Background: Polycomb Repressive Complex 2 (PRC2) methylates histone H3 at lysine 27.
Results: CDYL directly interacts with PRC2 and tri-methylated histone H3 lysine 27 (H3K27me3) and enhances the methyltransferase activity of PRC2.
Conclusion: CDYL is a molecular bridge between PRC2 and H3K27me3.
Significance: CDYL facilitates PRC2-mediated H3K27me3 modifications of the chromatin, leading to a repressive chromatin state that inhibits target gene expression.
Keywords: Chromatin Modification, Epigenetics, Histone Methylation, Polycomb, Transcription Regulation, CDYL
Abstract
Polycomb group proteins play essential roles in transcriptional regulation of multiple gene families involved in various pathophysiological processes. It is believed that Polycomb Repressive Complex 2 (PRC2) is targeted to chromatin by the EED subunit to methylate histone H3 lysine 27 (H3K27), leading to a repressive chromatin state that inhibits gene expression. Here we report that the chromodomain-containing protein CDYL specifically recognizes di- and tri-methylated H3K27 (H3K27me2 and H3K27me3) and directly interacts with EZH2, the catalytic subunit of PRC2. We show that CDYL dramatically enhances the methyltransferase activity of PRC2 toward oligonucleosome substrates in vitro. Genome-wide analysis of CDYL targets by ChIP sequencing revealed that CDYL and PRC2 share a number of genomic targets. CDYL is required for chromatin targeting and maximal enzymatic activity of PRC2 at their common target sites. Our experiments indicate that CDYL functions as a molecular bridge between PRC2 and the repressive chromatin mark H3K27me3, forming a positive feedback loop to facilitate the establishment and propagation of H3K27me3 modifications along the chromatin.
Introduction
Gene transcription in eukaryotic cells is a complex process that consists of a series of molecular events. Among these events, post-translational histone modifications alter the interaction of histones and DNA or histones and other nuclear proteins, and thus change local chromatin structures to affect gene transcription (1–4). Polycomb group proteins play essential roles in transcriptional regulation of multiple gene families involved in several key cellular functions such as development, stem cell maintenance, and carcinogenesis (5–8). Polycomb Repressive Complex 2 (PRC2),2 which contains EZH2, SUZ12, EED, RbAp48, and AEBP2, is responsible for the methylation (di- and tri-) of lysine 27 of histone H3 (H3K27me2/3) through the catalytic SET domain of EZH2 (9). H3K27me3 modifications are primarily associated with a repressive local chromatin state, leading to the inhibition of target gene expression (10, 11). It is thought that once established, H3K27me3 is maintained by Polycomb Repressive Complex 1 (PRC1), which binds specifically to H3K27me3 and monoubiquitylates lysine 119 of histone H2A (H2AK119ub) (12–14) and stabilizes the compact chromatin structures. However, accumulating evidence indicates that there are genes targeted by PRC2 that lack H2AK119ub and genes targeted by PRC1 in the absence of PRC2, suggesting that gene regulation does not always require both PRC1 and PRC2 (15–17). Previous studies indicated that PRC2 has a strong preference for H3 in oligonucleosome form, although the complex is also capable of methylating H3 existing alone, in octamers, or in mononucleosome form (9). The substrate preference strongly suggests that chromatin structures are critical for maximal PRC2 activity, and that pre-existing H3K27me3 may facilitate catalysis of the same modifications on adjacent nucleosomes. Nevertheless, the mechanism of this positive feedback regulation is not fully understood.
To decode the epigenetic language of covalent histone modifications, it is important to have “readers,” which recognize specific histone modification marks and entail downstream effects such as recruiting other chromatin remodeling complexes. The “writer-reader” interplay influences the dynamics of local chromatin structures and affects gene transcription, DNA repair, replication, and other chromatin-based events (2, 18, 19). Among “reader” proteins that have been identified, proteins containing tudor, the malignant brain tumor (MBT), and chromo domains could recognize methylated lysine residues within histones (20). One such protein, chromodomain Y-like (CDYL), contains a chromodomain and has been implicated in the repression of gene transcription (21). CDYL belongs to a multigene family called the CDY-related gene family, members of which have been shown to play important roles in mammalian spermatogenesis (22). In addition to the N-terminal chromodomain, CDYL has a C-terminal enoyl-coenzyme A (CoA) hydratase-isomerase catalytic domain, whose function remains unknown. Biochemical studies have identified CDYL as a component of repressor complexes CtBP (23) and REST/CDYL/G9a. In the latter case, CDYL bridges the transcription repressor REST and the histone methyltransferase G9a (24). Although CDYL is believed to play a role primarily in transcriptional repression, CDYL target genes are largely unknown and biological functions of this protein are poorly understood.
In our present study, we found that CDYL specifically recognizes H3K27me2 and H3K27me3 modifications and directly interacts with EZH2, the catalytic component of PRC2. CDYL drastically stimulates the histone methyltransferase activity of PRC2 toward oligonucleosome substrates. Genome-wide identification of CDYL target genes revealed that CDYL and PRC2 methyltransferase share a significant number of common binding sites. CDYL is required for the presence and maximal enzymatic activity of PRC2 at their common target promoters. Taken together, we propose CDYL functions as the missing link in the positive feedback regulation of PRC2 activity along the chromatin by bridging PRC2 and H3K27me3 modifications. Through modulating the enzymatic activity and chromatin recruitment of PRC2, CDYL could participate in various biological functions of the complex.
EXPERIMENTAL PROCEDURES
Histone Binding and Histone Peptide Binding Assays
Histone binding assays were performed essentially as described previously (25). Briefly, recombinant full-length or the chromodomain of CDYL (del 2)-GST fusion proteins were expressed in E. coli strain BL21 and purified by glutathione-affinity resin (Amersham Biosciences). Fusion proteins (10, 15, or 20 μg) were incubated with 10 μg of native calf thymus histones (Worthington) in binding buffer (50 mm Tris-HCl, pH 7.5, 1 m NaCl, 1% Nonidet P-40, 0.5 mm EDTA, 1 mm phenylmethyl sulfonyl fluoride (PMSF) plus protease inhibitors (Roche)) at 4 °C for 4 h. Alternatively, GST fusion proteins were incubated with 10 μg of recombinant histone octamers, which were prepared by mixing the four unfolded Xenopus laevis recombinant histones in equimolar amounts as previously described (26). Protein complexes were pulled down with glutathione beads, washed five times with the binding buffer, and subjected to Coomassie Blue staining (Fig. 1A) or Western blotting using anti-H3 antibodies (Abcam catalogue no. 1791, Fig. 1B). Histone peptide binding assays were performed according to a previous protocol (27). Unmodified H3 peptides (1–20 and 21–40) were synthesized by ChinaPeptides, and all modified H3 peptides were purchased from Millipore. Five micrograms of purified GST or GST-CDYL del2 proteins were incubated with 0.2 μg of biotinylated histone peptides in 100 μl of binding buffer (50 mm Tris-HCl, pH7.5, 150 mm NaCl, 0.05% Nonidet P-40, 0.3 mg/ml BSA plus protease inhibitors) overnight at 4 °C. Protein-peptide complexes were pulled down with streptavidin beads (Millipore), washed five times with the binding buffer, and subjected to Western blotting using anti-GST antibodies (MBL, Fig. 1C). More stringent conditions were used for the peptide pull-down assays in Fig. 4E. 2.5 μg of histone peptides were first incubated with streptavidin beads in the binding buffer containing 0.1% Nonidet P-40 and 300 mm NaCl overnight, and the beads were washed five times with the binding buffer before they were mixed with baculovirus-expressed FLAG-CDYL or FLAG-EED proteins. FLAG-CDYL (0.2, 0.4, or 1 μg) and FLAG-EED (0.6, 1.2, or 3 μg) proteins were used to determine the binding affinity for H3K27me3 of the two proteins. One μg of FLAG-CDYL or 3 μg of FLAG-EED proteins were incubated with unmodified H3 peptides (21–44) for the control reactions. After incubation, the beads were washed again five times with the binding buffer, and bound proteins were subjected to Western blotting using anti-FLAG antibodies.
