Background: IL-36 proteins are IL-1 family members with a key role in the skin.
Results: Truncation of IL-36 ligands and IL-36Ra is required for full activity. IL-36Ra binds IL-1Rrp2 and prevents signaling.
Conclusion: The mechanism of action of IL-36Ra is directly analogous to that of IL-1Ra.
Significance: Protease(s) that activate IL-36 cytokines could be excellent drug targets for psoriasis.
Keywords: Cytokine, Cytokine Action, Immunology, Interleukin, Skin, IL-1, IL-1F, IL-36, IL-36Ra
Abstract
IL-36α, IL-36β, and IL-36γ (formerly IL-1F6, IL-1F8, and IL-1F9) are IL-1 family members that signal through the IL-1 receptor family members IL-1Rrp2 (IL-1RL2) and IL-1RAcP. IL-36Ra (formerly IL-1F5) has been reported to antagonize IL-36γ. However, our previous attempts to demonstrate IL-36Ra antagonism were unsuccessful. Here, we demonstrate that IL-36Ra antagonist activity is dependent upon removal of its N-terminal methionine. IL-36Ra starting at Val-2 is fully active and capable of inhibiting not only IL-36γ but also IL-36α and IL-36β. Val-2 of IL-36Ra lies 9 amino acids N-terminal to an A-X-Asp motif conserved in all IL-1 family members. In further experiments, we show that truncation of IL-36α, IL-36β, and IL-36γ to this same point increased their specific activity by ∼103–104-fold (from EC50 1 μg/ml to EC50 1 ng/ml). Inhibition of truncated IL-36β activity required ∼102–103-fold excess IL-36Ra, similar to the ratio required for IL-1Ra to inhibit IL-1β. Chimeric receptor experiments demonstrated that the extracellular (but not cytoplasmic) domain of IL-1Rrp2 or IL-1R1 is required for inhibition by their respective natural antagonists. IL-36Ra bound to IL-1Rrp2, and pretreatment of IL-1Rrp2-expressing cells with IL-36Ra prevented IL-36β-mediated co-immunoprecipitation of IL-1Rrp2 with IL-1RAcP. Taken together, these results suggest that the mechanism of IL-36Ra antagonism is analogous to that of IL-1Ra, such that IL-36Ra binds to IL-1Rrp2 and prevents IL-1RAcP recruitment and the formation of a functional signaling complex. In addition, truncation of IL-36α, IL-36β, and IL-36γ dramatically enhances their activity, suggesting that post-translational processing is required for full activity.
Introduction
The IL-1 family of cytokines consists of 11 members, including the canonical IL-1 ligands, IL-1α (IL-1F1) and IL-1β (IL-1F2), and their natural receptor antagonist, IL-1Ra (IL-1F3). Other well studied ligands in this family are IL-18 (IL-1F4) and the recently characterized IL-33 (IL-1F11) (1, 2). IL-1 family ligands signal through heterodimeric receptors. IL-1α and IL-1β bind to their primary receptor, IL-1 receptor type 1 (IL-1R1), which allows for recruitment of a second receptor subunit, IL-1R accessory protein (IL-1RAP, IL-1RAcP). Formation of the receptor heterodimer induces signaling, which typically involves the activation of NF-κB and MAPK pathways (3). IL-1Ra functions through binding to IL-1R1, thereby inhibiting binding of IL-1α and IL-1β. However, unlike IL-1α and IL-1β, IL-1Ra binding to IL-1R1 does not allow recruitment of IL-1RAcP and does not generate a functional signaling receptor (4). IL-18 signaling occurs through a similar mechanism whereby IL-18 binds initially to IL-18R, which allows recruitment of a second receptor, IL-18RAP, which leads to signaling and activation of NF-κB and MAPKs (5). IL-18 also has a natural antagonist, IL-18-binding protein (IL-18BP). The mechanism of IL-18BP inhibition is distinct from that of IL-1Ra in that it inhibits IL-18 by binding to the ligand itself and preventing IL-18 binding to its receptor (6).
IL-1F5, IL-1F6, IL-1F8, and IL-1F9 were, for a long time, orphan ligands in the IL-1 family. In 2001, IL-1F9 was shown to activate signal transduction in an IL-1Rrp2 (IL-1RL2)-dependent manner (7), and the authors claimed that IL-1F5 antagonized this response. In 2004, these results were extended by the demonstration that IL-1F6 and IL-1F8 also signal through IL-1Rrp2 and that IL-1RAcP is a required co-receptor for IL-1F6, IL-1F8, and IL-1F9 (8). The receptor requirements for IL-1F6 were confirmed in vivo when IL-1F6 transgenic mice were crossed onto an IL-1Rrp2 or IL-1RAcP null background (9). Transgenic mice expressing IL-1F6 in basal keratinocytes driven by the K14 promoter had a neonatal skin phenotype characterized by acanthosis, hyperkeratosis, a mixed inflammatory cell infiltrate, and elevated cytokine and chemokine expression. The phenotype was most severe at postnatal day 7 and had overtly resolved by postnatal day 21. When K14/IL-1F6 mice were crossed onto the IL1RL2- or IL1RAP-deficient background, the phenotype was eliminated, demonstrating that both receptors are required for IL-1F6 signaling in vivo. IL-1F6 mice on an IL1F5 heterozygous knock-out background had an even more pronounced skin phenotype that did not resolve at weaning, and on a homozygous IL1F5-deficient background, the majority of the IL-1F6 transgenic mice did not survive. These results are consistent with IL-1F5 acting as an antagonist of IL-1F6 in vivo. However, we found that, in vitro, IL-1F5 was not a potent antagonist of IL-1F6 (or IL-1F8 and IL-1F9) (8). IL-1F5, IL-1F6, IL-1F8, and IL-1F9 have been recently renamed as IL-36Ra, IL-36α, IL-36β, and IL-36γ (10) and will be referred to as such throughout the rest of the study.
IL-36Ra, IL-36α, IL-36β, and IL-36γ lack a conventional signal sequence, and although they are presumed to act extracellularly, it is unclear how they are secreted (2). IL-1β and IL-18 have prodomains at their N termini that require cleavage by caspase-1 for activation and secretion. The cellular processes that control cleavage of pro-IL-1β and pro-IL-18 are highly regulated and dependent on activation of the inflammasome (11). IL-36α, IL-36β, and IL-36γ contain no obvious prodomains or caspase cleavage sites, and although they have biological activity as full-length molecules, relatively high concentrations are required to elicit maximal cellular responses (>1 μg/ml IL-36β is required for maximal activity, whereas IL-1β activity is maximal at <1 ng/ml in the same assay) (8).
