1A) (19).
FIGURE 1.
A, domain organization for eIF4G and eIFiso4G. Plant eIF4G and eIFiso4G have similar domain organization, except eIF4G has an extended N-terminal region (19). The eIF4E binding site and HEAT domains are indicated. The HEAT domains interact with eIF4A and eIF3 as indicated. Plant eIF4G and eIFiso4G lack the third HEAT domain present in mammalian eIF4G, and yeast eIF4G only has HEAT domain 1. B, sequence of wheat eIF4G gene. The coding region is shown in capital letters, and noncoding regions and introns are in lowercase. Differences between the gene sequence and expression clone are indicated (green, silent mutants; red, amino acid changes). The N terminus of the expression clone was altered to generate an NcoI site as indicated and has a single methionine, and the second amino acid was changed to glycine. The N-terminal peptide obtained from Edmund degradation of native protein is indicated by a double underlining, and internal peptides obtained from trypsin digests of native protein are indicated by single underlining. The naturally occurring NcoI and AgeI sites used to generate the expression clone are indicated. The eIF4E binding site (dotted underlining) and HEAT domains (yellow and blue) are indicated.
Plants contain a second form of eIF4F not found in other eukaryotes, eIFiso4F (9, 20). The amount of eIF4F present in wheat germ extracts is about 5–10 times less compared with eIFiso4F (21). eIFiso4F consists of two subunits, eIFiso4G and eIFiso4E, and has activities in vitro similar to those of eIF4F (9, 22). The eIFiso4G subunit contains an eIF4E binding site and the two HEAT domains found in plant eIF4G, but has lost most of the N-terminal region compared with eIF4G (see Fig. 1A). Plant eIF4E and eIFiso4E are similar in molecular weight and are about 50% similar in amino acid sequence (23). Mutations in eIF4G or eIFiso4G, as well as eIF4E or eIFiso4E, result in resistance to various plant viruses; however, the precise roles of these translation factor subunits in viral replication have not been fully elucidated (reviewed in Refs 24 and 25).
In the model plant Arabidopsis thaliana, there are two genes for eIFiso4G and a single gene for eIF4G. The functional deletion of the two eIFiso4G genes in Arabidopsis results in a phenotype that includes impaired growth/development, poor seed quality, lower fertility, altered stress responses, and lower chlorophyll (19). This phenotype suggests eIFiso4G has a significant role in proper plant growth/development and response to environmental stress. Interestingly, the deletion of the single gene for eIFiso4E does not result in a strong phenotype, only virus resistance to certain potyviruses (26, 27).
Having two forms of the cap-binding complex in plants presents the question of whether specific roles in the initiation of translation exist for eIF4F and eIFiso4F. To begin to address this question, we determined whether or not there is molecular specificity in forming the eIF4F or eIFiso4F complexes. We find that in vitro, mixed complexes of eIF4G-eIFiso4E or eIFiso4G-eIF4E are able to form spontaneously and function in the initiation of translation in a manner similar to the eIF4F or eIFiso4F complex comprised of the respective large subunit. We find that very low levels of mixed complexes are found in vivo, consistent with the biochemical evidence that there is strong specificity in binding affinity of the correct binding partners of the complexes compared with the mixed complexes.
EXPERIMENTAL PROCEDURES
N-terminal and Internal Peptide Analysis of Wheat eIF4G and eIF4E
eIF4F was purified from wheat germ as previously described (28). The eIF4F complex was resolved into eIF4G and eIF4E subunits by SDS-PAGE and transferred to PVDF or nitrocellulose. Trypsin digestion, N-terminal sequence analysis (Edmund degradation), and peptide sequence analysis of eIF4G were carried out by John Leszyk (University of Massachusetts Medical Center Proteomics Mass Spectrometry Center, Worchester, MA) or by William Lane (Harvard University Micro Sequencing Center, Cambridge, MA). The N terminus (Edmund degradation) of eIF4E was determined at the Protein Microanalysis Laboratory (Institute for Cellular and Molecular Biology, University of Texas at Austin).
Screening of the Wheat cDNA Library for eIF4G
The λZAP cDNA library preparation and screening were as previously described (22, 29). The expression library was screened using eIF4G affinity purified rabbit antibody to native wheat eIF4F (21).
Screening of a Wheat Genomic Library for eIF4G
The preparation and screening of a λFIX II (Stratagene) wheat (var. Chinese Spring) genomic library was as previously described (30). The genomic library was screened with a 32P-labeled probe made from the truncated cDNA template for eIF4G obtained from the λZAP cDNA (see above). The complete sequence of the eIF4G gene was obtained by primer walking on both DNA strands (GenBankTM accession number JN091779).
Analysis of Introns in eIF4G Gene
In silico prediction of splice sites was carried out using the NetPlant Gene Server (31). DNA amplification primers were designed to confirm the presence or absence of the predicted introns. Total RNA was extracted from 3-day-old seedlings (var. Chinese Spring) using a plant RNeasy extraction kit (Qiagen). Reverse transcription (Retroscript, Ambion) and DNA amplification and sequencing were carried out with the appropriate primer sets to check the presence of the predicted introns.