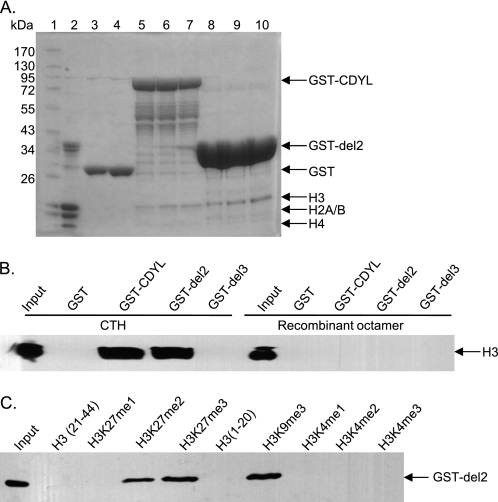
FIGURE 1.
CDYL recognizes histone lysine 27 methylation. A. CDYL interacts with native H3 histones. Histone binding assays were performed using native calf thymus histones (CTH) as substrates. Purified GST (10 μg and 20 μg for lanes 3 and 4, respectively), GST-CDYL (10, 15, and 20 μg for lanes 5–7, respectively), or GST-del 2 proteins (the CDYL chromodomain, 10, 15, and 20 μg for lanes 8–10, respectively) immobilized on glutathione beads were incubated with 10 μg of CTH in binding buffer. Bound protein complexes were subjected to SDS-PAGE and Coomassie staining after extensive washes. Lane 1: protein marker; lane 2: 10 μg of CTH. B, CDYL only binds H3 with post-translational modifications. Histone binding assays were performed as described in A. Either CTH or recombinant Xenopus histone octamers were used as substrates. Histones pulled down by GST-del2 were detected by Western blotting using anti-H3 antibodies. C, CDYL binds H3K9me3, H3K27me3, and H3K27me2 modifications. Histone peptide binding assays were performed with GST-del2 and 0.2 μg of biotinylated histone peptides with indicated modifications. Protein-peptide complexes were subjected to Western blotting using anti-GST antibodies.
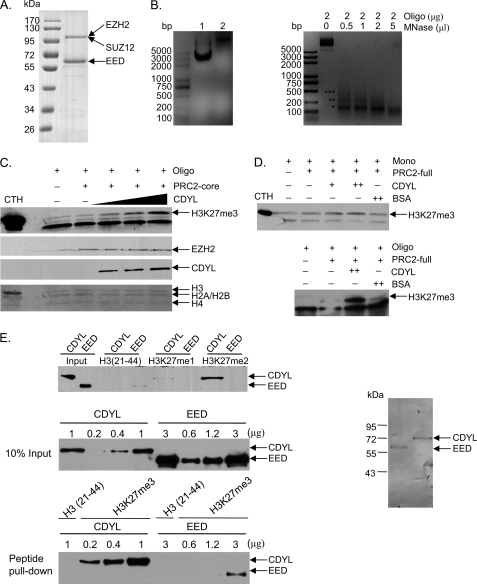
FIGURE 4.
CDYL enhances PRC2 activity in vitro. A, Coomassie Blue staining of PRC2 complexes (containing EZH2, SUZ12, and EED) purified from Sf9 cells. B, MNase digestion of reconstituted oligonucleosomes resolved by 2% agarose gel. Left panel: lane 1 shows the pure pG5E4 plasmid DNA, and lane 2 shows band shift of pG5E4 DNA assembly into oligonucleosomes. Right panel: equimolar amounts of reconstituted oligonucleosomes were digested with increasing amounts of MNase (Sigma). The DNA was isolated and subjected to electrophoresis on a 2% agarose gel in the presence of ethidium bromide. Partial MNase digestion (oligonucleosome: MNase = 2 μg: 0.5 μl) generated a nucleosomal DNA ladder with visible mono-, di-, and trinucleosomal fragments, which are indicated by corresponding numbers of asterisks. Mononucleosomal DNA runs as a 147 bp fragment. C, CDYL stimulates PRC2 activity in vitro. Reconstituted recombinant oligonucleosomes were incubated with EZH2/SUZ12/EED complexes (PRC2-core) in the absence or presence of increasing amounts of baculovirus generated CDYL and histone methyltransferase activity was determined by standard HMT assays. The reaction products were examined by Western blotting with the antibodies indicated on the right. Ponceau staining of histones is shown in the bottom panel to show equal amounts of substrates used in each reaction. D, CDYL only stimulates PRC2 methyltransferase activity toward oligonucleosome, but not mononucleosome substrates. Reconstituted Xenopus oligonucleosomes were digested with MNase (oligonucleosome: MNase = 1 μg: 1 μl) at room temperature for 5 min. This treatment yielded mainly mononucleosomes (see Fig. 4B). Equal amounts of mononucleosomes were used as substrates for the HMT assay in the top panel, whereas equal amounts of undigested oligonucleosomes were used as substrates in the bottom panel. Commercially available EZH2/EED/SUZ12/RbAp48/AEBP2 complexes (PRC2-full) were used to provide methyltransferase activity as indicated. The mild increase of PRC2 activity seen upon CDYL addition in the top panel was mainly due to incomplete digestion of oligonucleosomes (see Fig. 4B). E, binding affinity between CDYL and H3K27me3 is much stronger than the affinity between EED and H3K27me3. In the top panel, histone peptide binding assays show that CDYL, but not EED, binds to H3K27me2 when the same amounts of FLAG-tagged proteins (0.5 μg) were used in the assay. To compare the binding affinity for H3K27me3, 0.2, 0.4, or 1 μg of baculovirus-expressed FLAG-CDYL and 0.6, 1.2, or 3 μg of FLAG-EED proteins were used in the peptide binding assay. Ten percent of total proteins were used as loading controls (middle panel). Peptide-protein complexes were pulled down by streptavidin beads and bound proteins were examined by Western blotting using anti-FLAG antibodies (bottom panel). Ponceau staining of baculovirus-expressed FLAG-CDYL and FLAG-EED is shown in the right panel.