In this study, we show that IL-36Ra is indeed an antagonist not only of IL-36γ but also of IL-36α and IL-36β and demonstrate that this activity requires removal of the N-terminal methionine from IL-36Ra. More extensive N-terminal truncation of IL-36α, IL-36β, and IL-36γ also leads to a dramatic enhancement in their activity (>1000-fold). As expected from this result, the truncated ligands bind to the IL-1Rrp2/IL-1RAcP receptor heterodimer with greater affinity than the full-length ligands. IL-36Ra inhibits both the full-length and truncated ligands; the ratio of antagonist to agonist required for full inhibition is similar to that of IL-1Ra to IL-1β. We further show that IL-36Ra binds to IL-1Rrp2 and inhibits an IL-36β-mediated association of IL-1Rrp2 with IL-1RAcP in vitro. Therefore, IL-36Ra antagonizes IL-36α, IL-36β, and IL-36γ through a mechanism analogous to IL-1Ra inhibition of IL-1α and IL-1β.
EXPERIMENTAL PROCEDURES
Expression of IL-36 Protein
The following IL-36 constructs were generated, and protein was expressed in Escherichia coli. (a) Untagged human IL-36α, IL-36β, and IL-36γ and mouse IL-36α, IL-36β, and IL-36γ (Met-1–Phe-158, Glu-157, Asp-169, His-160, Lys-156, and Ser-164, respectively) were purified by conventional means. (b) N-terminal truncation series of mouse IL-36α (Lys-3, Glu-4, Lys-5, Glu-6, Leu-7, and Arg-8, all with a methionine added prior to the indicated N-terminal amino acid to enable translation initiation, and Ser-8 and Ala-9, which will not retain the initiating Met) were expressed as fusion proteins containing a C-terminal FLAG-polyhistidine tag (RSGSSDYKDDDDKGSSHHHHHH) and purified from lysates via affinity chromatography over nickel-nitrilotriacetic acid columns (Qiagen). (c) N-terminally truncated human IL-36α (Lys-6–Phe-158), human IL-36β (Arg-5–Glu-157), human IL-36γ (Ser-18–Asp-169), mouse IL-36α (Arg-8–His-160), mouse IL-36β (Ser-4–Lys-156), mouse IL-36γ (Gly-13–Ser-164), and mouse IL-36Ra (Val-2–Asp-155) were expressed in the vector pET SUMO (Invitrogen) and purified according to the manufacturer's instructions. (d) Two forms of human IL-36Ra (Met-1–Asp-155 and Val-2–Asp-155) were expressed containing both a C-terminal FLAG-polyhistidine tag and an N-terminal glutathione S-transferase fusion (separated from the IL-36Ra by a factor Xa cleavage sequence (IEGR)) (EMD Biosciences). The N-terminal tag was removed, and IL-36Ra was purified according to the manufacturer's instructions. In addition, human IL-36Ra (Met-1–Asp-155) with a C-terminal FLAG-polyhistidine tag was expressed in mammalian cells (COS) and purified via affinity chromatography over a nickel-nitrilotriacetic acid column.
Expression of IL-1Rrp2-Fc/IL-1RAcP-Fc Heterodimer
COS cells were cotransfected with the extracellular domains of human IL-1Rrp2 and IL-1RAcP, both fused at their C termini to the Fc domain of human IgG1, and bioactive material in the conditioned medium was purified by affinity chromatography on IL-36α(K6) (IL-36α starting at Lys-6) coupled to agarose, followed by elution at pH 3.0 and immediate neutralization. Some preparations were further purified by chromatography on protein A-Sepharose.
Expression Constructs
Constructs for full-length IL-1R1 and IL-1Rrp2 were described previously (8). The IL-1Rrp2out/IL-1R1in chimera fused Met-1–Tyr-337 of IL-1Rrp2 to Met-338–Gly-569 of IL-1R1. The IL-1R1out/IL-1Rrp2in chimera fused Met-1–Phe-359 of IL-1R1 to Lys-360–Gly-575 of IL-1Rrp2. The IL-8p-luciferase reporter contains 181 bp of the human IL-8 promoter (see supplemental Fig. S4 for a list of primers) in the pGL4.20- or pGL4.22-luciferase vector (Promega).
Reporter Cell Lines and Assays
Human Jurkat T cells (ATCC TIB-152) were stably transfected with the pGL4.20-IL-8p-luciferase reporter construct and a tetracycline-inducible expression vector (Invitrogen T-REx system) containing the human IL1Rrp2 gene to generate the Jurkat T-REx/IL-8p-luciferase reporter cell line. For assay of human IL-36 proteins, IL-1Rrp2 expression was induced with 1 μg/ml doxycycline and 1 mm sodium butyrate for 18–24 h. Cells were then seeded in 96-well tissue culture plates at 2 × 105 cells/well and treated with or without IL-36Ra for 15 min before the addition of agonist ligands for 5 h, after which cells were lysed, and luciferase1 IL-IR P2, activity was measured. Mouse IL-36 proteins were assayed using BaF3 pre-B cells stably expressing both murine (mu)2 IL-1Rrp2 and the IL-8p-luciferase reporter. Cells were seeded in 96-well tissue culture plates at 75 × 103 cells/well and treated with a dose titration of agonist ligands for 5 h before measurement of luciferase activity. Inhibition of IL-1R1, IL-1Rrp2, or chimeric receptors with IL-1Ra or IL-36Ra was assessed by transfecting parental Jurkat cells with the IL-8p-luciferase reporter and either IL-1R1, IL-1Rrp2, or a chimeric receptor construct using Lipofectamine 2000 transfection reagent (Invitrogen). 18–24 h later, washed cells were seeded in 96-well tissue culture plates at 2 × 105 cells/well and treated with or without inhibitors (IL-36Ra or IL-1Ra) for 15 min before the addition of agonist ligands (IL-1β or IL-36α) for 5 h, followed by measurement of luciferase activity.
Biacore Binding Assay
A Biacore T100 optical biosensor was used. Goat anti-human IgG (Jackson ImmunoResearch Laboratories) was immobilized (∼7000 resonance units) on flow cells 1 and 2 of a CM5 sensor chip (GE Healthcare), following which the IL-1Rrp2/IL-1RAcP heterodimer was captured (∼250 resonance units) onto flow cell 2, whereas flow cell 1 was used as a reference. IL-36 proteins (serial 3-fold dilutions) were run over the chip at 25 °C, and the association (4 min) and dissociation (10 min) rates were monitored. Data were fit to a 1:1 binding model (global Rmax) via Scrubber2 software (BioLogic Software Pty. Ltd). Data were fit using a mass transport model. The ligand concentrations used were as follows: IL-36α, 12.3–1000 nm; IL-36α(K6), 2.78–25 nm; IL-36β, 12.3–1000 nm; IL-36β(R5), 0.412–11.1 nm; IL-36γ, 4.12–1000 nm; IL-36γ(S18), 0.309–75 nm; and IL-36Ra(V2), 12.3–333.3 nm.