Preparation of Mouse Monoclonal and Rabbit Polyclonal Antibodies
Mouse monoclonal antibodies to native wheat eIF4F were prepared as previously described (32). Monoclonal antibodies were obtained to six distinct epitopes of wheat eIF4G, and one monoclonal was obtained to eIF4E. Mouse ascites fluid (1/1000) was used for Western blot analysis (21). Rabbit polyclonal antiserum was prepared to native eIF4F, native eIFiso4F, gel-purified native eIFiso4E, or eIFiso4G and to recombinant wheat eIFiso4E, eIFiso4G, eIF4G, or eIF4E at the University of Texas M. D. Anderson Cancer Center, Department of Veterinary Science (Bastrop, TX). Western blots were carried out as previously described (21).
Construction of Expression Clone for eIF4G
The N terminus of native wheat eIF4G was determined by Edmund degradation. The sequence obtained (see Fig. 1B) was identified on the deduced gene sequence. There are many potential translation start sites in this region of the sequence. It should be pointed out that any blocked N termini from other start sites would have been missed by the N-terminal sequencing, and thus other start sites may be used in vivo. However, for the purposes of biochemical analysis of eIF4G, the N-terminal sequence obtained was used as the start site for expression of recombinant protein.
The partial cDNA clone obtained from screening of the cDNA λZAP library was used as a template for DNA amplification using a forward primer containing an N-terminal NcoI site and a reverse primer containing a BamHI site. This DNA fragment was cloned into pET3d (Novagen) using the primer-derived NcoI and BamHI restriction sites. A second fragment was obtained by digestion of the genomic clone (a region devoid of introns) with NcoI (naturally occurring; see Fig. 1B) and AgeI (naturally occurring; see Fig. 1B). This fragment was inserted into pET3d containing the cDNA between the NcoI (vector) and AgeI (insert) restriction sites. This construct expresses an N-terminal truncated version of eIF4G that is fully functional in translation assays (data not shown). A third fragment was amplified by reverse transcription and DNA amplification of poly(A)+ mRNA (var. Chinese Spring) with a forward primer containing a NcoI site corresponding to the N terminus obtained from protein sequencing of eIF4G and a reverse primer complementary to the region 3′ to the naturally occurring NcoI site. This fragment was cloned into pET3d (containing fragments 1 and 2) at the NcoI restriction site located at the most 5′ end of the cDNA. Positive colonies were screened by PCR for the correct orientation. The complete reconstructed eIF4G cDNA for expression was sequenced (GenBankTM accession number EF190330).
Cloning of Wheat eIF4E
The cDNA previously obtained for eIF4E was not full length (29). The N-terminal protein sequence (AEDTETRPASAGAEEREEGEI) obtained by Edmund degradation was used to design an oligonucleotide primer to provide the missing sequence. The oligonucleotide was optimized for Escherichia coli expression but retained the correct wheat amino acid sequence (GenBankTM accession number Z12616).
Construction of Dicistronic eIF4F, eIF iso4F, and Mixed Complexes
A cap-binding subunit, eIF4E or eIFiso4E, was amplified using a primer containing a BamHI restriction site, an E. coli ribosome-binding site, and 12–15 nucleotides of the respective cap-binding protein 5′ coding region in the forward direction. The reverse direction primer contained a BamHI restriction site, a termination codon, and 12–15 nucleotides of the respective cap-binding protein 3′ coding region. The amplified DNA for eIF4E or eIFiso4E was cloned into the BamHI site that is 3′ to the termination codon of either the eIF4G (described above) or eIFiso4G (23) coding regions. The resulting plasmid constructs were screened for the correct orientation of the cap-binding protein by DNA amplification and confirmed by DNA sequencing.
Expression of eIF4F, eIFiso4F, and Mixed Complexes
The expression and purification of eIF4F and eIFiso4F was as previously described (33). The mixed complexes of eIF4G-eIFiso4E or eIFiso4G-eIF4E were purified similarly.
In Vitro Translation Assay
The abilities of the cap-binding complexes to support in vitro translation were determined as previously described (28, 34). The fractionated assay is dependent upon the addition of eIF4F or eIFiso4F for activity. Briefly, 100-μl reactions included 5 pmol of mRNA template (as indicated); recombinant eIF4F, eIFiso4F, or mixed complex as indicated; 0.6 μg (10 pmol) of wheat recombinant eIF4B; 5 μg (7 pmol) of native wheat eIF3; 10 μg (200 pmol) of recombinant wheat eIF4A; 24 mm HEPES-KOH, pH 7.6; 2.9 mm MgAc2; 100 mm KAc; 30 mm KCl; 2.4 mm DTT; 0.1 mm spermine; 1 mm ATP; 0.2 mm GTP; 34 μm [14C]leucine; 50 μm 19 amino acids; 7.8 mm creatine phosphate; 3 μg of creatine kinase; 0.75 A260 units of yeast tRNA; 1–2 A260 units of 1× washed wheat ribosomes; and 200 μg of wheat germ 40–70% ammonium sulfate fraction. Incubation was for 30 min at 27 °C, and the amount of [14C]leucine incorporated into protein was determined as previously described (28).