Western Blotting and Co-immunoprecipitation
MCF-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Western blotting and co-immunoprecipitation assays were performed as previously described (15, 28–31). Commercial antibodies used were CDYL (Abcam catalogue no. 5188), EZH2 (BD Biosciences catalogue no. 612667, ChIP Ab Millipore catalogue no. 17-662), SUZ12 (Cell Signaling catalogue no. 3737S), EED (Millipore catalogue no. 05-1320, ChIP Ab Millipore catalogue no. 17-663), FLAG (Sigma), REST (Santa Cruz catalogue no. 25398), H3K9me3 (Millipore catalogue no.05–1242), H3K27me1 (Millipore catalogue no. 07–448), H3K27me2 (Millipore catalogue no. 07–452), and H3K27me3 (Millipore catalogue no. 07–449).
GST Pull-down Assays
GST pull-down assays were performed as previously described (30, 32). Full-length EZH2 was transcribed/translated in vitro from the plasmid FLAG-EZH2/pcDNA3.1, as described in our previous work (30). Full-length SUZ12 and EED, and EZH2 deletion mutants were cloned into the plasmid pGBKT7, which contains a c-Myc epitope tag, and transcribed/translated in vitro. Anti-EZH2 and anti-Myc (MBL) antibodies were used to detect the respective translated proteins.
FPLC Chromatography
MCF-7 nuclear extracts were prepared and dialyzed against buffer D (20 mm HEPES, pH 8.0, 10% glycerol, 0.1 mm EDTA, 300 mm NaCl). FPLC was performed as previously described (33). Superose 6 or Superdex 200 10/300 GL Chromatographic Separation Columns (Amersham Biosciences) were used to purify protein complexes.
Baculovirus Production and Generation of CDYL and PRC2 Complexes
The baculovirus constructs were generated by insertion of the open reading frame of human EZH2, SUZ12, EED, or CDYL into the pFastBac HT A vector (Invitrogen) between the BamH1 and Xho1 sites. Each construct contained a C-terminal FLAG tag for further affinity purification. The viruses were generated and amplified according to the manufacturer's protocol. To purify CDYL proteins, sf9 cells were infected at a multiplicity of infection of 10 with viruses expressing FLAG-CDYL. Cells were harvested after 3 days and lysed by sonication, and the lysate was incubated for 4 h with M2 agarose beads (Sigma). Washes were performed with BC500 buffer containing 50 mm Tris, 2 mm EDTA, 500 mm KCl, 10% glycerol, and protease inhibitors. Proteins were eluted with FLAG peptide at a 0.2 mg/ml concentration. EZH2/EED/SUZ12 complexes were generated by co-infecting sf9 cells with viruses expressing FLAG-tagged EZH2, EED, and SUZ12.
Preparation of Recombinant Nucleosomes and Micrococcal Nuclease Digestion
Recombinant Xenopus histone octamers were prepared as described previously (26, 34). The plasmid pG5E4 (containing 10 5 S rDNA nucleosome positioning sequences) was mixed with recombinant histone octamers in equimolar amounts, and nucleosome arrays were assembled by stepwise salt dialysis according to a previous method (35). To examine the incorporation of histone octamers, the nucleosomal arrays were digested with micrococcal nuclease (MNase, Sigma catalogue no. N5386, dissolved the powder to make a 2 units/μl stock solution) and resolved by agarose electrophoresis as described previously (36). Mononucleosomes were generated by digesting 10 μg of oligonucleosomes with 10 μl of MNase at room temperature for 5 min.
Histone Methyltransferase (HMT) Assay
HMT assays were performed as described previously (37). For Fig. 4C, 0.5 μg of baculovirus-expressed PRC2 complexes (containing EZH2, SUZ12, and EED) plus 0.05, 0.1, or 0.15 μg of baculovirus-expressed CDYL proteins were added to the HMT reaction mixture. The reaction was stopped by adding SDS buffer. For Fig. 4D, 0.5 μg of recombinant human EZH2/EED/SUZ12/RbAp48/AEBP2 complexes (BPS Bioscience catalogue no. 51004) were used to provide methyltransferase activity and 2 μg of recombinant oligonucleosomes or mononucleosomes were used as substrates. CDYL proteins (0.03 or 0.1 μg) or BSA proteins (0.1 μg) were added to the reaction as indicated. The HMT assays with fractions of FPLC elutes (Fig. 2D) were performed by incubation of the fractions with 2 μg of recombinant Xenopus histone H3 proteins in a 50 μl reaction volume with HMT buffer (20 mm Tris-HCl, 4 mm EDTA, 1 mm PMSF, 0.5 mm DTT, pH 7.9) and 0.3 μm S-adenosyl-l-methionine for 1 h at 30 °C. The samples of HMT reactions were subjected to Western blotting using anti-H3K27me3 antibodies.
FIGURE 2.
CDYL is physically associated with the PRC2 complex. A, in vivo immunoprecipitation. Top two panels: endogenous IP of MCF-7 cell lysates using antibodies against CDYL. Antibodies (EZH2 or SUZ12) used for Western blotting are indicated on the right. Bottom panel: MCF-7 cells were transfected with a FLAG-CDYL construct and subjected to co-IP assays after 48 h. Cell protein extracts were immunoprecipitated with polyclonal antibodies against EED, and blotted with monoclonal anti-FLAG antibodies. An immunoglobulin G (IgG) control is included in each experiment. B, GST pull-down assays. Purified GST or GST-CDYL proteins immobilized on glutathione Sepharose 4B beads were incubated with in vitro translated EZH2, SUZ12, or EED. Bound proteins were detected with monoclonal anti-EZH2 antibodies (top panel) or anti-MYC tag antibodies (lower two panels). C, Superose 6 gel filtration analysis of the MCF-7 nuclear extracts. Migration of molecular markers is indicated above the panels and the antibodies for Western blotting are indicated on the right. Equal volumes from each fraction were analyzed. D, similar FPLC experiments as in C using a Superdex 200 10/300 GL column. The chromatographic fractions were analyzed by Western blotting using the indicated antibodies. Bottom panel: HMT assays were performed using the indicated eluted fractions. Recombinant Xenopus H3 proteins were used as substrates, and the reaction products were analyzed by Western blotting with anti-H3K27me3 antibodies.