Co-immunoprecipitation and Western Blotting
BaF3 cells stably overexpressing muIL-1Rrp2 were pretreated with various concentrations of muIL-36Ra or BSA (1 min, room temperature), followed by the addition of 0.1 μg/ml muIL-36β(S4) and incubation at 37 °C for 4 min. Cells were lysed on ice (Clontech phosphoprotein kit buffer A (635626) supplemented with Roche Complete protease inhibitors (11836170001)), and muIL-1RAcP was immunoprecipitated from cleared lysates with anti-muIL-1RAcP antibody M215 (Amgen) (12). Co-immunoprecipitation of muIL-1Rrp2 was detected on Western blots with a goat anti-muIL-1Rrp2 polyclonal antibody (R&D Systems (AF2354)).
FACS Competition Studies
IL-1Rrp2 expression was induced in Jurkat T-REx/IL-8p-luciferase reporter cells (see above). 1 × 106 cells were blocked for 15 min on ice with FACS buffer (PBS containing 3% fetal calf serum, 3% normal goat serum, and 3% normal rabbit serum) alone or containing IL-36α(K6), IL-36Ra, or IL-1Ra (all at 1.5 μm) as competitor. They were then incubated for 90 min on ice in the same buffer containing antibody at 6.7 nm (anti-muIL-1Rrp2 antibody M145, anti-muIL-1RAcP antibody M49, or mouse anti-FLAG control antibody M2 (all from Amgen)), washed, counterstained using a phycoerythrin-conjugated goat anti-mouse IgG F(ab′)2 fragment (Jackson ImmunoResearch Laboratories), and analyzed on a FACScan cell sorter (BD Biosciences).
RESULTS
IL-36Ra(V2) Inhibits IL-36α-, IL-36β-, and IL-36γ-stimulated IL-8 Promoter-driven Luciferase Activity in Jurkat Cells
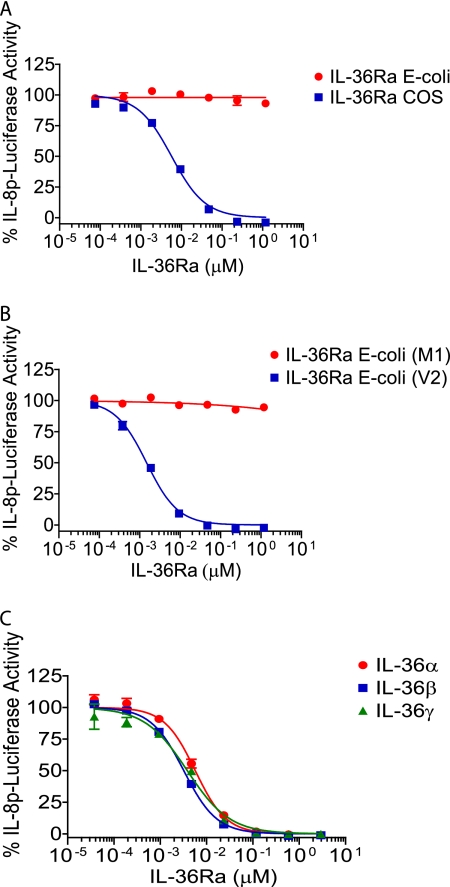
Previously, Debets et al. (7) demonstrated that IL-36Ra antagonizes IL-36γ activity in IL-1Rrp2-transfected Jurkat cells. We later reported that IL-36α, IL-36β, and IL-36γ are active in IL-1Rrp2-transfected Jurkat cells, but we were unable to demonstrate inhibition by IL-36Ra (8). In addition, although all three ligands were active, the agonist doses required both in our own study (8) and in that of Debets et al. (7) were much higher than is typically seen for cytokines. To address the discrepancy between our IL-36Ra in vitro results and those of Debets et al. (7), we characterized several different recombinant IL-36Ra preparations, including those produced in E. coli (as used previously) and in mammalian (COS) cells. IL-36Ra proteins were examined for their ability to inhibit IL-36γ activation of an IL-8 promoter-driven luciferase reporter in Jurkat cells stably expressing IL-1Rrp2. During characterization, a consistent difference in activity was observed between preparations generated in either COS cells or E. coli containing a free N terminus versus those generated in E. coli with a cleavable N-terminal GST domain that was removed post-purification via treatment with factor Xa to yield an N terminus beginning with Met-1. The latter were much less effective as antagonists of IL-36γ-induced signaling (Fig. 1A). To understand the basis for this differential activity, we compared the biochemical properties of two of these preparations, one made in mammalian cells and the other as an N-terminal GST fusion in E. coli. Both preparations behaved as monomers during size exclusion chromatography (estimated mass of 17–18 kDa), and both migrated with an apparent mobility of 18 kDa when reduced and subjected to SDS gel electrophoresis. Mass spectrometry analysis revealed the mass of the factor Xa-cleaved IL-36Ra preparations to be 19,480 Da, consistent with a theoretical mass of 19,486 Da (including the C-terminal FLAG-polyhistidine tag), whereas the mass of the IL-36Ra-FLAG-polyhistidine polypeptides expressed independently of the N-terminal GST tag was observed to be 19,348 Da, a value of 138 Da less than the theoretical value of 19,486 Da. Removal of the N-terminal initiator methionine from the non-GST-tagged IL-36Ra polypeptides by endogenous methionyl aminopeptidases would result in a loss of 131 Da. Edman degradation confirmed that the two IL-36Ra preparations differed with respect to the presence of the N-terminal methionine: the factor Xa-cleaved IL-36Ra construct began with Met-Val-Leu-Ser-Gly-Ala-Leu, whereas the N terminus of the non-N-terminally tagged IL-36Ra construct yielded Val-Leu-Ser-Gly-Ala-Leu. Thus, removal of the N-terminal methionine from IL-36Ra appeared to correlate with biological activity.
FIGURE 1.
Antagonistic activity of IL-36Ra requires removal of its N-terminal methionine. A, Jurkat reporter cells were incubated with a dose titration of huIL-36Ra (starting at 1.18 μm, 1:5 dilutions) expressed in E. coli or COS cells before stimulation with huIL-36γ (used at EC90 ∼ 0.138 μm). B, cells were incubated with a dose titration of huIL-36Ra beginning with Met-1 (M1) or Val-2 (V2) (starting at 1.18 μm, 1:5 dilutions) before stimulation with huIL-36γ (used at EC90 ∼ 0.138 μm). C, cells were incubated with a dose titration of huIL-36Ra(V2) (starting at 2.94 μm, 1:5 dilutions) before stimulation with huIL-36α (used at EC90 ∼ 0.576 μm), huIL-36β (used at EC90 ∼ 0.209 μm), or huIL-36γ (used at EC90 ∼ 0.435 μm). In A–C, all cells were treated with the huIL-36Ra proteins for 15 min before the addition of agonist for 5 h, although in other experiments, we have shown that simultaneous addition of IL-36Ra with the IL-36 agonist is equally effective as preincubation with IL-36Ra prior to agonist addition. IL-8p-luciferase activity was measured and is expressed as a percentage of the luciferase activity in the absence of IL-36Ra (set at 100%). IL-36Ra(V2) IC50 values were similar and ranged from 0.001 to 0.006 μm. Data are means ± S.D. from one experiment (in duplicate or triplicate) that is representative of at least three similar experiments with comparable results.
