Abstract
Many secreted proteins are synthesized as precursors with propeptides that must be cleaved to yield the mature functional form of the molecule. In addition, various growth factors occur in extracellular latent complexes with protein antagonists and are activated upon cleavage of such antagonists. Research in the separate fields of embryonic patterning and extracellular matrix formation has identified members of the BMP1/Tolloid-like family of metalloproteinases as key players in these types of biosynthetic processing events in species ranging from Drosophila to humans.
Keywords: Bone Morphogenetic Protein (BMP), Collagen, Development, Extracellular Matrix Proteins, Insulin-like Growth Factor (IGF), Metalloprotease, Transforming Growth Factor β (TGFβ), BMP1, Tolloid, Astacin-like Proteinases
Introduction
Bone morphogenic proteins (BMPs)2 were first defined by the ability to induce de novo bone formation and were first identified in bone extracts (1). Although all other BMPs are members of the TGFβ superfamily of growth factors, BMP1 is a metalloproteinase, the first demonstrated role of which was as a procollagen C-proteinase (pCP) (2) that cleaves C-propeptides from procollagen precursors to produce mature monomers of the major fibrillar collagens I–III. This activity is crucial to bone biology, as collagen I is the major protein component of bone and is essential to bone structure/function. After initial cloning of mammalian BMP1, Tolloid (TLD), the protein product of a zygotically active gene involved in dorsoventral patterning of Drosophila embryos, was shown to have a domain structure resembling that of BMP1 (3) and was later shown to exert patterning effects by activating the TGFβ-like BMP decapentaplegic (DPP) (4). Subsequently, BMP1 and TLD have become prototypes of the BMP1/TLD-like proteinase (B/TP) family. B/TPs in a broad range of species are now implicated in processes that include extracellular matrix (ECM) formation and mineralization, embryonic patterning, growth factor activation, and generation of anti-angiogenic protein fragments, all via cleavage of a growing list of substrates.
B/TP Structure
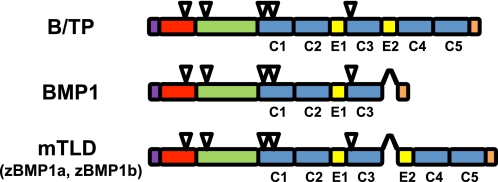
There are four mammalian B/TPs: BMP1, mammalian TLD (mTLD), and mammalian TLD-like 1 (mTLL1) and 2 (mTLL2). mTLD is a longer splice variant encoded by the same gene that encodes BMP1 (5), whereas TLL1 and TLL2 are genetically distinct. BMP1, mTLD, mTLL1, and mTLL2 homologs have been identified in Xenopus, whereas Drosophila has two B/TP family members distinct from those found in vertebrates (6). B/TPs share a similar protein domain structure (Fig. 1) comprising an N-terminal prodomain, followed by a conserved protease domain characteristic of the astacin M12A family of the metzincin subclan of metalloproteinases (7, 8) and then by non-catalytic EGF (EGF-like) and CUB motifs, the latter named after the first three proteins in which it was described (complement/Uef/BMP1) and thought to be involved in protein-protein interactions (9).
FIGURE 1.
B/TP protein domain structures. The upper schematic labeled B/TP represents the general protein domain structure shared by the majority of B/TPs across species. In the middle and lower schematics, arrowheads denote alternative splicing that produces BMP1 and mTLD variants form the same gene. Purple, red, green, blue, yellow, and pink boxes represent signal peptides, prodomains, metalloprotease, and CUB and EGF domains and domains unique to each protein, respectively. Triangles denote the sites of potential Asn-linked glycosylation, conserved in B/TPs across a broad range of species. CUB domains 1–5, and EGF domains 1 and 2 are labeled C1–5 and E1 and E2, respectively. z, zebrafish.
The prodomain confers latency and is cleaved in the Golgi of some cell types by subtilisin-like proprotein convertases (SPCs) (10), whereas in other cell types, B/TPs are secreted with prodomains intact (11, 12), suggesting extracellular activation. In Drosophila embryos, most TLD occurs as a prodomain-retaining form, suggesting an activation limited by either inefficient or regulated processing (4). BMP1/mTLD prodomain sequences, which co-purify with TGFβ-like BMPs from osteoinductive bone extracts (1), can bind BMP2 and BMP4 with high affinity and may participate in regulating their activity in vivo (12).
Crystal structure analysis indicates that the BMP1 protease domain, as in the prototypical protease astacin, has a deep active site cleft, within which three conserved histidines bind the catalytic zinc, but it differs from the astacin protease domain in that a conserved tyrosine does not participate in zinc binding (13). The specificity of B/TP active sites differs from that of the prototypic protease astacin but is similar to that of other astacin family members in having a strong preference for aspartate in the P1′ position of substrate cleavage sites (6, 14). Crystal structure analysis has identified a basic arginine in the S1′ pocket of BMP1, consistent with this preference for P1′ aspartates, whereas a bulky vicinal disulfide may contribute to a restricted S1 pocket, helping to explain a preference of B/TPs for small aliphatic resides in substrate P1 positions (6, 13). Only five cleavage sites of known B/TP substrates lack P1′ aspartates, and these all have glutamines in the P2 position (15), although the significance of this observation remains to be determined.