ChIP Sequencing
MCF-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum. Approximately 5 × 107 cells were used for each ChIP-seq assay. ChIP assays were performed according to a previous protocol proportionally scaled up for each step (38). The chromatin DNA precipitated by either normal rabbit IgG (control) or polyclonal CDYL antibodies (Abcam, ab5188) was purified with the Qiagen PCR purification kit. In-depth whole genome DNA sequencing was performed by the Beijing Genome Institute (BGI). The raw sequencing image data were examined by the Illumina analysis pipeline, aligned to the unmasked human reference genome (NCBI v36, hg18) using ELAND (Illumina), and further analyzed by MACS. Enriched binding peaks of CDYL were generated after filtering through the control IgG data. Genomic distribution of CDYL binding sites was analyzed by using the cis-regulatory element annotation system. De novo motif screening was performed on sequences ±125 bp from the centers of CDYL binding peaks by using the CEAS and MEME systems. ChIP-seq density of histone modification enrichment profiles were obtained through GEO. Heatmaps and correlation maps were generated by R.
RNA Interference
siRNAs were synthesized by Shanghai GenePharma Co., Ltd. The sequences were as follows: CDYL-siRNA#3 sense: GAGAUAUUGUGGUCAGGAAtt; antisense: UUCCUGACCACAAUAUCUCtt; CDYL-siRNA#4 sense: GGUACAUCUCCGUUCAUGGtt; antisense: CCAUGAACGGAGAUGUACCtt; non-silencing siRNA sense: UUCUCCGAACGUGUCACGUtt, antisense: ACGUGACACGUUCGGAGAAtt. siRNAs were transfected into MCF7 cells using LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's instructions.
RESULTS
CDYL Recognizes Histone Lysine 27 Methylation
Previous studies have suggested that CDYL functions mainly as a transcription corepressor in somatic cells (21, 39, 40). CDYL contains an N-terminal chromodomain, which often serves as a recognition module for methylated histone lysine residues. To elucidate the mechanism of CDYL action in transcription regulation, we first characterized the histone binding preference of CDYL. Histone binding assays were performed using native calf thymus total histones (CTH) as substrates. Recombinant CDYL protein mainly interacted with H3, but not H4, H2A, or H2B (Fig. 1A, lanes 5–7). The chromodomain of CDYL is responsible for the histone binding ability of the protein, as the CDYL truncation mutant containing only the chromodomain (CDYL del2)-bound histone H3 as efficiently as the full-length protein (Fig. 1A, lanes 8–10). To examine whether the binding between CDYL and H3 is dependent on post-translational modifications of H3, we compared the binding affinity of CDYL to native CTHs, which retain multiple in vivo modifications, and to recombinant Xenopus octamers, which possess no post-translational modifications. CDYL was found to interact only with native H3 but not recombinant H3 proteins (Fig. 1B), suggesting that post-translational modifications of H3 are critical for the recruitment of CDYL. As expected, deletion of the chromodomain of CDYL (CDYL del3) completely abolished the interaction between CDYL and native H3 proteins (Fig. 1B, compare GST-del2 and GST-del3). We further examined the binding preference of CDYL for histone modifications by using histone peptide binding assays. CDYL was found to strongly interact with the repressive H3 lysine methylation marks, including H3K9me3, H3K27me2, and H3K27me3 (Fig. 1C), whereas no binding could be detected for H3K4 methylation modifications, which are largely associated with transcriptional activation. Our observations are consistent with previous reports (24, 41, 42) and support the role of CDYL in regulating transcriptional repression.
CDYL Is Physically Associated with PRC2
CDYL was previously found to be a component of the REST/CDYL/G9a complex (24). CDYL interacts with both methylated H3K9 residues and G9a, a histone methyltransferase for H3K9, suggesting that CDYL may bridge histone methylation and the modification enzyme. The strong interaction between CDYL and H3K27me2/H3K27me3 prompted us to examine whether CDYL was able to directly interact with the H3K27 methyltransferase, PRC2. To examine the in vivo interaction between CDYL and PRC2, total proteins from MCF-7 cells were extracted and immunoprecipitated with anti-CDYL antibodies. The immunoprecipitates were then immunoblotted with antibodies against the core components of PRC2. The results showed that endogenous EZH2 and SUZ12 could be efficiently co-immunoprecipitated with CDYL (Fig. 2A, top two panels). Because EED runs close to the immunoglobulin heavy chain in SDS-PAGE, we performed the EED-CDYL co-immunoprecipitation assay using MCF-7 cells transfected with a FLAG-CDYL construct. Cell lysates were collected 48 h after transfection and immunoprecipitated with the polyclonal antibody against EED. The immunoprecipitates were then immunoblotted with monoclonal anti-FLAG antibodies. CDYL and EED clearly interacted with each other in vivo (Fig. 2A, bottom panel).
Next, GST pull-down assays were performed with bacterially expressed GST-CDYL proteins and components of PRC2 transcribed/translated in vitro. CDYL interacted directly with only the EZH2 component of PRC2, whereas no direct binding between CDYL and SUZ12 or EED was detected (Fig. 2B). These data suggested that the recruitment of PRC2 by CDYL in vivo is mediated by an interaction of CDYL with EZH2, the catalytic subunit of PRC2.
To further consolidate the association between CDYL and PRC2, protein fractionation experiments were carried out through a high salt extraction and size exclusion approach by fast protein liquid chromatography (FPLC). We first used the Superose 6 size column (optimal separation range for proteins is from 5 to 5000 kDa). The experimental results revealed that native CDYL in MCF-7 cells could be eluted in chromatographic fractions with apparent molecular masses much greater than that of the monomeric protein, and the elution pattern of CDYL in chromatographic fractions with high molecular masses largely overlapped with that of PRC2 components, especially EZH2 and SUZ12 (Fig. 2C). In addition, we performed FPLC chromatography using a Superdex 200 column (optimal separation range for proteins is from 10 to 600 kDa). The results confirmed that CDYL could be eluted from the same fractions as the components of PRC2 (Fig. 2D, upper panels). We then examined whether the Superdex 200 eluted fractions had any methyltransferase activity. Histone methyltransferase assays (HMT) were performed using these fractions and recombinant Xenopus histone H3 proteins. Western blotting of the reaction products using H3K27me3 antibodies clearly indicated methyltransferase activity in fractions containing both PRC2 and CDYL (Fig. 2D, bottom panel), suggesting that CDYL is physically and functionally associated with PRC2 in vivo.
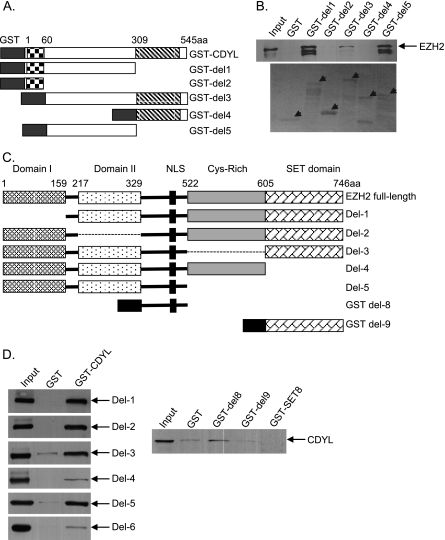
We constructed a series of CDYL and EZH2 deletion mutants to map the domains mediating the interaction between CDYL and EZH2. GST pull-down assays using these deletion mutants revealed that the region from 61–309 aa in CDYL and the region from 329–522 aa in EZH2 were essential for their interaction (Fig. 3). The chromodomain of CDYL and the SET domain of EZH2 are not present at their interface, allowing the two proteins to perform their respective histone reading and writing actions properly upon binding.