To confirm that the absence of the N-terminal residue and not the host cell or some other uncharacterized difference was the important variable, IL-36Ra forms starting at either Met-1 (M1) or Val-2 (V2) were generated in E. coli by use of a removable N-terminal GST tag. The V2 version potently inhibited IL-36γ activity, whereas the M1 version was completely inactive (Fig. 1B). Because IL-36α and IL-36β signal through the same heterodimeric receptor complex as IL-36γ, we tested whether IL-36Ra could inhibit IL-36α and IL-36β activity. Jurkat reporter cells were preincubated with truncated IL-36Ra for 15 min, followed by stimulation with IL-36α, IL-36β, and IL-36γ at the EC90 concentration. IL-36Ra inhibited IL-36α and IL-36β activity with comparable potency to inhibition of IL-36γ. Therefore, IL-36Ra is an antagonist of IL-36α, IL-36β, and IL-36γ, and removal of the N-terminal methionine is key to IL-36Ra activity.
Specific Truncation of IL-36α, IL-36β, and IL-36γ Leads to Dramatic Enhancement of Their Potency
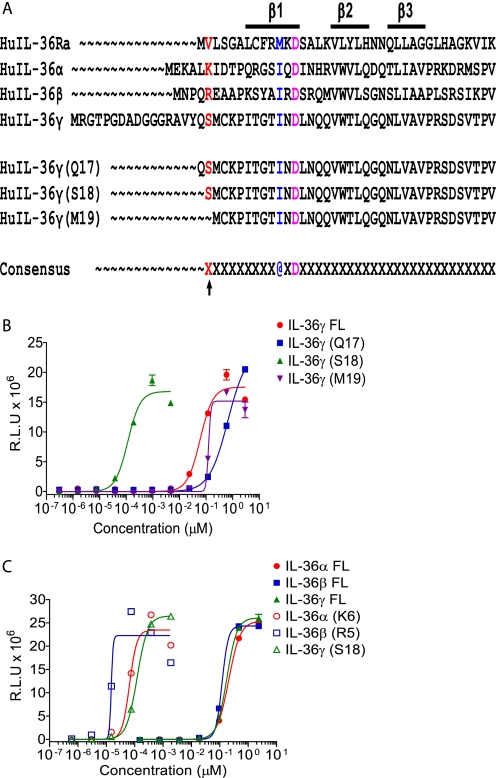
As alluded to above, unusually high doses of the agonist ligands IL-36α, IL-36β, and IL-36γ are required to demonstrate biological activity (7, 8). We wondered whether N-terminal truncation of these molecules would lead to increased specific activity, similar to N-terminal truncation of IL-36Ra. Accordingly, deletion series were constructed for IL-36 proteins in which increasing numbers of amino acids were removed from the N termini and replaced with a single methionine to allow translation initiation. For each IL-36 protein, we found a particular N-terminal truncation that led to substantially increased activity (1000–10,000-fold increase), whereas shorter or longer truncations had activity similar to the full-length protein (Fig. 2 (B and C), Table 1, and supplemental Figs. S1 and S2). Interestingly, the amino acid motif A-X-Asp (where A is an aliphatic amino acid) is conserved in IL-1 family members because it plays a structural role in forming the β-sheet structure, and if this motif is used to anchor the sequence, then the N-terminal truncations leading to dramatically increased specific activity of all IL-36 cytokines (IL-36Ra, IL-36α, IL-36β, and IL-36γ) have their new N terminus 9 residues N-terminal to the aliphatic residue (Fig. 2A). Interestingly, despite being more potent, the magnitude of the response elicited by truncated IL-36α, IL-36β, and IL-36γ was comparable to that induced by full-length cytokines (Fig. 2C and supplemental Fig. S2). Similar truncations in murine IL-36Ra, IL-36α, IL-36β, and IL-36γ also greatly enhanced the activity of these cytokines (supplemental Fig. S3). Preliminary experiments suggest that mouse truncated ligands have very low activity on human cells, whereas human ligands act on mouse cells with an ∼2–3 log decrease in activity compared with human ligands on human cells or mouse ligands on mouse cells.
FIGURE 2.
Truncating IL-36α, IL-36β, and IL-36γ enhances their activity. A, alignment of the protein sequences of human IL-36Ra, IL-36α, IL-36β, and IL-36γ highlighting the A-X-Asp (@XD) motif, the position 9 amino acids N-terminal to it, and the huIL-36γ truncations (positions −8, −9, and −10) generated for activity testing. B, Jurkat reporter cells were stimulated with a dose titration of full-length (FL) huIL-36γ (starting at 2.94 μm, 1:5 dilutions) or huIL-36γ truncated at position −8, −9, or −10 (with respect to the A-X-Asp motif) (huIL-36γ(Q17) starting at 2.94 μm, huIL-36γ(S18) starting at 0.005 μm, and huIL-36γ(M19) starting at 2.94 μm; 1:5 dilutions). C, Jurkat reporter cells were stimulated with titrations of full-length (starting at 2.35 μm) or appropriately truncated (starting at 0.002 μm) huIL-36α, IL-36β, and IL-36γ (1:5 dilutions) for comparison. In B and C, all cells were stimulated with the IL-36 proteins for 5 h before measurement of luciferase activity. Data are expressed as relative light units (R.L.U) and are means ± S.D. from one experiment (in duplicate) that is representative of at least three similar experiments with comparable results.
TABLE 1.