C-terminal to the protease domain are the CUB and EGF domains. A subset of CUB domains appears to require Ca2+ for optimum binding activity (16). The most N-terminal BMP1 CUB domain (C1) may play a role in imparting “chordinase” activity, or ability to cleave chordin (17), a substrate described below. EGF domains bind Ca2+ and may confer structural rigidity to portions of B/TPs (18). BMP1, the most proteolytically active vertebrate B/TP against a number of substrates, has the fewest C-terminal non-catalytic domains, and deletion of EGF domains from mTLD enhances its pCP activity and imparts an otherwise absent chordinase activity (19). Evidence suggests that the reduced proteolytic activity of mTLD, relative to BMP1, involves Ca2+-dependent homodimerization via its extra CUB and EGF domains, in particular the more C-terminal EGF domain, E2, leading to decreased proteolytic activity by partial occlusion of the active site by the more C-terminal CUB domains, C4 and C5 (20). A similar mechanism seems to apply for mTLL1 (21). B/TPs have a number of Asn-linked glycosylation sites, many of which are conserved among family members (Fig. 1). Glycosylation at such sites can affect BMP1 secretion, thermostability, and pCP activity (22).
B/TP Distributions and General Functions
All four mammalian B/TPs are expressed in mouse gastrulas, consistent with roles in dorsoventral patterning, whereas in later development, BMP1, mTLD, and mTLL1 are expressed at relatively high levels in areas of bone formation, consistent with roles in this process, and mTLL2 expression localizes to skeletal muscle (23). mTLL2 appears to serve a non-redundant function in muscle, as mTLL2-null mice have a small reduction in muscle mass (24). Xenopus BMP1, mTLL2, and mTLL1 homologs are designated BMP1, Xolloid, and Xolloid-related, respectively. BMP1 and Xolloid are expressed ubiquitously in Xenopus early embryos, whereas Xolloid-related is up-regulated in ventral regions by BMP signaling (25). In Drosophila embryos, which have inverted dorsoventral axes compared with vertebrates, TLD is localized dorsally (4). Studies in Xenopus and Drosophila were the first to demonstrate B/TP roles in embryonic dorsoventral patterning (4, 26). Expression domains of a second Drosophila B/TP, TLD-related (TLR; also known as Tolkin), partially overlap those of TLD in embryos, but TLR functional importance appears to lie mainly in larvae, in which TLD is not expressed (27–29). Mammalian B/TP expression is at relatively high levels in the developing and adult central nervous systems (5, 30–32), suggesting roles in development and homeostasis of this tissue. Expression levels of mTLL1, in particular, are high in the adult central nervous system in areas of high synaptic plasticity (31, 32). Relatedly, B/TPs appear to play roles in synaptic plasticity in the mollusc Aplysia (33), whereas TLR participates in axon guidance in Drosophila (27, 29). Mice null for the Bmp1 gene, which encodes BMP1 and mTLD, are perinatal lethal, with failure of ventral body wall closure and persistent gut herniation, likely due to defective ECM and limited disruption of dorsoventral patterning (34). Consistent with a loss of pCP activity, Bmp1-null mice have abnormal collagen fibrils. Surprisingly, however, given the importance of collagen I to bone, gross skeletal abnormalities are not observed, perhaps due to functional redundancy with mTLL1 in this tissue (34). In mice, mTLL1 expression is limited to the cardiovascular system until 10 days post-conception (dpc) and, at early times, co-localizes in the heart field with the cardiac-specific transcription factor Nkx-2.5 (30), which may participate in driving mTLL1 expression in this tissue (35). After 10 dpc, mTLL1 is more broadly expressed, but mTLL1-null mice die at ∼13.5 dpc from deficits apparently confined to the cardiovascular system (30). In humans, non-synonymous polymorphisms have been reported in mTLL1 coding sequences in several patients with atrial septal defects (36), although the significance of these findings remains to be determined.
Recently, mTLD was reported to as a component of human plasma (37), although with a size of 95 kDa, smaller than the 130 kDa previously reported for mTLD (11, 23). Thus, mTLD or an mTLD derivative may exert systemic effects, in addition to localized effects exerted by mTLD produced in individual tissues. Circulating mTLD appears to be involved in bone fracture repair and in the fibrosis associated with chronic kidney disease (37, 39). Thus, inhibition or provision of circulating mTLD may represent future approaches to therapeutic interventions in these and related conditions.
Roles in ECM Formation
B/TPs appear to play important roles in regulating ECM deposition by proteolytic trimming of precursors of various ECM-related proteins, including collagens, small leucine-rich proteoglycans (SLRPs), small integrin-binding ligand N-linked glycoproteins (SIBLINGs), lysyl oxidase (LOX), and basement membrane components perlecan and laminin-332.