FIGURE 3.
Mapping the domains responsible for the interaction between CDYL and EZH2. A, schematic drawing of CDYL protein. CDYL deletion mutants including del1 (1–309 aa), del2 (1–60 aa, the chromodomain), del3 (61–545 aa), del4 (310–545 aa, the coAP domain), and del5 (61–309 aa) were fused to GST. B, GST pull-down experiments were performed with in vitro translated FLAG-EZH2 and purified GST or GST-CDYL deletion mutants. The precipitated complexes were examined by Western blotting using monoclonal anti-EZH2 antibodies (the upper panel). Only CDYL mutants containing the middle region from 61–309 aa (del1, del3, del5) efficiently pulled down EZH2. The lower panel shows the Ponceau staining of purified GST fusion proteins added to the reaction. The arrows indicate the positions of the respective GST fusion proteins as labeled on the top. C, schematic drawing of EZH2 protein. EZH2 deletion mutants (del1 to del5) were cloned into the pGBKT7 plasmid, which contains a c-Myc epitope tag and can be transcribed/translated in vitro. Del8 and Del9 were fused to GST. D, GST pull-down experiments were performed with in vitro translated Myc-EZH2 deletion mutants and purified GST or GST-CDYL in the left panels. Right panel: GST pull-down assays were performed with in vitro translated Myc-CDYL, which was incubated with purified GST, GST-del8, GST-del9, or GST-SET8 (negative control protein). Bound proteins were examined by Western blotting using monoclonal anti-Myc antibodies.
CDYL Enhances Enzymatic Activity of PRC2 in Vitro
PRC2 preferentially methylates histone H3 in oligonucleosome form, indicating that pre-existing H3K27 methylation modifications may facilitate PRC2 catalysis of the same modification on adjacent nucleosomes. This positive feedback regulation of PRC2 enzymatic activity could be achieved by reader protein recognition of pre-existing H3K27me3 and subsequent recruitment of more PRC2, allowing rapid and efficient propagation of H3K27me3 to neighboring nucleosomes. CDYL serves as an ideal candidate for a bridging factor in this positive feedback loop because it directly interacts with both H3K27me3 and the EZH2 component of PRC2. To examine whether CDYL is able to enhance the methyltransferase activity of PRC2, we purified CDYL and PRC2 independently from Sf9 cells (Fig. 4A). Recombinant Xenopus oligonucleosomes were reconstituted by salt dialysis procedures (36, 43) and were used as substrates for PRC2. Successful assembly of oligonucleosomes was demonstrated by partial micrococcal nuclease (MNase) digestion, which generated a nucleosomal DNA ladder with visible mono-, di-, and trinucleosomal fragments (Fig. 4B). CDYL was added to PRC2, and histone methyltransferase assays (HMT) were performed. Western blotting of the reaction products using anti-H3K27me3 antibodies showed that while three-component PRC2 (containing EZH2, SUZ12, and EED) only weakly methylated H3K27, adding CDYL to the reaction drastically stimulated the enzymatic activity of PRC2 (Fig. 4C). We also obtained commercially available five-component PRC2 (EZH2/EED/SUZ12/RbAp48/AEBP2) and repeated the HMT assays. CDYL significantly enhanced the methyltransferase activity of five-component PRC2 toward oligonucleosome substrates (Fig. 4D, bottom panel). We digested reconstituted oligonucleosomes with MNase (oligonucleosome: MNase = 1 μg: 1 μl) to generate mononucleosomes (Fig. 4B). Notably, CDYL showed little stimulatory effect on PRC2 when mononucleosomes were used as substrates (Fig. 4D, top panel). In addition, the stimulation observed did not result from a contaminant protein associated with CDYL and appeared to be specific, as the addition of bovine serum albumin (BSA) did not affect the methylation activity. The molar ratio of CDYL to PRC2 that resulted in detectable stimulatory effect was ∼1:10 to 1:3, and within this range, increasing amounts of CDYL exhibited stronger activity. These data indicate that CDYL is not necessarily an integral component of PRC2, but rather acts as a positive regulator of PRC2 activity by bridging the pre-existing H3K27me3 and newly recruited PRC2 on neighboring nucleosomes.
Two recent studies reported that the EED component of PRC2 specifically binds to histone tails carrying trimethyl-lysine residues via its carboxyl-terminal WD40 repeats. EED is therefore thought to be responsible for the propagation of H3K27me3 marks to neighboring nucleosomes or the sister chromatid (44, 45). Researchers have noted, however, that the affinity of EED for H3K27me3 is significantly weaker than that of HP1 and Pc chromodomains for the H3K9me3 or H3K27me3 peptides, suggesting additional readers may contribute to the recruitment of PRC2 to pre-existing H3K27me3 (45). We compared the binding affinity of CDYL and EED for H3K27me3 through semi-quantitative histone peptide binding assays. Increasing amounts of baculovirus-expressed FLAG-CDYL or FLAG-EED proteins were incubated with H3K27me3 or control peptides immobilized on streptavidin beads. After extensive washes, bound proteins were detected by Western blotting using anti-FLAG antibodies. In our experimental conditions, to get similar signal strength on Western blot, the amounts of EED needed were at least 15–20 times more than the amounts of CDYL (Fig. 4E). We estimated the affinity for H3K27me3 of CDYL was ∼20-fold higher than EED. Therefore, CDYL recognizes H3K27me3 much more efficiently than EED. CDYL also strongly bound to H3K27me2 in peptide binding assays, whereas the interaction between EED and H3K27me2 was hardly detected (Fig. 4E, top panel). It has been proposed that H3K27me2 is an intermediate H3K27 methylation state that marks genes as being potentially repressible by PRC2 (37). Through efficient binding to both H3K27me2 and H3K27me3, CDYL serves as a more competent bridging factor than EED to connect PRC2 and histone modifications in chromatin.
Genomic Landscape of CDYL Target Genes in MCF-7 Cells
Rather than being a core component, CDYL appears to be an associate protein of PRC2, because previous biochemical approaches to purify PRC2 did not identify CDYL in the complex (9). It is conceivable that CDYL regulates the function of PRC2 only on selected target genes or under special cellular conditions. We sought to explore the functional relationship between CDYL and PRC2 by identifying their common target genes in vivo.