Truncating IL-36 ligands enhances their activity
EC50 values compare the activities of full-length and truncated human IL-36α, IL-36β, and IL-36γ stimulation of Jurkat reporter cells (data are from the same experiment as depicted in Fig. 2C). EC50 values were obtained using a 4-parameter curve fit in GraphPad Prism.
| Ligand | EC50 | Increase in activity |
|---|---|---|
| nm | -fold | |
| IL-36α | 206 | |
| IL-36α(K6) | 0.066 | ∼3000 |
| IL-36β | 120 | |
| IL-36β(R5) | 0.015 | ∼8000 |
| IL-36γ | 177 | |
| IL-36γ(S18) | 0.122 | ∼1500 |
To understand the mechanism of increased activity of the IL-36α, IL-36β, and IL-36γ truncations, we examined their receptor binding affinity. For unknown reasons, we have not been able to express the extracellular domain of the IL-1Rrp2 receptor in a form that is active in binding ligand, but we have managed to generate a small amount of IL-1Rrp2/IL-1RAcP heterodimer that is capable of binding. The IL-1Rrp2/IL-1RAcP heterodimer was immobilized on a Biacore flow cell, and the binding of full-length and truncated ligands was measured. The truncated ligands bound to the receptor heterodimer with 1–35 × 103-fold higher affinity compared with the full-length ligands (Table 2). Similar to the cell-based activity data, full-length IL-36Ra was unable to bind to the receptor, whereas IL-36Ra(V2) bound with an affinity intermediate between the full-length and truncated agonist IL-36 ligands. Thus, it appears that truncation of IL-36α, IL-36β, IL-36γ, and IL-36Ra leads to increased biological activity due to increased affinity of the truncated ligands for the heterodimeric receptor.
TABLE 2.
Truncated IL-36 ligands display enhanced binding to an IL-1Rrp2/IL-1RAcP heterodimer
The binding affinities of full-length and truncated ligands for human IL-36α, IL-36β, and IL-36γ and truncated IL-36Ra(V2) to an IL-1Rrp2/IL-1RAcP heterodimer were determined in a Biacore binding assay. Full-length IL-36Ra did not bind. IL-36Ra(V2) was tested in a separate experiment from the rest.
| Ligand | ka on-rate | kd off-rate | KD | Improvement with truncation |
|---|---|---|---|---|
| m−1s−1 | s−1 | nm | -fold | |
| IL-36α | 2.54 × 103 | 1.94 × 10−3 | 762 | |
| IL-36α(K6) | 8.59 × 105 | 1.83 × 10−5 | 0.021 | ∼36,000 |
| IL-36β | 4.66 × 104 | 4.33 × 10−3 | 92.9 | |
| IL-36β(R5) | 3.52 × 106 | 2.58 × 10−5 | 0.007 | ∼13,000 |
| IL-36γ | 4.67 × 103 | 6.76 × 10−4 | 144 | |
| IL-36γ(S18) | 1.87 × 105 | 2.74 × 10−5 | 0.147 | ∼980 |
| IL-36Ra(V2) | 8.33 × 105 | 8.47 × 10−3 | 10.2 |
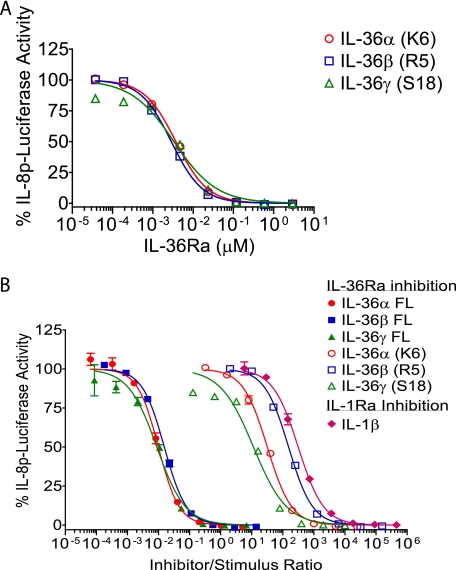
IL-36Ra Inhibits Full-length and Truncated Ligands with Similar Potency, but a Higher Molar Excess of IL-36Ra Is Required for Inhibition of Truncated IL-36 Activity
IL-36Ra(V2) is a potent inhibitor of full-length IL-36α, IL-36β, and IL-36γ (Fig. 1C). When full-length or truncated ligands were tested in the Jurkat assay at their EC90 concentrations, IL-36Ra(V2) inhibited full-length and truncated proteins with comparable IC50 values (between 3 and 6 nm) (Figs. 1C and 3A). However, when the data were plotted as a ratio of inhibitor to stimulus (IL-36Ra to IL-36α, IL-36β, and IL-36γ), it was apparent that a much greater molar excess of IL-36Ra was necessary to inhibit the truncated ligands (Fig. 3B). When IL-1Ra inhibition of IL-1β was examined in a similar assay (Jurkat cells transfected with IL-1R1 and the IL-8p-luciferase reporter), it was clear that the stoichiometry of IL-36Ra inhibition of the truncated ligands was similar to the stoichiometry of IL-1Ra inhibition of IL-1β activity, whereas the molar excess of IL-36Ra inhibition of the full-length ligands was comparable to that reported by Debets et al. (7) with significant inhibition at a 1:1 ratio of IL-36Ra to the full-length ligands.
FIGURE 3.
Large molar excess of IL-36Ra is required to block truncated IL-36 agonist activity. A, Jurkat reporter cells were treated with a dose titration of huIL-36Ra(V2) (starting at 2.94 μm, 1:5 dilutions) for 15 min before the addition of human IL-36α(K6), IL-36β(R5), and IL-36γ(S18) at their EC90 concentrations (IL-36α(K6), ∼0.118 nm; IL-36β(R5), ∼0.019 nm; and IL-36γ(S18), ∼0.294 nm) for 5 h. IL-8p-luciferase activity was measured and is expressed as a percentage of the luciferase activity in the absence of IL-36Ra (set at 100%). IL-36Ra IC50 values were similar for inhibition of IL-36α, IL-36β, and IL-36γ and ranged from 0.003 to 0.006 μm. B, Jurkat cells were transiently transfected with huIL-1R1 and the IL-8p-luciferase reporter. Cells were treated with a dose titration of huIL-1Ra (starting at 0.294 μm, 1:5 dilutions) for 15 min before the addition of huIL-1β at the EC90 concentration (IL-1β, ∼0.647 pm). IL-8p-luciferase activity was measured and is expressed as a percentage of the luciferase activity in the absence of inhibitor. The ratios of IL-1Ra to IL-1β and of IL-36Ra to IL-36 agonist were calculated and are plotted versus the percentage of IL-8p-luciferase activity. The ratios of IL-36Ra(V2) to full-length (FL) and truncated IL-36 agonists were calculated from Fig. 1C and panel A. Data are means ± S.D. from one experiment (in duplicate) that is representative of at least three similar experiments with comparable results.