Collagens
The major fibrillar collagens I–III are synthesized as procollagens with N- and C-terminal peptides that must be removed to produce mature triple helical monomers capable of forming fibrils (40). The C-propeptides are cleaved by B/TPs (2, 41) intracellularly or extracellularly in a tissue- and developmental stage-specific manner (42). Prior to secretion, procollagens form intracellular aggregates (42), which may be processed more efficiently by B/TPs than are single procollagen molecules (43). It has been reported that retained C-propeptides preclude monomer incorporation into fibrils in vitro and in cultures of normal fibrogenic cells (44, 45), although collagen monomers with uncleaved C-propeptides do appear to be incorporated into fibrils of cells and tissues of embryonic lethal mice doubly null for the genes that together encode BMP1, mTLD, and mTLL1 (45, 46). Mutations at collagen I C-propeptide cleavage sites that result in partial impairment of cleavage by B/TPs result in mild cases of the brittle bone disease osteogenesis imperfecta (47), although it is likely that complete inability to cleave major fibrillar collagen C-propeptides is incompatible with viability. Recently, it was shown that meprins, astacin family members related to the B/TPs, can cleave both N- and C-propeptides of procollagen III in vitro and that the C-propeptide cleavage site is the same as that used by B/TPs (48). However, it is unknown whether such cleavages by meprins occur in vivo, and the physiological relevance of such findings remains to be determined.
Minor fibrillar collagens V and XI are incorporated into and are thought to regulate the geometries of fibrils of the more abundant collagens I and II, respectively (49, 50). The pro-α2(V) chain of collagen V and the major fibrillar collagen I–III procollagen chains share an identical protein domain structure and constitute the clade A procollagen chains, whereas the pro-α1(V) and pro-α3(V) chains of collagen V and the pro-α1(XI) and pro-α2(XI) chains of collagen XI constitute the clade B procollagen chains (51). An obvious difference between clade A and B procollagens is in the configuration of their N-terminal globular regions (Fig. 2). As with major fibrillar procollagens, B/TPs cleave the pro-α2(V) C-propeptide (52). In contrast, clade B procollagen C-propeptides are cleaved by SPCs, whereas B/TPs cleave within the large N-terminal globular domains of the pro-α1(V), pro-α1(XI), and pro-α2(XI) chains (45, 52). Pro-α3(V) N-terminal sequences can be cleaved by B/TPs in vitro and in cell culture systems (53), although such processing may not occur in at least some tissues in vivo (54). Proteolytic trimming defines the portions of clade B N-terminal globular regions that project beyond fibril surfaces and that may participate in regulating fibril geometry (55).
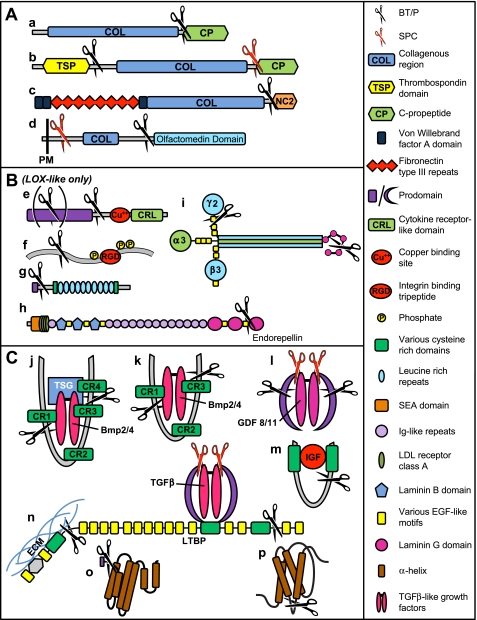
FIGURE 2.
B/TP substrates. Schematics are shown for known B/TP substrates. A, collagens: a, clade A fibrillar procollagens; b, clade B fibrillar procollagens; c, procollagen VII; d, gliomedin. B, non-collagenous ECM-related proteins: e, pro-LOX and pro-LOX-like (B/TPs cleave the former once and the latter at two sites); f, SIBLING proteins (cleaved by B/TPs into N- and C-terminal fragments, the latter more highly phosphorylated and containing integrin-binding RGD sequences); g, SLRPs; h, perlecan; i, laminin-332 (cleaved in both the α3 and γ2 chains by B/TPs). C, non-ECM-related substrates: j, chordin and SOG (each cleaved by B/TPs at two corresponding locations, with SOG cleaved at a third additional site (not shown) just N-terminal to CR2; TSG binds BMP2/4-chordin and DPP-SOG complexes, thereby modulating chordin/SOG cleavage by B/TPs); k, CHL1 and Chl (each has three cysteine-rich (CR) domains with homology to those of chordin/SOG); l, GDF8/11; m, IGFBP3; n, LTBP (cleaved by B/TPs to release the TGFβ “large latent complex” from the ECM); o, pro-ApoA1; p, prolactin/growth hormone.
The C-terminal globular NC2 domain of non-fibrillar collagen VII is cleaved upon formation of collagen VII antiparallel dimers that self-associate to form anchoring fibrils, important in securing the epidermis to the underlying stroma, and B/TPs can cleave the NC2 domain in vitro (56). Moreover, mutations that eliminate this cleavage site result in procollagen VII accumulation at the dermal-epidermal junction in vivo and in the blistering skin disease dystrophic epidermolysis bullosa (56). Although procollagen VII processing appears undiminished in Bmp1-null mice, this may result from functional compensation by mTLL1 and/or mTLL2 (57).