SPR Analysis
SPR (Biacore) experiments were carried out at the Center for Biomolecular Interaction Analysis (Dr. David Myszka, Director; University of Utah). Briefly, reactions were performed at 25 °C using a Biacore 3000 optical biosensor equipped with a CM4 sensor chip and equilibrated with running buffer (10 mm HEPES, pH 7.4, 150 mm NaCl). eIF4G or eIFiso4G were amine-coupled in individual flow cells to densities of 580 and 750 resonance units, respectively. For each immobilization, the surface was activated for 5 min, the protein was coupled, and any remaining active sites on the surface were blocked with 1 m ethanolamine, pH 8.9. eIFiso4E and eIF4E were tested for binding to the immobilized proteins in 20 mm HEPES, pH 7.6, 100 mm KCl, 1.0 mm DTT, 0.1 mm EDTA, 100 μm m7GTP, 5% glycerol, 0.01% Tween 20, 0.1 mg/ml BSA. Starting at 50 nm, a 3-fold dilution series of eIFiso4E or eIF4E were tested in triplicate. The response data were globally fit using Scrubber2 (Biologic Software Pty Ltd) to a 1:1 interaction model to extract binding constants.
Immunoprecipitation of eIF4E or eIFiso4E from Wheat Germ Extracts
Magnetic protein A beads (40 μl, Genscript) were incubated with 100 μl of rabbit sera to recombinant wheat eIF4G or eIFiso4G for 1 h at room temperature and washed according to the manufacturer's instructions. The beads were then incubated for 3 h at 4 °C on a rotating platform with 1.0 ml of wheat germ extract supplemented with Complete protease inhibitor (40 μl of a 25× solution made from a tablet; Roche Applied Science). The beads were washed three times with IP buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.1% Nonidet P-40, 1 mm PMSF) and eluted with 50 μl of 2× SDS sample buffer (35). Eluted proteins (20 μl) from the beads were separated on 10–20% gradient gels (Invitrogen) and transferred to nitrocellulose (Genscript). Monoclonal mouse antibody to eIF4E (monoclonal 8F7) or rabbit antisera raised to native gel purified eIFiso4E were used to probe the Western blot using a 1-h Western IP kit (Genscript) according to the manufacturer's instructions.
Gel Shift Assay for Displacement of Cap-binding Protein from Mixed Complexes
Correct or mixed complexes (2.5 pmol) were presented cap-binding proteins (0, 2.5, 7.5 or 20 pmol) in a 10-μl reaction containing 20 mm HEPES, pH 7.6, 100 mm KCl, 0.1 mm EDTA, 1 mm DTT, 10% glycerol. 0.1 μg/μl BSA and 100 μm m7GTP. The reactions were incubated on ice for 10 min, and then 10 μl of native gel buffer (Invitrogen) was added and loaded on a 4–12% nondenaturing NOVEX gel (Invitrogen) and electrophoresed at 100 volts at 4 °C for ∼2.5 h. The gel was blotted to PVDF and probed with mouse monoclonal antibody to eIF4E (8F7, 1/1000) and then stripped and probed with rabbit anti-native eIFiso4E (1/1,000). The second antibody used was goat anti-mouse or goat anti-rabbit HRP (1/10,000; Kirkegaard-Perry), respectively.
RESULTS
Construction of cDNA Expression Clones
Construction of full-length cDNA clones of wheat eIF4E and eIF4G was necessary to analyze their interactions and compare them with those of eIFiso4E and eIFiso4G that were previously constructed (22). A combination of protein sequence data, cDNA, and genomic DNA sequencing and intron analysis was used as described under “Experimental Procedures” to construct the expression clones for eIF4E and eIF4G.
The genomic DNA sequence for eIF4G indicates that there are numerous potential start sites in addition to the dual ATGs immediately preceding the N-terminal amino acid sequence obtained (Fig. 1B). It is not known at this time whether any additional upstream start sites are used in the expression of wheat eIF4G in vivo. If the N termini of alternative start sites were blocked, then Edmund degradation would not have identified them. However, because the goal was to obtain recombinant protein, the N terminus identified by Edmund degradation was selected for construction of the recombinant protein.
The complete reconstructed cDNA was sequenced (GenBankTM accession number EF190330), and eight differences (three silent; five amino acid changes) were identified between the gene sequence and the expression construct as shown in Fig. 1B. These differences represent mutations that either arose from the amplification process used to generate the N-terminal fragment or are allelic/cultivar differences in the source materials.
Dicistronic Expression of Wheat eIF4F
To facilitate preparation of recombinant eIF4F or eIFiso4F for biochemical analysis, both subunits of the respective complex were placed into the same pET expression vector, coexpressed, and purified as described (33, 34). Recombinant wheat eIF4F or eIFiso4F prepared in this manner is highly pure (Fig. 2, second and fifth lanes, respectively). The monoclonal antibodies obtained to native wheat eIF4F react with the recombinant forms of eIF4G and eIF4E (Fig. 3), indicating that all of the epitopes represented in the native form are present in the recombinant form of eIF4G.
FIGURE 2.