Mechanism of Action of IL-36Ra Is Similar to That of IL-1Ra
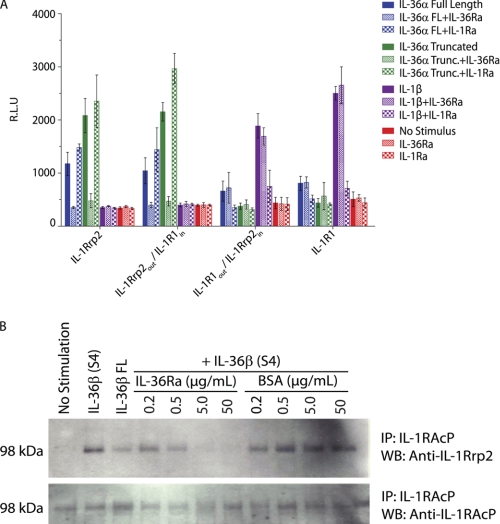
To further determine the mechanism of IL-36Ra antagonism of IL-36α, IL-36β, and IL-36γ, we utilized chimeric receptors containing either the IL-1Rrp2 extracellular domain fused to the IL-1R1 transmembrane and cytoplasmic domains or the IL-1R1 extracellular and transmembrane domains fused to the IL-1Rrp2 cytoplasmic domain as described previously (13). IL-1Ra inhibits IL-1 signaling by binding to the same site on IL-1R1 that is used by IL-1α and IL-1β, thereby preventing the agonist ligands from binding to the receptor. If IL-36Ra acts via a similar mechanism, then antagonism by IL-36Ra should require only the extracellular domain of IL-1Rrp2, and thus, IL-36Ra inhibition should be observed with the chimeric construct containing the IL-1Rrp2 extracellular domain and the IL-1R1 cytoplasmic domain (IL-1Rrp2out/IL-1R1in). To test this hypothesis, we transfected Jurkat cells, which endogenously express IL-1RAcP (but not IL-1R1 or IL-1Rrp2), with full-length IL-1R1, full-length IL-1Rrp2, or the chimeric receptors (IL-1Rrp2out/IL-1R1in or IL-1R1out/IL-1Rrp2 in) and the IL-8p-luciferase reporter. Cells were stimulated with IL-36α (full-length or truncated) or IL-1β in the presence or absence of IL-36Ra or IL-1Ra. IL-36Ra inhibited activation of the IL-8p-luciferase reporter only in cells transfected with full-length IL-1Rrp2 or IL-1Rrp2out/IL-1R1in constructs (Fig. 4A). Likewise, IL-1Ra inhibited cells transfected only with IL-1R1 or IL-1R1out/IL-1Rrp2in constructs, demonstrating that only the extracellular domain of the respective receptors is required for inhibition by IL-36Ra and IL-1Ra. This suggests that IL-36Ra inhibits IL-36α, IL-36β, and IL-36γ through a mechanism similar to that of IL-1Ra inhibition of IL-1α and IL-1β.
FIGURE 4.
IL-36Ra blocks IL-36 ligand binding to the IL-1Rrp2 receptor and the subsequent recruitment of IL-1RAcP. A, Jurkat cells were transfected with the IL-8p-luciferase reporter along with various receptor constructs. Cells were treated with 1.18 μm huIL-1Ra or huIL-36Ra(V2) for 15 min before the addition of full-length (FL) huIL-36α (0.059 μm), truncated (Trunc.) huIL-36α (0.006 μm), or huIL-1β (0.059 nm) for 5 h. Luciferase activity was measured and is expressed as relative light units (R.L.U). Data are means ± S.D. from one experiment (in quadruplicate) that is representative of at least three similar experiments with comparable results. B, murine BaF3 cells stably overexpressing muIL-1Rrp2 were pretreated with 0.2, 0.5, 5, or 50 μg/ml muIL-36Ra(V2) or the BSA control for 1 min before the addition of 0.1 μg/ml muIL-36β(S4) for 4 min at 37 °C. Control samples receiving no muIL-36Ra(V2) or BSA were treated with 50 μg/ml full-length muIL-36β, 0.1 μg/ml muIL-36β(S4), or medium alone. muIL-1RAcP was immunoprecipitated from cell lysates overnight with an anti-muIL-1RAcP antibody. Co-immunoprecipitation of IL-1Rrp2 was detected with an anti-muIL-1Rrp2 antibody by Western blotting. Data are representative of at least three similar experiments with comparable results.
IL-1α or IL-1β binding to IL-1R1 leads to subsequent recruitment of the second subunit, IL-1RAcP, which is necessary for signal transduction. IL-1Ra not only blocks binding of IL-1α or IL-1β to the receptor but also fails to enable IL-1RAcP recruitment. We asked whether IL-36Ra acts similarly. Murine BaF3 B cells transfected with IL-1Rrp2 were stimulated with full-length or truncated muIL-36β(S4) in the presence of IL-36Ra or a control protein, BSA. After stimulation, cell lysates were immunoprecipitated with an anti-IL-1RAcP antibody, and recruitment of IL-1Rrp2 into a heterodimeric receptor complex was monitored by Western blotting of the immunoprecipitate with an anti-IL-1Rrp2 antibody. muIL-36β promoted the association of IL-1Rrp2 with IL-1RAcP as evidenced by the coprecipitation of IL-1Rrp2 with IL-1RAcP (Fig. 4B). Truncated muIL-36β(S4) at 0.1 μg/ml promoted this association more efficiently than full-length muIL-36β at 50 μg/ml. Pretreatment of cells with muIL-36Ra(V2) inhibited muIL-36β(S4)-mediated IL-1Rrp2 association with IL-1RAcP in a dose-dependent manner, whereas the BSA control had no effect (Fig. 4B). Therefore, IL-1Rrp2 and IL-1RAcP associate in an IL-36β-dependent manner, and IL-36Ra prevents this association. This result is consistent with IL-36Ra binding to IL-1Rrp2 and preventing the binding of IL-36α, IL-36β, and IL-36γ and the subsequent recruitment of IL-1RAcP to generate a competent signaling receptor.