Gliomedin, a transmembrane collagen important in formation of the nodes of Ranvier, is shed from Schwann cell surfaces by SPCs but can be further processed by B/TPs (58). The latter cleavage results in free olfactomedin domains with enhanced ability to aggregate, which may help stabilize nodes of Ranvier (58).
Non-collagenous ECM-related Proteins
LOX and LOX-like are extracellular enzymes necessary for the formation of covalent cross-links that provide collagen and elastic fibers with much of their tensile strength. Both are secreted as zymogens that are activated by B/TPs via cleavage of prodomains (59, 60). Dentin matrix protein 1 (DMP1) and DSPP (dentin sialophosphoprotein), SIBLING family members, are highly acidic proteins that can be cleaved by B/TPs to produce fragments involved in initiating mineralization of hard tissues (61, 62). Observations that DMP1-processing activity is decreased in cells null for BMP1, mTLD, and mTLL1 (62) and that expression of both BMP1 and DMP1 increases coincident with mineralization (63) are supportive of the physiological relevance for B/TP cleavage of DMP1. Osteoglycin, which is believed to regulate collagen fibril diameters, and biglycan and decorin, which appear to play roles in regulating both collagen fibrillogenesis and TGFβ signaling, are SLRPs that are synthesized as precursors and cleaved by B/TPs to mature forms (64–66). B/TPs also process basement membrane proteins laminin-332 (also known as laminin-5), in which the γ2 and α3 chains are trimmed, and perlecan, a proteoglycan (67, 68). B/TP cleavage of perlecan liberates the anti-angiogenic fragment endorepellin (67). However, peptides that inhibit mTLD in vitro may reduce angiogenesis in some systems (69). Thus, B/TPs may balance regulation of angiogenesis by assisting in blood vessel growth while releasing anti-angiogenic factors to prevent excessive angiogenesis.
Non-ECM-related Substrates: Growth Factors
In addition to direct roles in ECM formation, B/TPs affect development and homeostasis via effects on various non-ECM proteins, including a subset of growth factors such as BMP2 and BMP4. Aside from roles as osteoinductive factors (1), BMP2 and BMP4 generate signaling gradients that are major determinants of dorsoventral patterning in vertebrate embryogenesis, a mechanism conserved in Drosophila by signaling gradients of the BMP2/4 homolog DPP (70). BMP2 and BMP4 are bound and inhibited by the extracellular antagonist chordin, whereas DPP is bound and inhibited by the Drosophila chordin homolog short gastrulation (SOG). In vertebrates, B/TPs provide chordinase activity that cleaves chordin, thus freeing BMP2/4 to bind cell surface receptors, whereas SOG cleavage by TLD serves a similar purpose in Drosophila (4, 26, 45). A difference is that SOG is efficiently cleaved only when bound to DPP, whereas chordin cleavage is BMP2/4-independent (4, 26). Dependence of SOG cleavage on DPP as a co-substrate apparently bolsters a long-range DPP diffusion function of SOG, thus contributing to a steeper and more stable DPP signaling gradient (71). In vertebrates and Drosophila, the protein twisted gastrulation (TSG) binds BMP2/4-chordin or DPP-SOG complexes, thus modulating chordin/SOG cleavage by B/TPs (72). The mammalian protein chordin-like 1 (CHL1) and the zebrafish protein chordin-like (Chl), with similarities of protein domain structure to chordin, bind and inhibit BMP2/4 and are both cleaved by B/TPs (73). Interestingly, the BMP1/mTLD prodomain, which co-purifies from bone extracts with BMP2/4 (1), can avidly bind BMP2/4 and thus inhibit signaling (12). Although BMP1 prodomain-BMP4 complexes are found in tissues (12), the roles that such interactions may play in vivo are yet to be determined.
TGFβ1–3, which play important roles in regulating cell behaviors, are synthesized as inactive proproteins. Upon cleavage by SPCs, TGFβ1–3 remain noncovalently bound to their prodomains as latent complexes. Most often, these complexes are covalently linked via their prodomains to latent TGFβ-binding proteins (LTBPs), which tether them to the ECM (74). B/TPs cleave LTBP1, thus releasing from the ECM a processed form of the complex with increased susceptibility to further activation by matrix metalloproteinases (75). However, this method of TGFβ1–3 activation is one of several reported mechanisms for TGFβ1–3 activation and may thus be limited to a subset of physiological circumstances. TGFβ-related growth and differentiation factors (GDFs) 8 and 11, negative regulators of skeletal muscle growth and neurogenesis, respectively, also form noncovalent latent complexes with their SPC-cleaved prodomains, and in both cases, these latent complexes are activated by B/TP cleavage of prodomains (76, 77). Similarly, in Drosophila, TLD and TLR can cleave prodomains of TGFβ-like factors activin, dawdle, and myoglianin (27), the latter a homolog of mammalian GDF8. TLR cleavage of dawdle appears to play a role in axon guidance and fasciculation (27).