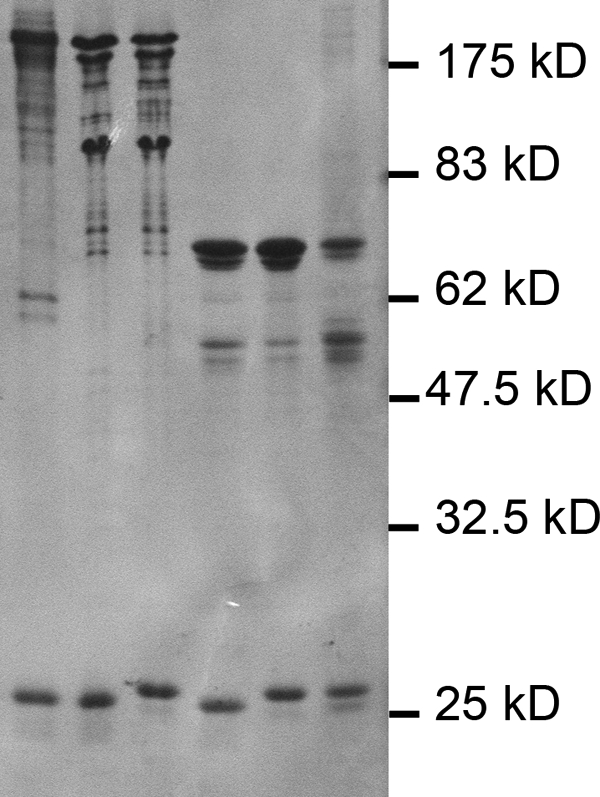
SDS-PAGE analysis of eIF4F, eIFiso4F, and mixed complexes. SDS-PAGE was carried out on a 12.5% acrylamide gel and stained with Coomassie Brilliant Blue. Each lane contains 25 pmol of the indicated complex. First lane, native eIF4F; second lane, recombinant eIF4F; third lane, eIF4G-eIFiso4E; fourth lane, eIFiso4G-eIF4E; fifth lane, recombinant eIFiso4F; sixth lane, native eIFiso4F.
FIGURE 3.
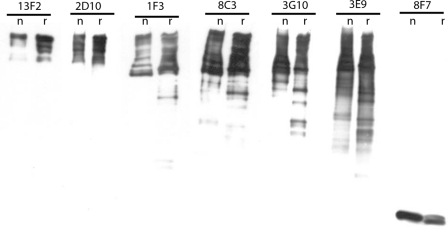
Monoclonal antibodies to wheat eIF4F. Mouse monoclonal antibodies were raised to wheat native eIF4F. The ability of individual monoclonal antibodies to react with native (n) eIF4F and recombinant (r) eIF4F was tested by Western blotting. Mouse ascites fluid (1/1000) was incubated overnight at 4 °C, and the second antibody was goat anti-mouse HRP (1/20,000, Kirkegaard-Perry). The chemiluminescent substrate was Super Signal West Pico (Thermo-Pierce).
Recombinant wheat eIF4F or eIFiso4F has similar activity to native eIF4F or eIFiso4F in in vitro translation (Fig. 4). These results suggest that the expressed dicistronic forms of eIF4F and eIFiso4F complexes are fully functional. Interestingly, eIFiso4F displays sigmoidal behavior with some mRNA templates such as β-hemoglobin mRNA (shown in Fig. 4) or yeast polysomal mRNA (9). Other plant mRNA templates also display this behavior in vitro (data not shown), and this behavior may reflect a difference in the interaction of some mRNAs with eIFiso4F compared with eIF4F.
FIGURE 4.

Comparison of recombinant and native eIF4F and eIFiso4F in translation. Each 100-μl translation reaction contained 5 pmol of capped rabbit β-hemoglobin mRNA and the indicated amounts of eIF4F or eIFiso4F complexes (■, solid line, native eIF4F; □, dashed line, recombinant eIF4F; ●, solid line, native eIFiso4F; △, dashed line, recombinant eIFiso4F) as described under “Experimental Procedures.” The amount of [14C]leucine incorporated in the absence of cap-binding complexes was 3.8 pmol. Each point represents the average of three experiments, and the error bars are indicated.
Do “Mixed” Complexes Form?
Preliminary experiments using the yeast two-hybrid system showed that wheat eIFiso4G could interact with either eIFiso4E or eIF4E and eIF4G could interact with either eIF4E or eIFiso4E, suggesting that mixed complexes were possible (data not shown). To determine whether stable mixed complexes could form in vitro and to test their ability to function in translation, dicistronic expression clones of eIF4G-eIFiso4E and eIFiso4G-eIF4E were prepared, and the complexes were expressed and purified similarly to eIF4F or eIFiso4F. As shown in Fig. 2 (third and fourth lanes), stable mixed complexes were obtained and purified from E. coli.
In Vitro Translation Activity of Mixed Complexes
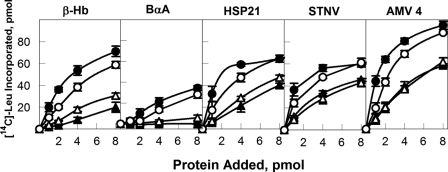
To determine whether the mixed complexes were able to support initiation of translation in vitro, the mixed complexes were compared with recombinant eIF4F and eIFiso4F in the presence of five different mRNAs. The mRNAs selected include both capped cellular mRNAs (barley α-amylase, A. thaliana HSP21, and β-hemoglobin), capped viral RNA (AMV 4 RNA), and a noncapped viral RNA (STNV). As shown in Fig. 5, the mixed complexes are able to fully support initiation of translation. Interestingly, the translational efficiency appears to correlate more closely with the large subunit rather than the cap-binding subunit. That is, the complex containing eIF4G-eIFiso4E has activity more similar to eIF4F, and the complex containing eIFiso4G-eIF4E is more similar to eIFiso4F. These results suggest that the large subunit provides some level of specificity of mRNA translation by eIF4F or eIFiso4F.