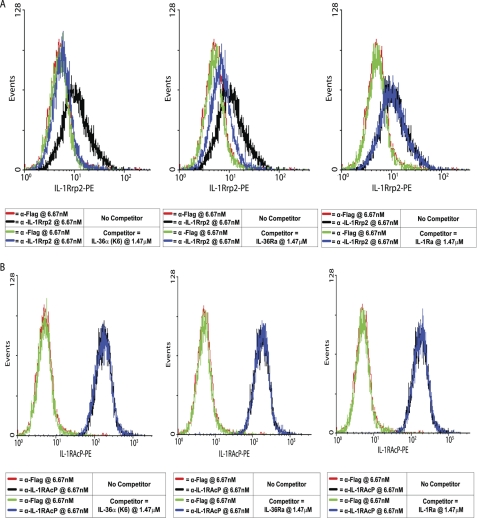
IL-36α and IL-36Ra Bind to IL-1Rrp2 on the Surface of Cells
For unknown reasons, we have been unable to detect binding of IL-36 proteins to IL-1Rrp2-expressing cells via FACS. However, we raised an antibody to human (hu) IL-1Rrp2, M145, which can both detect IL-1Rrp2 cell surface expression and inhibit IL-36α, IL-36β, and IL-36γ activity in vitro (data not shown). Because M145 is a neutralizing antibody, it likely inhibits binding of IL-36α, IL-36β, and IL-36γ to the receptor. We used this antibody to investigate to which receptor subunit IL-36 proteins bind. Jurkat reporter cells were preincubated with or without IL-36α(K6), IL-36Ra(V2), or IL-1Ra as a control for 15 min before M145 antibody staining. An irrelevant anti-FLAG antibody did not stain the Jurkat cells and was used as a negative control. As shown in Fig. 5A, IL-36α(K6) competed with M145 completely at a 220:1 molar ratio. Similar data were obtained for truncated IL-36β and IL-36γ (data not shown). IL-36Ra(V2) also competed with M145 staining, but the inhibition was not complete. IL-1Ra did not compete with M145 binding to IL-1Rrp2. The same experiment was conducted with an anti-IL-1RAcP antibody, M49. IL-36α(K6), IL-36Ra(V2), and IL-1Ra did not compete with the anti-IL-1RAcP antibody (Fig. 5B). These results demonstrate that IL-36α and IL-36Ra bind to IL-1Rrp2 but not to IL-1RAcP. Taken together, the results suggest that IL-36Ra inhibits IL-36α, IL-36β, and IL-36γ activity by binding to the IL-1Rrp2 receptor and preventing IL-36α, IL-36β, and IL-36γ binding, without itself leading to the recruitment of IL-1RAcP to form a functional signaling complex. This is exactly parallel to the mechanism used by IL-1Ra to inhibit IL-1α and IL-1β.
FIGURE 5.
IL-36Ra(V2) and IL-36α(K6) can compete with anti-IL-1Rrp2 antibody binding. A, Jurkat reporter cells were preincubated with or without human IL-36α(K6), IL-36Ra(V2), or IL-1Ra for 15 min before staining with anti-huIL-1Rrp2 antibody or anti-FLAG control antibody. B, Jurkat reporter cells were preincubated with or without human IL-36α(K6), IL-36Ra(V2), or IL-1Ra for 15 min before staining with anti-huIL-1RAcP antibody or anti-FLAG control antibody. Data are representative of at least two similar experiments with comparable results. PE, phycoerythrin.
DISCUSSION
IL-1 family cytokines play critical roles in the function of the innate and adaptive immune system (2). IL-36α, IL-36β, and IL-36γ signal through a shared receptor to activate NF-κB and MAPKs and have important roles in skin pathology. A number of mysteries have surrounded the biology of the IL-36 ligands. For instance, unusually high levels of cytokine have been required in previous studies to elicit a biological response. Moreover, IL-36Ra was claimed to antagonize IL-36γ (7), but we were unable to replicate these results (8). We have shown here that IL-36Ra is indeed an antagonist of not only IL-36γ but also IL-36α and IL-36β. Antagonist activity requires removal of the N-terminal methionine present in the primary translation product. The mechanism of antagonism by IL-36Ra is exactly analogous to that used by IL-1Ra to inhibit IL-1. Finally, we have demonstrated that like IL-36Ra, IL-36α, IL-36β, and IL-36γ have dramatically enhanced activity when N-terminally truncated to a specific position.
Most likely, our earlier failure to reproduce claims of IL-36Ra antagonism was due to our use of an N-terminal tag to facilitate purification, followed by cleavage in vitro to leave the intact full-length protein. Methionyl aminopeptidases in cells normally cleave the initiating methionine when it is followed by a small amino acid residue (14) such as the valine at position 2 in IL-36Ra. We have shown that IL-36Ra(V2) is a potent antagonist of not only IL-36γ, as demonstrated earlier (7), but also IL-36α and IL-36β. We used chimeric receptors to demonstrate that the extracellular domain of IL-1Rrp2 is necessary and sufficient for IL-36Ra inhibition of IL-36β. We further demonstrated that IL-1Rrp2 and IL-1RAcP associate in a ligand-dependent manner and that IL-36Ra is able to prevent this association. Finally, we demonstrated using FACS that both IL-36Ra and the agonist ligand IL-36α bind to the IL-1Rrp2 subunit of the receptor rather than to IL-1RAcP. All of these results lead to the conclusion that IL-36Ra inhibits IL-36α, IL-36β, and IL-36γ action in a manner parallel to that used by IL-1Ra to inhibit IL-1α and IL-1β activity.
Having found that removal of the N-terminal methionine is key to IL-36Ra activity, we wondered whether N-terminal truncation of the IL-36 agonist ligands could also lead to enhanced specific activity. Full-length IL-36α, IL-36β, and IL-36γ had been shown to induce NF-κB and MAPKs as well as IL-6 and IL-8 secretion; however, high concentrations (EC50 values in the μm range) were required for these activities (8). Because other IL-1 family members such as IL-1α, IL-1β, and IL-18 have EC50 values in the low nm range, it seemed that the activity of IL-36α, IL-36β, and IL-36γ was not optimal. Indeed, truncation of IL-36α, IL-36β, and IL-36γ to a specific position 9 amino acids N-terminal to a conserved A-X-Asp motif resulted in a dramatic enhancement of their activity, resulting in EC50 values in the low nm range. In addition, IL-36Ra inhibition of the truncated ligands looks much more similar to IL-1Ra inhibition of IL-1β in that a substantial molar excess is required for inhibition.
One remaining unanswered question concerns the mechanism of secretion of the IL-36 ligands. Similar to IL-1α, IL-1β, and IL-18, IL-36α, IL-36β, and IL-36γ are translated from mRNAs that do not encode a signal sequence (15, 16). The cellular processes that control post-translational processing and the export of IL-1β and IL-18 are complex and involve activation of the inflammasome and cleavage of the full-length molecule by caspase-1 (11, 17). It is unclear whether truncation of IL-36 is coupled to release of the cytokines, but it is unlikely that IL-36 processing occurs via caspase-1, as the amino acid sequences surrounding the truncation sites do not resemble a caspase-1 site. Moreover, no evidence of IL-36α processing was observed in transfected mouse macrophages under conditions in which caspase-1 activation of pro-IL-1β occurred (18). The protease responsible for cleaving IL-36α, IL-36β, and IL-36γ is unknown. It is also unclear whether a single protease is responsible for cleavage of all three cytokines. The sequences of the three ligands surrounding the truncation sites bear little similarity to one another, and therefore, it is possible that distinct proteases are responsible for cleaving the individual ligands.