Insulin-like growth factors (IGFs), which have important roles in development and metabolism, are bound by IGF-binding proteins (IGFBPs), which modulate IGF activity. B/TPs can cleave IGFBP3, one of six mammalian IGFBPs, in vitro and are responsible for most IGFBP3 processing in mouse embryo fibroblasts (15). This processing appears to lower the ability of IGFBP3 to block IGF cell signaling while enhancing some IGF-independent IGFBP3 effects on cells (15).
Additional Non-ECM-related Substrates
When secreted by endothelia, prolactin and growth hormone have angiogenic effects, whereas naturally occurring N-terminal cleavage fragments of the same hormones are anti-angiogenic. B/TPs can cleave prolactin and growth hormone in vitro and in cell culture, creating N-terminal fragments similar in size to those found in vivo and with similar anti-angiogenic effects (78). Thus, as with perlecan (see above), B/TPs can generate anti-angiogenic fragments, in this case via cleavage of pro-angiogenic hormones. Consistent with possible B/TP roles in angiogenesis is the finding that mTLD mRNA is among the transcripts most strongly induced by transition of resting endothelia to the activated endothelia associated with tumors (79).
ApoA1, the major protein component of HDL, is secreted as a proprotein unable to bind lipids. BMP1-neutralizing antibodies or siRNA blocks pro-ApoA1 propeptide cleavage, whereas recombinant BMP1 can cleave the propeptide (80). Also, the physiological pro-ApoA1 cleavage site resembles those found in known B/TP substrates. Thus, B/TPs may be responsible for cleaving pro-ApoA1, perhaps enhancing ApoA1 conversion to a conformation able to bind phospholipids (80).
B/TP Regulators
A growing number of protein regulators of B/TP activities have been reported that, due to their modulation of B/TP activities, may play similarly important roles in morphologic and homeostatic events.
pCP Enhancers
pCP enhancers 1 and 2 (PCPE1 and PCPE2; also known as PCOLCE1 and PCOLCE2), proteins that can markedly enhance B/TP pCP activity, each consist of two N-terminal CUB domains and a C-terminal netrin-like (NTR) domain (81, 82). The CUB domains of PCPE1 bind procollagen (82) in a cooperative manner (83), and its NTR domain can bind BMP1 and mTLL1 (84, 85), suggesting that PCPE1 may act as a linker that enhances procollagen-B/TP interactions. Moreover, enhancement of pCP activity by PCPE1 is potentiated by heparin or heparan sulfate, both of which bind the PCPE1 NTR domain, procollagen, and BMP1 (85, 86), suggesting that heparan sulfate proteoglycans (HSPGs) may foster procollagen processing in vivo by bolstering formation of PCPE-procollagen-B/TP complexes (85, 86). HSPGs may also bind PCPEs to cell surfaces (86). PCPE1 enhancement of B/TPs seems specific to pCP activity, as PCPE1 failed to enhance cleavage of a number of other substrates in vitro (87). However, the extent of collagen fibril abnormalities in tissues of PCPE1-null mice (46) suggests possible additional roles for PCPEs. Suggestive but inconclusive genetic studies have implicated PCPE2 in modulating serum levels of HDL, whereas biochemical studies have shown PCPE2 to be associated with serum HDL and to be capable of binding both pro-ApoA1 and BMP1 and perhaps enhancing pro-ApoA1 processing by BMP1 in vitro (88). In vivo roles for PCPE2 in modulating HDL levels and pro-ApoA1 processing are supported by recent findings of decreased pro-ApoA1 processing and changes to HDL levels and properties in PCPE2-null mice (89). PCPE1 is also found in serum, and differential glycosylation of serum PCPE1 has been reported as a potential marker for levels of collagen remodeling in humans (90). PCPE1 can bind β2-microglobulin (91), although the significance of this finding remains to be elucidated.
Scaffold Proteins
PCPEs and HSPGs are not the only molecules able to bind both B/TPs and their substrates, thus fostering interactions. In Xenopus, the secreted olfactomedin family protein ONT1 binds both B/TPs and chordin, thereby facilitating chordin degradation (92). Expressed dorsally in embryos, ONT1 seems important in stabilizing dorsoventral patterning, as its loss sensitizes patterning to disruption upon manipulation of levels of chordin or other factors involved in regulating BMP signaling (92). Fibronectin (FN), a non-collagenous ECM protein, binds BMP1 non-protease domains via multiple FN sites (93). FN also binds various B/TP substrates, including LOX, chordin, biglycan, fibrillar collagens, and IGFBP3; and proteolytic processing of all these substrates is markedly reduced in FN-null mouse fibroblasts (15, 93, 94). Thus, FN appears able to act as a scaffold that facilitates B/TP-substrate interactions. Although FN can directly enhance BMP1 cleavage of chordin, procollagen I, and biglycan in vitro, effects of FN on BMP1 activity are more striking and consistent in cell cultures (15, 93). Thus, other factors may be important to FN enhancement of processing. Periostin, a secreted protein expressed primarily in collagen-rich connective tissues, binds BMP1 and FN and enhances both deposition of BMP1 into FN ECM and processing/activation of LOX (95). Thus, periostin may act as a scaffold that, by facilitating BMP1, LOX, and FN interactions, enhances formation of covalent cross-links that bolster the tensile strength of dense connective tissues.