FIGURE 5.
Translation Assay of eIF4F, eIFiso4F, and mixed complexes. Each 100-μl translation reaction contained 5 pmol of the indicated mRNA (capped rabbit β-hemoglobin, capped barley α-amylase, capped AtHSP21, STNV RNA (uncapped), and capped AMV RNA 4) and the indicated amounts of recombinant eIF4F, eIFiso4F or mixed complex (●, eIF4F); (○, eIF4G-eIFiso4E); (▴, eIFiso4F); or (△, eIFiso4G-eIF4E) as described under “Experimental Procedures.” The amount of [14C]leucine incorporated in the absence of cap-binding complexes was as indicated: β-hemoglobin (β-Hb, 4.0 pmol); barley α-amylase (BαA, 4.1 pmol); AtSHSP 21 (6.6 pmol); STNV RNA (16.8 pmol); and AMV RNA 4 (5.3 pmol). Each point represents the average of three experiments, and the error bars are indicated.
IP Analysis
It was clear that the subunits are able to form functional mixed complexes in vitro. The question then was whether or not mixed complexes occur in planta because the predominant forms purified from wheat germ are the eIF4F and eIFiso4F complexes. To address this issue, wheat germ extracts were immunoprecipitated using rabbit antibodies to recombinant eIF4G or eIFiso4G. The resulting immunoprecipitated proteins were probed with antibodies to eIF4E or eIFiso4E to determine whether mixed complexes are present in vivo. The data shown in Fig. 6 suggest that very low levels of eIFiso4E are detected when eIF4G is immunoprecipitated. Conversely, no eIF4E was detected when eIFiso4G was immunoprecipitated. It is not clear whether this means that there is not any eIFiso4G-eIF4E mixed complex or whether it reflects a detection limit, because eIF4E is present in 5–10-fold lower amounts than eIFiso4E (21). These results show that very low levels of mixed complexes, at least of eIF4G-eIFiso4E, are present in plants.
FIGURE 6.
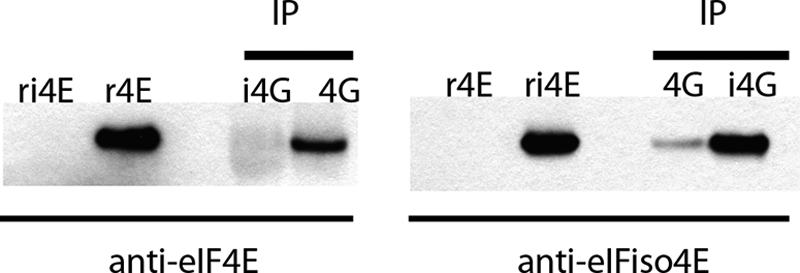
Immunoprecipitation of wheat germ extracts. Wheat germ extract (1 ml) was immunoprecipitated (IP) with rabbit antisera raised to either recombinant eIF4G or eIFiso4G bound to protein A magnetic beads (Genscript). The precipitated proteins were eluted with 50 μl of 2× SDS-PAGE sample buffer, 20 μl of which was separated on a 10–20% acrylamide gel (Bio-Rad) and transferred to nitrocellulose. Controls of recombinant wheat eIF4E (0.05 μg) and eIFiso4E (0.05 μg) were also separated on the gel. The blots were probed with either mouse monoclonal antibody for eIF4E (8F7) or rabbit polyclonal serum raised to native gel-purified eIFiso4E using a mouse or rabbit 1-h complete IP Western kit (Genscript).
Binding Affinity and Displacement Analysis
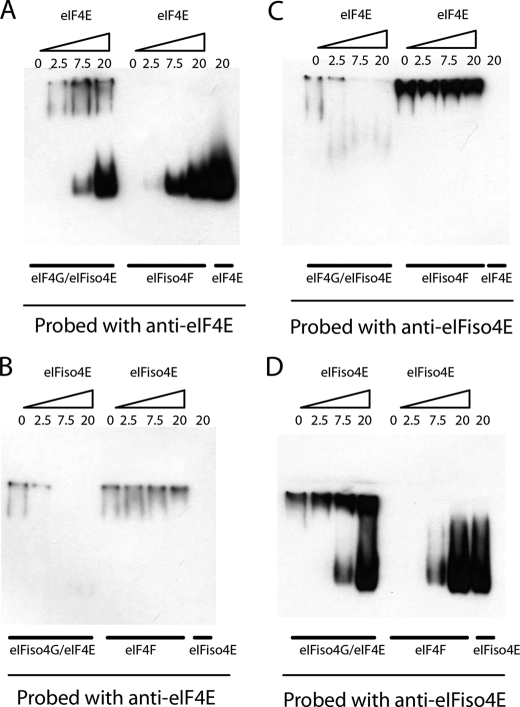
Although it is possible to form mixed complexes in vitro and in vivo, it is expected that there may be some level of specificity for complex formation. The binding affinity for association of the cap-binding subunits with the large subunits was measured to assess the level of specificity of complex formation. Initial experiments with isothermal calorimetry indicated that there were significant differences in the binding affinity for correct and mixed complexes; however, the apparent subnanomolar binding affinities observed for the correct complexes are not accurately measured by isothermal calorimetry.3 To obtain more accurate measurements of the binding constants, SPR (BiaCore) was carried out on the correct and mixed complexes. The SPR data clearly support a “preferred” binding partner (Fig. 7 and Table 1) and show that the correct complexes have subnanomolar KD values. The KD for the eIF4G-eIF4E complex is ∼80-fold tighter than for the eIF4G-eIFiso4E mixed complex. Similarly, the eIFiso4G-eIFiso4E complex is ∼148-fold tighter than the eIFiso4G-eIF4E mixed complex. The subnanomolar KD values for the correct complexes suggest that if a mixed complex is presented with the correct binding partner, then the correct binding partner should displace the incorrect binding partner, preferentially forming the correct complex.