Transgenic overexpression of IL-36α in mouse epidermis results in an inflammatory skin condition at birth that resolves by 2–3 weeks of age (9). The skin of young adult IL-36α transgenic mice is histologically normal, but application of phorbol ester rapidly elicits a pronounced inflammatory state that strongly resembles human plaque psoriasis in its histological and molecular characteristics and responsiveness to therapeutic agents (19). It is unclear why the skin phenotype resolves in transgenic mice at weaning or why the adult transgenic mice are hyper-responsive to irritants, but it does not appear to be simply due to IL-36α or IL-36Ra expression levels, as there are only modest changes in expression in either condition that are unlikely to account for such a dramatic change in phenotype. It is possible that the key change is in the expression or activation of the protease required for truncation of IL-36α into its active form, which could be regulated temporally and/or by phorbol ester. The antagonistic role of IL-36Ra in vivo is clearly demonstrated by the successively more severe inflammation seen when the IL-36α transgene is crossed onto IL-36Ra+/− and IL-36Ra−/− backgrounds (9). IL-36 cytokines also appear to be important in human psoriasis, as they are strongly elevated in psoriatic lesional tissue, and the phenotype of human psoriasis lesional skin transplanted onto immunodeficient mice is normalized when the mice are treated with an anti-huIL-1Rrp2 antibody (9, 19). In addition, a rare life-threatening form of psoriasis, generalized pustular psoriasis, has recently been shown to be caused by mutations in IL-36Ra (20, 21). Together, these data suggest that IL-36α, IL-36β, and IL-36γ play a significant role in human psoriasis and that the protease(s) involved in activation of these cytokines would be excellent drug targets.
Acknowledgments
We thank Dirk Smith, G. Duke Virca, Unja Martin, and Hal Blumberg for useful discussions; Mike Brown and Tim Vanden Bos for Biacore work; and Dave Meininger, Ai Ching Lim, Rob Kegel, Shanon Turnbaugh, and Kurt Edelman for reagent generation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- mu
- murine
- hu
- human.
REFERENCES
- 1. Dunn E., Sims J. E., Nicklin M. J., O'Neill L. A. (2001) Trends Immunol. 22, 533–536 [DOI] [PubMed] [Google Scholar]
- 2. Sims J. E., Smith D. E. (2010) Nat. Rev. Immunol. 10, 89–102 [DOI] [PubMed] [Google Scholar]
- 3. O'Neill L. A. (2008) Immunol. Rev. 226, 10–18 [DOI] [PubMed] [Google Scholar]
- 4. Greenfeder S. A., Nunes P., Kwee L., Labow M., Chizzonite R. A., Ju G. (1995) J. Biol. Chem. 270, 13757–13765 [DOI] [PubMed] [Google Scholar]
- 5. Born T. L., Thomassen E., Bird T. A., Sims J. E. (1998) J. Biol. Chem. 273, 29445–29450 [DOI] [PubMed] [Google Scholar]
- 6. Novick D., Kim S. H., Fantuzzi G., Reznikov L. L., Dinarello C. A., Rubinstein M. (1999) Immunity 10, 127–136 [DOI] [PubMed] [Google Scholar]
- 7. Debets R., Timans J. C., Homey B., Zurawski S., Sana T. R., Lo S., Wagner J., Edwards G., Clifford T., Menon S., Bazan J. F., Kastelein R. A. (2001) J. Immunol. 167, 1440–1446 [DOI] [PubMed] [Google Scholar]
- 8. Towne J. E., Garka K. E., Renshaw B. R., Virca G. D., Sims J. E. (2004) J. Biol. Chem. 279, 13677–13688 [DOI] [PubMed] [Google Scholar]
- 9. Blumberg H., Dinh H., Trueblood E. S., Pretorius J., Kugler D., Weng N., Kanaly S. T., Towne J. E., Willis C. R., Kuechle M. K., Sims J. E., Peschon J. J. (2007) J. Exp. Med. 204, 2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dinarello C., Arend W., Sims J., Smith D., Blumberg H., O'Neill L., Goldbach-Mansky R., Pizarro T., Hoffman H., Bufler P., Nold M., Ghezzi P., Mantovani A., Garlanda C., Boraschi D., Rubartelli A., Netea M., van der Meer J., Joosten L., Mandrup-Poulsen T., Donath M., Lewis E., Pfeilschifter J., Martin M., Kracht M., Muehl H., Novick D., Lukic M., Conti B., Solinger A., Kelk P., Peyman K., van de Veerdonk F., Gabel C. (2010) Nat. Immunol. 11, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martinon F., Mayor A., Tschopp J. (2009) Annu. Rev. Immunol. 27, 229–265 [DOI] [PubMed] [Google Scholar]
- 12. Smith D. E., Hanna R., Della Friend Moore H., Chen H., Farese A. M., MacVittie T. J., Virca G. D., Sims J. E. (2003) Immunity 18, 87–96 [DOI] [PubMed] [Google Scholar]
- 13. Born T. L., Smith D. E., Garka K. E., Renshaw B. R., Bertles J. S., Sims J. E. (2000) J. Biol. Chem. 275, 29946–29954 [DOI] [PubMed] [Google Scholar]
- 14. Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar S., McDonnell P. C., Lehr R., Tierney L., Tzimas M. N., Griswold D. E., Capper E. A., Tal-Singer R., Wells G. I., Doyle M. L., Young P. R. (2000) J. Biol. Chem. 275, 10308–10314 [DOI] [PubMed] [Google Scholar]
- 16. Smith D. E., Renshaw B. R., Ketchem R. R., Kubin M., Garka K. E., Sims J. E. (2000) J. Biol. Chem. 275, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 17. Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 18. Martin U., Scholler J., Gurgel J., Renshaw B., Sims J. E., Gabel C. A. (2009) J. Immunol. 183, 4021–4030 [DOI] [PubMed] [Google Scholar]
- 19. Blumberg H., Dinh H., Dean C., Jr., Trueblood E. S., Bailey K., Shows D., Bhagavathula N., Aslam M. N., Varani J., Towne J. E., Sims J. E. (2010) J. Immunol. 185, 4354–4362 [DOI] [PubMed] [Google Scholar]
- 20. Onoufriadis A., Simpson M. A., Pink A. E., Di Meglio P., Smith C. H., Pullabhatla V., Knight J., Spain S. L., Nestle F. O., Burden A. D., Capon F., Trembath R. C., Barker J. N. (2011) Am. J. Hum. Genet. 89, 432–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marrakchi S., Guigue P., Renshaw B. R., Puel A., Pei X. Y., Fraitag S., Zribi J., Bal E., Cluzeau C., Chrabieh M., Towne J. E., Douangpanya J., Pons C., Mansour S., Serre V., Makni H., Mahfoudh N., Fakhfakh F., Bodemer C., Feingold J., Hadj-Rabia S., Favre M., Genin E., Sahbatou M., Munnich A., Casanova J. L., Sims J. E., Turki H., Bachelez H., Smahi A. (2011) N. Engl. J. Med. 365, 620–628 [DOI] [PubMed] [Google Scholar]