Secreted Frizzled-related Proteins (sFRPs)
sFRPs consist of a C-terminal NTR domain and an N-terminal frizzled domain. The latter has homology to Wnt ligand-binding proteins, and most sFRPs participate in regulating Wnt signaling. However, sizzled, an sFRP expressed ventrally in Xenopus embryos, does not bind Wnt ligands, but it affects dorsoventral patterning by binding B/TPs and competitively inhibiting their chordinase activity (96). Crescent, a Xenopus sFRP expressed dorsally in embryos, also inhibits B/TP chordinase activity and binds Wnt ligands (25), perhaps providing cross-talk between the BMP and Wnt signaling pathways. Together, these two sFRPs may help stabilize the dorsoventral BMP signaling gradient, as sizzled may provide negative feedback that limits ventral BMP signaling, whereas crescent may help maintain maximum chordin levels dorsally (25). The mammalian protein sFRP2 also binds B/TPs but does not have anti-chordinase activity (97). Instead, it seems capable of enhancing B/TP pCP activity in vitro, and sFRP2-null fibroblasts show decreased procollagen processing (97). In addition, sFRP2-null mice have decreased fibrosis and improved cardiac function following myocardial infarction (MI) (97), presumably because B/TP procollagen processing is a rate-limiting step in the collagen deposition underlying fibrosis and because fibrosis in MI impairs cardiac function. sFRP2, which binds BMP1 non-protease domain sequences, appears to enhance binding of BMP1 to procollagen, suggesting that pCP activity enhancement may occur via formation of a BMP1-sFRP2-procollagen complex (97). A subsequent study (98) confirmed sFRP2-BMP1 binding and sFRP2 enhancement of pCP activity at physiological sFRP2 concentrations similar to those used by Kobayashi et al. (97) but found that 10-fold higher concentrations of sFRP2 can inhibit pCP activity and that direct injection of sFRP2 into rat heart with MI can inhibit collagen deposition. Attempting to explain the apparently contradictory observations of decreased fibrosis in sFRP2-null mice (97) and decreased fibrosis upon injection of sFRP2 into rat heart with MI, He et al. (98) suggested that reduced MI fibrosis in sFRP2-null mice may result from apoptotic loss of fibrogenic cells due to loss of sFRP2 anti-apoptotic activity. A third study reported no effect of sFRP2 on the in vitro pCP activity of BMP1 or mTLD (61).
Endogenous B/TP Inhibitors
α2-Macroglobulin, a serum protein also produced by various cell types, traps and irreversibly inhibits some proteases, including the B/TPs (99). Interestingly, another B/TP inhibitor is BMP4, which can non-competitively inhibit chordinase activity by binding B/TP CUB domains, thus providing negative feedback that limits BMP signaling in regions of high BMP2/4 and B/TP concentrations (38).
Perspectives
B/TPs are implicated in manifold interactions likely to affect key morphogenetic and homeostatic processes. Future studies employing tissue-specific knockdown/knock-out of B/TPs, singly or in combination, to remove redundancy should provide insights into the actual importance of such interactions in vivo. However, this approach may be complicated by B/TPs in the general circulation, as tissues in which B/TP expression is ablated may nevertheless be infused with B/TPs produced elsewhere. It will be of interest to determine whether circulating B/TPs are complexed to other proteins that affect their stability/activity. It seems reasonable to assume that such complexes exist, if only to protect B/TPs from α2-macroglobulin, which is at relatively high levels in serum and which would otherwise irreversibly inactivate circulating B/TPs.
This work was supported, in whole or in part, by National Institutes of Health Grant AR53815 (to D. S. G.). This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- BMP
- bone morphogenetic protein
- pCP
- procollagen C-proteinase
- TLD
- Tolloid
- DPP
- decapentaplegic
- B/TP
- BMP1/TLD-like proteinase
- ECM
- extracellular matrix
- mTLD
- mammalian TLD
- mTLL
- mammalian TLD-like
- SPC
- subtilisin-like proprotein convertase
- TLR
- TLD-related
- dpc
- days post-conception
- SLRP
- small leucine-rich proteoglycan
- SIBLING
- small integrin-binding ligand N-linked glycoprotein
- LOX
- lysyl oxidase
- DMP
- dentin matrix protein
- SOG
- short gastrulation
- TSG
- twisted gastrulation
- CHL1
- chordin-like 1
- Chl
- chordin-like
- LTBP
- latent TGFβ-binding protein
- GDF
- growth and differentiation factor
- IGF
- insulin-like growth factor
- IGFBP
- IGF-binding protein
- PCPE
- pCP enhancer
- NTR
- netrin-like
- HSPG
- heparan sulfate proteoglycan
- FN
- fibronectin
- sFRP
- secreted frizzled-related protein
- MI
- myocardial infarction.