FIGURE 7.
SPR binding analysis of correct and mixed complexes. eIF4E (A and D) or eIFiso4E (B and C) were tested for binding to the respective immobilized proteins eIF4G (A and B) or eIFiso4G (C and D). The running buffer contained 20 mm HEPES, 100 mm KCl, 1.0 mm DTT, 0.1 mm EDTA, 100 μm m7GTP, 5% glycerol, 0.01% Tween 20, and 0.1 mg/ml BSA, pH 7.6. Starting at 50 nm, a 3-fold dilution series of eIFiso4E or eIF4E were tested in triplicate. The response data were globally fit using Scrubber2 (Biologic Software Pty Ltd) to a 1:1 interaction model to extract binding constants.
TABLE 1.
Binding affinities of correct and mixed complexes measured by SPR
| KD | ||
|---|---|---|
| nm | ||
| eIF4G | eIF4E | 0.181 ± 0.002 |
| eIFiso4E | 14.3 ± 0.2 | |
| eIFiso4G | eIF4E | 11.8 ± 0.1 |
| eIFiso4E | 0.080 ± 0.002 |
Displacement assays were performed using eIF4F, eIFiso4F, and mixed complexes. As shown in Fig. 8, when the mixed complex of eIF4G-eIFiso4E was presented with increasing levels of eIF4E, the eIFiso4E was displaced by the eIF4E, and eIF4E appears in the complex (Fig. 8A). Conversely, eIFiso4E disappears from the complex (Fig. 8C). Similarly, when eIFiso4G-eIF4E is presented with eIFiso4E, eIF4E is displaced from the complex (Fig. 8B), and eIFiso4E appears in the complex (Fig. 8D). However, increasing amounts of eIF4E could not displace eIFiso4E from the eIFiso4F complex (Fig. 8C, right side), and eIF4E remains in the unbound state (Fig. 8A, right side); nor did eIFiso4E displace eIF4E from the eIF4F complex (Fig. 8B, right side) and eIFiso4E remains unbound (Fig. 8D, right side). These results suggest preferential binding when the appropriate binding partner is present. Thus the low level of mixed complex of eIF4G-eIFiso4E in vivo demonstrated by immunoprecipitation likely reflects a very small excess of eIFiso4E over eIFiso4G that may be bound by any excess eIF4G not in complex with eIF4E. Interestingly, it was recently shown by an extensive proteomic analysis of mammalian cells that eIF4G and eIF4E were present in roughly equal molar amounts (36), which is consistent with the plant immunoprecipitation data indicating very low levels of excess subunits that form mixed complexes.
FIGURE 8.
Displacement assays. To determine whether the correct cap-binding protein is able to displace a mismatched cap-binding protein in a mixed complex, the indicated amounts of eIF4E or eIFiso4E were added to 2.5 pmol of mixed complexes, eIF4F, or eIFiso4F. A and B were probed with mouse monoclonal antibody (8F7) to eIF4E (1/1000). The blots were stripped and probed with rabbit polyclonal antibody raised to native gel-purified eIFiso4E (1/1000) shown in C and D. The antibody to native eIFiso4E displays a small amount of cross-reactivity with eIFiso4G (see D, eIFiso4G-eIF4E with no eIFiso4E added).
DISCUSSION
The discovery of a plant-specific form of eIF4F, eIFiso4F, suggests that there may be specialized functions for these complexes in plant initiation of translation (9, 20). Biochemical analysis of these complexes should be able to reveal important information about the specificity of binding of the subunits to form complexes and their interactions with mRNAs. We constructed functional wheat discistronic expression clones and have shown that the recombinant eIF4F and eIFiso4F complexes are functionally similar to native complexes in in vitro translation (Fig. 4).
To assess the ability of the subunits of eIF4F and eIFiso4F to form mixed complexes, additional dicistronic plasmids were constructed that contained eIF4G-eIFiso4E and eIFiso4G-eIF4E, and they were used to purify mixed complexes. As shown in Fig. 5, these complexes are functional and support the initiation of translation in vitro. Interestingly, the large subunit appears to be dominant in determining the activity of the mixed complex. The mixed complex containing eIF4G-eIFiso4E behaved more similarly to eIF4F, whereas the mixed complex containing eIFiso4G-eIF4E was more like eIFiso4F. These results suggest that the large subunit is most likely responsible for discrimination among groups of plant mRNAs.