Although several groups, including ours, have described the transcriptional corepressor function of CDYL, these studies were mainly based on the artificial Gal4-luciferase reporter system (21, 24, 39) and the in vivo target genes of CDYL were poorly defined. In order to determine the genomic landscape of CDYL target genes, we performed chromatin immunoprecipitation coupled with genomic sequencing (ChIP-seq) in MCF-7 cells with control or CDYL-specific polyclonal antibodies. Using MACS program (28), we identified over 2000 high confidence CDYL-specific binding peaks from pre-processed sequencing reads with a p value cutoff of 10−5. Genomic distribution analysis indicated that the majority of the CDYL binding sites (58%) were distant from proximal promoter regions. The second largest group of CDYL binding sites was located within gene bodies (introns, 34%) and about 3% of the binding sites were located at promoter regions (Fig. 5A).
FIGURE 5.
Genomic landscape of CDYL binding sites determined by ChIP sequencing. A, genomic distribution of CDYL binding regions determined by CEAS. B, consensus CDYL binding motif is nearly identical to the REST binding motif. Motif screening was performed using MEME suite. C, examples of ChIP-seq identified CDYL binding to two REST target genes, BDNF (upper panels), and STMN2 (lower panels). Blue blocks represent REST binding sites as reported previously (48, 49). ChIP-seq density of CDYL is shown in red. The chromosome number and position of loci are shown below. D, ChIP-seq density heatmaps of histone H3K27 trimethylation (H3K27me3, red), H3K9me3 (H3K9me3, green), and H3K14 acetylation (H3K14ac, yellow) for CDYL binding sites and randomly distributed genomic regions (control). CDYL binding sites were aligned vertically along the center of each peak region called by MACS (indicated by triangle), and randomly selected genomic regions were used as controls. Each row represents a unique ChIP-seq peak region. The average ChIP-seq density of H3K27me3, H3K9me3, or H3K14ac was calculated within 250 bp bins around the genomic regions from −2.5 kb to +2.5 kb relative to each peak center. Heatmaps were arranged in descending order by the average ChIP-seq density per row. E, demonstration of ChIP-seq density of CDYL (blue), H3K27me3 (red), and H3K9me3 (green) at specific regions of chromosome 16. The position of loci is shown below the ChIP-seq density track.
We next screened for consensus CDYL binding motifs on sequences from ±125 bp relative to each CDYL-binding peak center. We performed an unbiased search against two datasets, CEAS (cis-regulatory element annotation system (46)) and MEME (multiple Em for motif elicitation (47)). The results revealed a recurrent appearance of the neuron-restrictive silencer element (NRSE, also known as RE1) in about 500 CDYL-binding sites (p < 1.7E-244 and p < 8E-2097, estimated by CEAS and MEME respectively) (Fig. 5B). RE1 is the binding sequence for the transcriptional repressor REST. Importantly, we found significant enrichment of CDYL-binding signals surrounding the RE1 sites of known REST target genes, such as superior cervical ganglion-10 (SCG, STMN2) and brain-derived neurotrophic factor (BDNF) (Fig. 5C) (48, 49). Our finding that the REST-binding motif is associated with CDYL target sites is consistent with a previous report showing CDYL and REST coexist in the transcriptional repressor complex REST/CDYL/G9a (24).
To compare the genomic landscape between CDYL and PRC2 methyltransferase activity, we intercrossed our CDYL ChIP-seq data with previously reported histone modification ChIP-seq results in MCF-7 cells (50). We compared characteristic enrichment of H3K27me3, H3K9me3, and H3K14 acetylation between CDYL binding sites and randomly selected genomic control regions. Compared with the control regions, H3K27me3 and H3K9me3 were found to be significantly enriched in regions surrounding the CDYL genomic binding sites, whereas little difference in H3K14 acetylation could be found between CDYL binding sites and the nonspecific regions (Fig. 5, D and E).
CDYL Is Required for Targeting PRC2 to Chromatin
The above data indicate that genomic distributions of CDYL-binding sites and PRC2 activity are closely related. To validate the CDYL ChIP-seq results as well as to examine the co-occupancy of CDYL and PRC2 at their common target promoters, we performed quantitative ChIP assays (qChIP). In addition to a number of CDYL targets identified by ChIP-seq, we also included promoters of HoxA3 and FBXO32, two previously reported PRC2 target loci (51, 52). qChIP assays were performed in MCF-7 cells with antibodies against CDYL, EZH2, H3K27me3, or control IgG. In accordance with the ChIP-seq results, we found that CDYL and EZH2 were both present at the promoters of DBC1, MYT1, NEUROD1, JIP1, BASE, and SCG, whereas CDYL was absent from HoxA3 and FBXO32 promoters. Significant amounts of CDYL were detected at the VGF promoter, whereas little, if any, EZH2 was present at this region (Fig. 6A). These results confirmed the CDYL ChIP-seq data and indicated that CDYL and EZH2 could regulate overlapping target genes in vivo, although there are also genes regulated by only one of them.
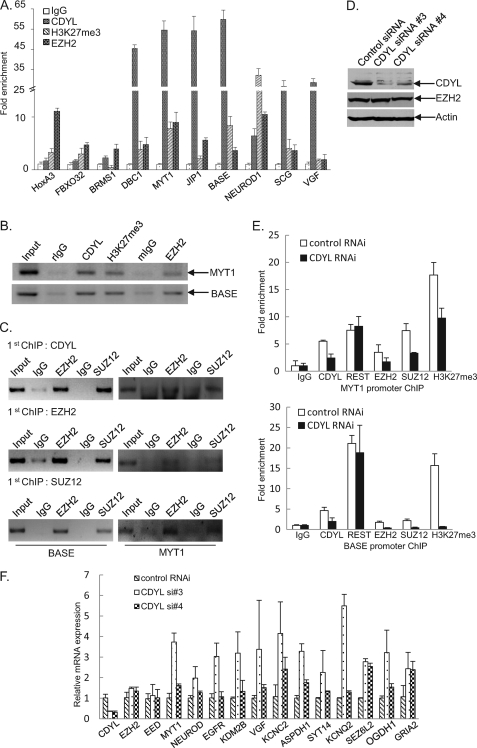
FIGURE 6.
Validation of common target genes of CDYL and PRC2. A, quantitative ChIP assays were performed in MCF-7 cells with primer pairs specific to indicated gene promoters (see supplemental Table S1). Normal rabbit IgG, as well as polyclonal antibodies against CDYL, EZH2, and H3K27me3 were used to immunoprecipitate the protein-DNA complex. B, conventional semi-quantitative ChIP assays performed at the MYT1 and BASE promoters. C, CDYL and PRC2 exist in the same protein complex at the MYT1 and BASE promoters. ChIP and re-ChIP experiments were performed with the indicated antibodies and primer pairs. D, CDYL expression was efficiently knocked down by specific siRNAs. Non-silencing or CDYL specific siRNAs were transfected into MCF-7 cells. Total proteins were extracted and the expression of CDYL and EZH2 proteins were examined by Western blotting. Actin protein levels were measured to indicate equal loading of protein lysates. E, CDYL is required for PRC2 chromatin targeting at the MYT1 and BASE promoters. MCF-7 cells were transfected with control siRNA or CDYL-specific siRNA. 48 hours after the transfection, cell lysates were collected, and ChIP experiments were performed using the indicated antibodies. Real-time PCR assays were performed for the measurement. F, CDYL mainly represses the expression of target genes. MCF-7 cells were transfected with control or CDYL-specific siRNAs. Total RNAs were prepared and the mRNA levels of the indicated genes were examined by real-time RT-PCR. The data were normalized against the expression of GAPDH. Each bar represents the mean ± S.D. for triplicate measurements.