REFERENCES
- 1. Wozney J. M., Rosen V., Celeste A. J., Mitsock L. M., Whitters M. J., Kriz R. W., Hewick R. M., Wang E. A. (1988) Science 242, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 2. Kessler E., Takahara K., Biniaminov L., Brusel M., Greenspan D. S. (1996) Science 271, 360–362 [DOI] [PubMed] [Google Scholar]
- 3. Shimell M. J., Ferguson E. L., Childs S. R., O'Connor M. B. (1991) Cell 67, 469–481 [DOI] [PubMed] [Google Scholar]
- 4. Marqués G., Musacchio M., Shimell M. J., Wünnenberg-Stapleton K., Cho K. W., O'Connor M. B. (1997) Cell 91, 417–426 [DOI] [PubMed] [Google Scholar]
- 5. Takahara K., Lyons G. E., Greenspan D. S. (1994) J. Biol. Chem. 269, 32572–32578 [PubMed] [Google Scholar]
- 6. Hopkins D. R., Keles S., Greenspan D. S. (2007) Matrix Biol. 26, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bond J. S., Beynon R. J. (1995) Protein Sci. 4, 1247–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomis-Rüth F. X. (2009) J. Biol. Chem. 284, 15353–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bork P., Beckmann G. (1993) J. Mol. Biol. 231, 539–545 [DOI] [PubMed] [Google Scholar]
- 10. Leighton M., Kadler K. E. (2003) J. Biol. Chem. 278, 18478–18484 [DOI] [PubMed] [Google Scholar]
- 11. Lee S., Solow-Cordero D. E., Kessler E., Takahara K., Greenspan D. S. (1997) J. Biol. Chem. 272, 19059–19066 [DOI] [PubMed] [Google Scholar]
- 12. Jasuja R., Ge G., Voss N. G., Lyman-Gingerich J., Branam A. M., Pelegri F. J., Greenspan D. S. (2007) J. Biol. Chem. 282, 9053–9062 [DOI] [PubMed] [Google Scholar]
- 13. Mac Sweeney A., Gil-Parrado S., Vinzenz D., Bernardi A., Hein A., Bodendorf U., Erbel P., Logel C., Gerhartz B. (2008) J. Mol. Biol. 384, 228–239 [DOI] [PubMed] [Google Scholar]
- 14. Becker-Pauly C., Barré O., Schilling O., Auf dem Keller U., Ohler A., Broder C., Schütte A., Kappelhoff R., Stöcker W., Overall C. M. (2011) Mol. Cell. Proteomics 10, M111.009233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim B., Huang G., Ho W. B., Greenspan D. S. (2011) J. Biol. Chem. 286, 29014–29025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanc G., Font B., Eichenberger D., Moreau C., Ricard-Blum S., Hulmes D. J., Moali C. (2007) J. Biol. Chem. 282, 16924–16933 [DOI] [PubMed] [Google Scholar]
- 17. Petropoulou V., Garrigue-Antar L., Kadler K. E. (2005) J. Biol. Chem. 280, 22616–22623 [DOI] [PubMed] [Google Scholar]
- 18. Werner J. M., Knott V., Handford P. A., Campbell I. D., Downing A. K. (2000) J. Mol. Biol. 296, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 19. Garrigue-Antar L., François V., Kadler K. E. (2004) J. Biol. Chem. 279, 49835–49841 [DOI] [PubMed] [Google Scholar]
- 20. Berry R., Jowitt T. A., Ferrand J., Roessle M., Grossmann J. G., Canty-Laird E. G., Kammerer R. A., Kadler K. E., Baldock C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8561–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berry R., Jowitt T. A., Garrigue-Antar L., Kadler K. E., Baldock C. (2010) FEBS Lett. 584, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garrigue-Antar L., Hartigan N., Kadler K. E. (2002) J. Biol. Chem. 277, 43327–43334 [DOI] [PubMed] [Google Scholar]
- 23. Scott I. C., Blitz I. L., Pappano W. N., Imamura Y., Clark T. G., Steiglitz B. M., Thomas C. L., Maas S. A., Takahara K., Cho K. W., Greenspan D. S. (1999) Dev. Biol. 213, 283–300 [DOI] [PubMed] [Google Scholar]
- 24. Lee S. J. (2008) PLoS ONE 3, e1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ploper D., Lee H. X., De Robertis E. M. (2011) Dev. Biol. 352, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piccolo S., Agius E., Lu B., Goodman S., Dale L., De Robertis E. M. (1997) Cell 91, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Serpe M., O'Connor M. B. (2006) Development 133, 4969–4979 [DOI] [PubMed] [Google Scholar]
- 28. Serpe M., Ralston A., Blair S. S., O'Connor M. B. (2005) Development 132, 2645–2656 [DOI] [PubMed] [Google Scholar]
- 29. Meyer F., Aberle H. (2006) Development 133, 4035–4044 [DOI] [PubMed] [Google Scholar]
- 30. Clark T. G., Conway S. J., Scott I. C., Labosky P. A., Winnier G., Bundy J., Hogan B. L., Greenspan D. S. (1999) Development 126, 2631–2642 [DOI] [PubMed] [Google Scholar]