The binding constants were determined using SPR for the interaction of eIF4F, eIFiso4F, and mixed complex subunits. The KD values for binding of eIF4G to eIF4E and eIFiso4G to eIFiso4E determined by SPR were in the subnanomolar range (0.18 and 0.08 nm, respectively), whereas the mixed complexes were in the ∼12–14 nm range. This represents a ∼80–148-fold difference in binding affinity and is supported by the displacement data in Fig. 8 showing that the correct binding partner readily displaces an incorrect binding partner. Mammalian and yeast KD values for eIF4G and eIF4E binding are reported in the 2.5–27 and 2–15 nm ranges, respectively (supplemental Table A). The wheat mixed complexes (∼12–14 nm) are more similar in KD to those of the yeast and mammalian complexes. This may reflect that in contrast to mammalian and yeast systems, plants do not appear at this time to have the 4E-binding proteins that participate in the regulation of the accessibility of eIF4E to bind eIF4G. Because such a system does not appear to function in plants, there is no apparent need for the subunits to readily dissociate as indicated by the strong binding constants and displacement data (Table 1 and Fig. 8). Although there are reports of plant proteins that interact with eIF4E in the yeast two-hybrid system (37, 38), these interactions have not been studied biochemically, and their biological role is unknown. One might speculate that there is a rich and varied set of protein interactions for the subunits of eIF4F or eIFiso4F that may regulate translation in plants; however, given the strength of the interactions of the subunits (subnanomolar) of the eIF4F or eIFiso4F complexes, there would most likely need to be a significant input of energy to dissociate these complexes in the form of ATP hydrolysis or another similar biochemical process. However, such a system has not yet been detected. It needs to be determined whether plant eIF4F or eIFiso4F dissociation occurs and/or has a role in regulation.
The precise eIF4F or eIFiso4F subunit requirement for initiation of translation in plants remains to be determined. Preliminary evidence from generating multiple Arabidopsis T-DNA insertion mutants for the subunits of eIF4F or eIFiso4F suggests that a mixed complex of eIFiso4G and eIF4E is quite sufficient for normal growth and development; however, plants with only the mixed complex of eIF4G and eIFiso4E fail to thrive or proceed through normal development to flowering. This suggests that certain mRNAs may need eIFiso4G to be properly translated. We have recently shown that a double mutant lacking both eIFiso4G genes in Arabidopsis shows an impaired growth phenotype that includes less chlorophyll, reduced fertility, and loss of long term seed viability (19). These results suggest that plant eIFiso4G is very important in plant growth and development, whereas eIF4G is not required or at least the lack of eIF4G does not present an obvious impairment. It remains to be determined whether eIFiso4F provides optimal translation for a specific subset of mRNAs necessary for plant growth and development or if, as recently shown for the two genes for yeast eIF4G, it is the total amount of eIF4G (and/or eIFiso4G) present, not the particular gene product (39). It is also possible that plants have evolved different cap-binding complexes for the pioneer round versus steady-state translation (40). There is still much to be learned about the roles for eIF4F and eIFiso4F in the initiation of plant translation and its regulation. The further elucidation of the roles of eIF4F and eIFiso4F subunits in plant initiation of translation may illuminate how other cap-binding complexes form and function in organisms that have multiple genes for eIF4G and eIF4E.
Supplementary Material
Acknowledgments
We thank Dr. John Leszyk (University of Massachusetts Medical Center Proteomics Mass Spectrometry Center, Worchester, MA) and Dr. William Lane (Harvard University Micro Sequencing Center, Cambridge, MA) for protein sequence analysis; Drs. Verna Frasca and Lung-Nan Lin (GE Lifesciences, Northampton, MA) for the isothermal calorimetry analysis and comments on the manuscript; and Dr. David Myszka (Director, Center for Biomolecular Interaction Analysis, University of Utah) for the BiaCore analysis and comments on the manuscript.
This work was supported by National Science Foundation Grants MCB1052530 and MCB0745146 (to K. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table A.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s)JN091779.
V. Frasca, personal communication.
- m7G
- 7-methyl guanosine
- SPR
- surface plasmon resonance
- AMV
- alfalfa mosaic virus
- STNV
- satellite tobacco necrosis virus.