Next, we performed ChIP/re-ChIP assays to further demonstrate that CDYL and PRC2 interact and exist in the same protein complex at their common target sites. We selected representative promoters of two genes, MYT1 (a classical EZH2 target gene) (37) and BASE (a newly identified CDYL/EZH2 target gene by our ChIP-seq). The MYT1 and BASE promoters that were immunoprecipitated with antibodies against CDYL could be re-immunoprecipitated with antibodies against EZH2 and SUZ12 (Fig. 6C, top panel). The same results were obtained when the initial ChIP was performed with antibodies against EZH2 or SUZ12 (Fig. 6C, lower two panels). These data strongly suggest that CDYL and PRC2 physically interact at their common target promoters.
To examine whether CDYL is necessary for chromatin targeting and maximal enzymatic activity of PRC2 in vivo, we transfected MCF-7 cells with CDYL-specific or mock siRNAs and performed ChIP analysis using antibodies against CDYL, EZH2, or SUZ12. Knocking down CDYL expression led to a decreased occupancy of CDYL and subsequent deprivation of EZH2 and SUZ12 from the promoters of MYT1 and BASE (Fig. 6E), indicating that CDYL is required for efficient chromatin binding of PRC2 at their common target promoters. Furthermore, transfection of CDYL specific siRNA to cells led to a drastic decrease of H3K27me3 levels at the MYT1 promoter and the BASE promoter (Fig. 6E), suggesting CDYL is necessary for maximal PRC2 activity at these regions. Consistent with the corepressor function of CDYL in cells, the presence of CDYL at target promoters mainly functions in transcriptional repression of the corresponding genes, as knocking down CDYL expression by specific siRNAs resulted in an increased expression of the majority of target genes (Fig. 6F).
DISCUSSION
Faithful transmission of histone modifications is a fundamental theme in epigenetics. In quiescent cells, efficient propagation of histone modifications to adjacent nucleosomes is critical to establish a proper local chromatin environment and achieve desired regulation of gene expression. Rapid spreading to adjacent nucleosomes is especially important for repressive histone marks, which are often found to extend thousands of nucleosomes in chromatin (13). Specific chromatin structures can also be inherited following DNA replication, therefore maintaining cell identity over generations. The current model of chromatin inheritance posits that histone modifications can be re-established by complexes that recognize a specific modification on an inherited parental histone and catalyze the same type of modification on adjacent newly deposited nucleosomes (53–55). CDYL specifically recognizes H3K27me2 and H3K27me3 modifications. CDYL also interacts with the H3K27 methyltransferase PRC2. In vitro HMT assays clearly showed that CDYL significantly enhances the methyltransferase activity of PRC2 toward oligonucleosome substrates, but not mononucleosomes. By bridging PRC2 and the histone modifications, CDYL serves as a linking factor for the positive feedback loop regulating PRC2 enzymatic activity in chromatin. Although alteration of the cellular levels of CDYL by overexpression or siRNA-based gene silencing does not lead to major changes of the overall H3K27me3 levels (data not shown), CDYL ChIP-seq and subsequent validation experiments clearly demonstrated that CDYL and PRC2 share a number of common genomic binding targets, where CDYL is required for the recruitment and optimal enzymatic activity of PRC2.
Recently, two studies reported that EED, a component of PRC2, specifically binds to H3K27me3 via its C-terminal WD40 repeats and is responsible for the propagation of H3K27me3 to neighboring nucleosomes (44, 45). We found that CDYL exhibits much stronger binding affinity for H3K27me3 than EED. Our observation is also supported by a recent report describing genome-wide quantitative interaction proteomics of histone modifications and their readers. CDYL was identified as one of the strongest binders of H3K27me3 modifications according to the quantitative mass spectrometry analysis (42). CDYL is not present at some classical PRC2 target promoters, such as those of the HoxA family genes, and previous biochemical purification approaches did not identify CDYL in the H3K27 methyltransferase protein complex (9). Therefore, CDYL is likely a peripheral component of PRC2 and modulates PRC2 activity when needed. Compared with EED, whose homologs can be found throughout eukaryotic species ranging from Drosophila to plants to mammals, homologs of CDYL exist only in mammals (39). It is conceivable that CDYL provides additional means for the regulation of PRC2 function under different cellular conditions, allowing more complex biological activities in advanced organisms.
We demonstrated that the consensus binding motifs of CDYL and REST are almost identical. CDYL potentially shares extensive common genomic targets with the transcription repressor REST. It is believed that REST mediates gene silencing through two distinct effector arms: one via LSD1-CoREST and separately via the adaptor protein CDYL and the H3K9 methyltransferase G9a (24). While genome-scale analysis in MCF-7 cells showed a mild increase of H3K27me3 and H3K9me3 correlation within CDYL binding regions compared with randomly selected genomic regions (Fig. 7A), conflicting data were obtained when we examined the concurrence of H3K9me3 and H3K27me3 at individual CDYL target sites (Fig. 7, B and C). At this point it is hard to conclude whether the presence of CDYL leads to increased coordination of H3K9 and H3K27 methyltransferase activity.
FIGURE 7.
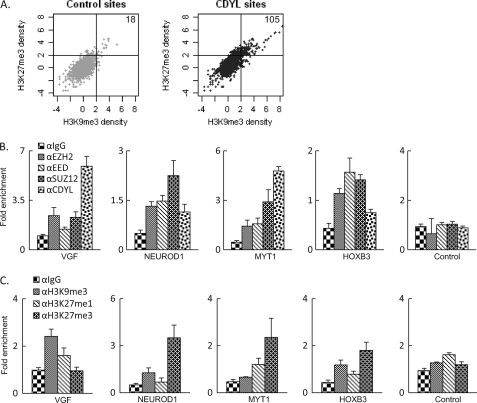
Correlation of H3K9me3 and H3K27me3 at CDYL target promoters. A, correlation of H3K27me3 and H3K9me3 within CDYL binding sites (right panel) and randomly selected genomic regions (left panel). Numbers in the upper right quadrant indicate total sites with both H3K27me3 and H3K9me3 present (the cut-off value is 2). B and C, ChIP-coupled real-time RT PCR assays were performed with the indicated antibodies in MCF-7 cells. Among the regions examined, the VGF promoter apparently enriched more H3K9me3 modifications and the MYT1 promoter enriched more H3K27me3 modifications, while significant amounts of CDYL were present at both regions. Both NEUROD1 and HOXB3 promoters showed coexistence of H3K9me3 and H3K27me3 modifications, with mild enrichment of CDYL at the NEUROD1 promoter and undetectable CDYL at the HOXB3 promoter. The transcriptional active PS2 promoter was used as a control in these experiments.
Taken together, we have identified CDYL as a bridging factor for H3K27me3 and the modifying enzyme PRC2. CDYL stimulates PRC2 activity in vitro and is required for maximal activity of PRC2 at their common target sites in vivo. Because of the CDYL/PRC2 interplay at the molecular level, it will be interesting to study how CDYL participates in the biological functions of PRC2 and how CDYL contributes to inheritance of H3K27me3 modifications in animal development.
Supplementary Material
This work was supported by Grants 30971637 and 90919045 (to J. L.) and 30830032 and 30921062 (to Y. S.) from the National Natural Science Foundation of China and Grants 973 Program: 2011CB504204 (to J. L. and Y. S.) from the Ministry of Science and Technology of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- PRC
- polycomb repressive complex
- CTH
- calf thymus histones
- HMT
- histone methyltransferase
- CDYL
- chromodomain Y-like
- aa
- amino acids.
REFERENCES
- 1. Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 2. Campos E. I., Reinberg D. (2009) Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 3. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 4. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 5. Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., Ghosh D., Sewalt R. G., Otte A. P., Hayes D. F., Sabel M. S., Livant D., Weiss S. J., Rubin M. A., Chinnaiyan A. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Margueron R., Reinberg D. (2011) Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 10. Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002) Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y., Cao R., Wang L., Jones R. S. (2004) Cold Spring Harb. Symp. Quant. Biol. 69, 309–317 [DOI] [PubMed] [Google Scholar]
- 12. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M. (2006) Nat. Genet. 38, 694–699 [DOI] [PubMed] [Google Scholar]
- 14. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 15. Schoeftner S., Sengupta A. K., Kubicek S., Mechtler K., Spahn L., Koseki H., Jenuwein T., Wutz A. (2006) EMBO J. 25, 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sing A., Pannell D., Karaiskakis A., Sturgeon K., Djabali M., Ellis J., Lipshitz H. D., Cordes S. P. (2009) Cell 138, 885–897 [DOI] [PubMed] [Google Scholar]
- 17. Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S., Adli M., Kasif S., Ptaszek L. M., Cowan C. A., Lander E. S., Koseki H., Bernstein B. E. (2008) PLoS Genet. 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldberg A. D., Allis C. D., Bernstein E. (2007) Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 20. Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caron C., Pivot-Pajot C., van Grunsven L. A., Col E., Lestrat C., Rousseaux S., Khochbin S. (2003) EMBO Rep. 4, 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dorus S., Gilbert S. L., Forster M. L., Barndt R. J., Lahn B. T. (2003) Hum. Mol. Genet. 12, 1643–1650 [DOI] [PubMed] [Google Scholar]
- 23. Shi Y., Sawada J., Sui G., Affar el B., Whetstine J. R., Lan F., Ogawa H., Luke M. P., Nakatani Y., Shi Y. (2003) Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
- 24. Mulligan P., Westbrook T. F., Ottinger M., Pavlova N., Chang B., Macia E., Shi Y. J., Barretina J., Liu J., Howley P. M., Elledge S. J., Shi Y. (2008) Mol. Cell 32, 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 27. Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H. H., Whetstine J. R., Bonni A., Roberts T. M., Shi Y. (2007) Cell 128, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 30. Shi B., Liang J., Yang X., Wang Y., Zhao Y., Wu H., Sun L., Zhang Y., Chen Y., Li R., Zhang Y., Hong M., Shang Y. (2007) Mol. Cell Biol. 27, 5105–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yin N., Wang D., Zhang H., Yi X., Sun X., Shi B., Wu H., Wu G., Wang X., Shang Y. (2004) Cancer Res. 64, 5870–5875 [DOI] [PubMed] [Google Scholar]
- 33. Li R., Zhang H., Yu W., Chen Y., Gui B., Liang J., Wang Y., Sun L., Yang X., Zhang Y., Shi L., Li Y., Shang Y. (2009) EMBO J. 28, 2763–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 35. Sims R. J., 3rd, Trojer P., Li G., Reinberg D. (2006) Methods 40, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loyola A., Reinberg D. (2003) Methods 31, 96–103 [DOI] [PubMed] [Google Scholar]
- 37. Sarma K., Margueron R., Ivanov A., Pirrotta V., Reinberg D. (2008) Mol. Cell Biol. 28, 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 39. Li X., Liang J., Yu H., Su B., Xiao C., Shang Y., Wang W. (2007) Trends Genet. 23, 427–431 [DOI] [PubMed] [Google Scholar]
- 40. Kuppuswamy M., Vijayalingam S., Zhao L. J., Zhou Y., Subramanian T., Ryerse J., Chinnadurai G. (2008) Mol. Cell Biol. 28, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franz H., Mosch K., Soeroes S., Urlaub H., Fischle W. (2009) J. Biol. Chem. 284, 35049–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 43. Shi L., Sun L., Li Q., Liang J., Yu W., Yi X., Yang X., Li Y., Han X., Zhang Y., Xuan C., Yao Z., Shang Y. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J., 3rd, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J. (2009) Nature 461, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu C., Bian C., Yang W., Galka M., Ouyang H., Chen C., Qiu W., Liu H., Jones A. E., MacKenzie F., Pan P., Li S. S., Wang H., Min J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19266–19271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ji X., Li W., Song J., Wei L., Liu X. S. (2006) Nucleic Acids Res. 34(Web Server issue), W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bailey T. L., Elkan C. (1994) Proc. Int. Conf Intell Syst Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 48. Mori N., Schoenherr C., Vandenbergh D. J., Anderson D. J. (1992) Neuron 9, 45–54 [DOI] [PubMed] [Google Scholar]
- 49. Mortazavi A., Leeper Thompson E. C., Garcia S. T., Myers R. M., Wold B. (2006) Genome Res. 16, 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Joseph R., Orlov Y. L., Huss M., Sun W., Kong S. L., Ukil L., Pan Y. F., Li G., Lim M., Thomsen J. S., Ruan Y., Clarke N. D., Prabhakar S., Cheung E., Liu E. T. (2010) Mol. Syst Biol. 6, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan J., Yang X., Zhuang L., Jiang X., Chen W., Lee P. L., Karuturi R. K., Tan P. B., Liu E. T., Yu Q. (2007) Genes Dev. 21, 1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cao R., Wang H., He J., Erdjument-Bromage H., Tempst P., Zhang Y. (2008) Mol. Cell Biol. 28, 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moazed D. (2011) Cell 146, 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dodd I. B., Micheelsen M. A., Sneppen K., Thon G. (2007) Cell 129, 813–822 [DOI] [PubMed] [Google Scholar]
- 55. Kaufman P. D., Rando O. J. (2010) Curr. Opin. Cell Biol. 22, 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.