REFERENCES
- 1. Marintchev A., Wagner G. (2004) Q. Rev. Biophys. 37, 197–284 [DOI] [PubMed] [Google Scholar]
- 2. Pestova T. V., Lorsch J. R., Hellen C. U. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds) pp. 87–128, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 3. Mathews M. B., Sonenberg N., Hershey J. W. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds.) pp. 1–40, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 4. Livingstone M., Atas E., Meller A., Sonenberg N. (2010) Phys. Biol. 7, 021001 [DOI] [PubMed] [Google Scholar]
- 5. Merrick W. C. (2010) J. Biol. Chem. 285, 21197–21201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park E. H., Zhang F., Warringer J., Sunnerhagen P., Hinnebusch A. G. (2011) BMC Genomics 12, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogers G. W., Jr., Komar A. A., Merrick W. C. (2002) Prog. Nucleic Acid Res. Mol. Biol. 72, 307–331 [DOI] [PubMed] [Google Scholar]
- 8. Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., Sonenberg N. (2011) Nat. Rev. Mol. Cell Biol. 12, 235–245 [DOI] [PubMed] [Google Scholar]
- 9. Lax S., Fritz W., Browning K., Ravel J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernández G., Vazquez-Pianzola P. (2005) Mech. Dev. 122, 865–876 [DOI] [PubMed] [Google Scholar]
- 11. Prévôt D., Darlix J. L., Ohlmann T. (2003) Biol. Cell 95, 141–156 [DOI] [PubMed] [Google Scholar]
- 12. Buxade M., Parra-Palau J. L., Proud C. G. (2008) Front. Biosci. 13, 5359–5373 [DOI] [PubMed] [Google Scholar]
- 13. Korneeva N. L., Lamphear B. J., Hennigan F. L., Rhoads R. E. (2000) J. Biol. Chem. 275, 41369–41376 [DOI] [PubMed] [Google Scholar]
- 14. Marcotrigiano J., Lomakin I. B., Sonenberg N., Pestova T. V., Hellen C. U., Burley S. K. (2001) Mol. Cell 7, 193–203 [DOI] [PubMed] [Google Scholar]
- 15. Marintchev A., Wagner G. (2005) Biochemistry 44, 12265–12272 [DOI] [PubMed] [Google Scholar]
- 16. Oberer M., Marintchev A., Wagner G. (2005) Genes Dev. 19, 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bellsolell L., Cho-Park P. F., Poulin F., Sonenberg N., Burley S. K. (2006) Structure. 14, 913–923 [DOI] [PubMed] [Google Scholar]
- 18. Schütz P., Bumann M., Oberholzer A. E., Bieniossek C., Trachsel H., Altmann M., Baumann U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lellis A. D., Allen M. L., Aertker A. W., Tran J. K., Hillis D. M., Harbin C. R., Caldwell C., Gallie D. R., Browning K. S. (2010) Plant Mol. Biol. 74, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Browning K. S., Webster C., Roberts J. K., Ravel J. M. (1992) J. Biol. Chem. 267, 10096–10100 [PubMed] [Google Scholar]
- 21. Browning K. S., Humphreys J., Hobbs W., Smith G. B., Ravel J. M. (1990) J. Biol. Chem. 265, 17967–17973 [PubMed] [Google Scholar]
- 22. van Heerden A., Browning K. S. (1994) J. Biol. Chem. 269, 17454–17457 [PubMed] [Google Scholar]
- 23. Allen M. L., Metz A. M., Timmer R. T., Rhoads R. E., Browning K. S. (1992) J. Biol. Chem. 267, 23232–23236 [PubMed] [Google Scholar]
- 24. Robaglia C., Caranta C. (2006) Trends Plant Sci. 11, 40–45 [DOI] [PubMed] [Google Scholar]
- 25. Truniger V., Aranda M. A. (2009) Adv. Virus Res. 75, 119–159 [DOI] [PubMed] [Google Scholar]
- 26. Duprat A., Caranta C., Revers F., Menand B., Browning K. S., Robaglia C. (2002) Plant J. 32, 927–934 [DOI] [PubMed] [Google Scholar]
- 27. Lellis A. D., Kasschau K. D., Whitham S. A., Carrington J. C. (2002) Curr. Biol. 12, 1046–1051 [DOI] [PubMed] [Google Scholar]
- 28. Lax S. R., Lauer S. J., Browning K. S., Ravel J. M. (1986) Methods Enzymol. 118, 109–128 [DOI] [PubMed] [Google Scholar]
- 29. Metz A. M., Timmer R. T., Browning K. S. (1992) Nucleic Acids Res. 20, 4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Metz A. M., Wong K. C., Malmström S. A., Browning K. S. (1999) Biochem. Biophys. Res. Commun. 266, 314–321 [DOI] [PubMed] [Google Scholar]
- 31. Hebsgaard S. M., Korning P. G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. (1996) Nucleic Acids Res. 24, 3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lauer S. J., Browning K. S., Ravel J. M. (1985) Biochemistry 24, 2928–2931 [DOI] [PubMed] [Google Scholar]
- 33. Mayberry L. K., Dennis M. D., Allen M. L., Ruud Nitka K., Murphy P. A., Campbell L., Browning K. S. (2007) Methods Enzymol. 430, 397–408 [DOI] [PubMed] [Google Scholar]
- 34. Mayberry L. K., Allen M. L., Dennis M. D., Browning K. S. (2009) Plant Physiol. 150, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Serino G., Deng X. W. (2007) Cold Spring Harbor Prot. pdb.prot4683 doi: 10.1101/pdb.prot4683 [DOI] [Google Scholar]
- 36. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 37. Freire M. A., Tourneur C., Granier F., Camonis J., El Amrani A., Browning K. S., Robaglia C. (2000) Plant Mol. Biol. 44, 129–140 [DOI] [PubMed] [Google Scholar]
- 38. Freire M. A. (2005) Gene 345, 271–277 [DOI] [PubMed] [Google Scholar]
- 39. Clarkson B. K., Gilbert W. V., Doudna J. A. (2010) PLoS ONE 5, e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maquat L. E., Tarn W. Y., Isken O. (2010) Cell 142, 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.