- 31. Takahara K., Brevard R., Hoffman G. G., Suzuki N., Greenspan D. S. (1996) Genomics 34, 157–165 [DOI] [PubMed] [Google Scholar]
- 32. Scott I. C., Steiglitz B. M., Clark T. G., Pappano W. N., Greenspan D. S. (2000) Dev. Dyn. 217, 449–456 [DOI] [PubMed] [Google Scholar]
- 33. Liu Q. R., Hattar S., Endo S., MacPhee K., Zhang H., Cleary L. J., Byrne J. H., Eskin A. (1997) J. Neurosci. 17, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suzuki N., Labosky P. A., Furuta Y., Hargett L., Dunn R., Fogo A. B., Takahara K., Peters D. M., Greenspan D. S., Hogan B. L. (1996) Development 122, 3587–3595 [DOI] [PubMed] [Google Scholar]
- 35. Sabirzhanova I., Sabirzhanov B., Bjordahl J., Brandt J., Jay P. Y., Clark T. G. (2009) Dev. Growth Differ 51, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stańczak P., Witecka J., Szydło A., Gutmajster E., Lisik M., Auguciak-Duma A., Tarnowski M., Czekaj T., Czekaj H., Sieroń A. L. (2009) Eur. J. Hum. Genet. 17, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grgurevic L., Macek B., Mercep M., Jelic M., Smoljanovic T., Erjavec I., Dumic-Cule I., Prgomet S., Durdevic D., Vnuk D., Lipar M., Stejskal M., Kufner V., Brkljacic J., Maticic D., Vukicevic S. (2011) Biochem. Biophys. Res. Commun. 408, 25–31 [DOI] [PubMed] [Google Scholar]
- 38. Lee H. X., Mendes F. A., Plouhinec J. L., De Robertis E. M. (2009) Genes Dev. 23, 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grgurevic L., Macek B., Healy D. R., Brault A. L., Erjavec I., Cipcic A., Grgurevic I., Rogic D., Galesic K., Brkljacic J., Stern-Padovan R., Paralkar V. M., Vukicevic S. (2011) J. Am. Soc. Nephrol. 22, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prockop D. J., Kivirikko K. I. (1995) Annu. Rev. Biochem. 64, 403–434 [DOI] [PubMed] [Google Scholar]
- 41. Li S. W., Sieron A. L., Fertala A., Hojima Y., Arnold W. V., Prockop D. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5127–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Canty E. G., Lu Y., Meadows R. S., Shaw M. K., Holmes D. F., Kadler K. E. (2004) J. Cell Biol. 165, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hojima Y., Behta B., Romanic A. M., Prockop D. J. (1994) Anal. Biochem. 223, 173–180 [DOI] [PubMed] [Google Scholar]
- 44. Prockop D. J., Kuivaniemi H., Tromp G. (1994) Clin. Plast. Surg. 21, 407–413 [PubMed] [Google Scholar]
- 45. Pappano W. N., Steiglitz B. M., Scott I. C., Keene D. R., Greenspan D. S. (2003) Mol. Cell. Biol. 23, 4428–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steiglitz B. M., Kreider J. M., Frankenburg E. P., Pappano W. N., Hoffman G. G., Meganck J. A., Liang X., Höök M., Birk D. E., Goldstein S. A., Greenspan D. S. (2006) Mol. Cell. Biol. 26, 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lindahl K., Barnes A. M., Fratzl-Zelman N., Whyte M. P., Hefferan T. E., Makareeva E., Brusel M., Yaszemski M. J., Rubin C. J., Kindmark A., Roschger P., Klaushofer K., McAlister W. H., Mumm S., Leikin S., Kessler E., Boskey A. L., Ljunggren O., Marini J. C. (2011) Hum. Mutat. 32, 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kronenberg D., Bruns B. C., Moali C., Vadon-Le Goff S., Sterchi E. E., Traupe H., Böhm M., Hulmes D. J., Stöcker W., Becker-Pauly C. (2010) J. Invest. Dermatol. 130, 2727–2735 [DOI] [PubMed] [Google Scholar]
- 49. Birk D. E., Fitch J. M., Babiarz J. P., Doane K. J., Linsenmayer T. F. (1990) J. Cell Sci. 95, 649–657 [DOI] [PubMed] [Google Scholar]
- 50. Mendler M., Eich-Bender S. G., Vaughan L., Winterhalter K. H., Bruckner P. (1989) J. Cell Biol. 108, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boot-Handford R. P., Tuckwell D. S., Plumb D. A., Rock C. F., Poulsom R. (2003) J. Biol. Chem. 278, 31067–31077 [DOI] [PubMed] [Google Scholar]
- 52. Unsöld C., Pappano W. N., Imamura Y., Steiglitz B. M., Greenspan D. S. (2002) J. Biol. Chem. 277, 5596–5602 [DOI] [PubMed] [Google Scholar]
- 53. Gopalakrishnan B., Wang W. M., Greenspan D. S. (2004) J. Biol. Chem. 279, 30904–30912 [DOI] [PubMed] [Google Scholar]
- 54. Huang G., Ge G., Wang D., Gopalakrishnan B., Butz D. H., Colman R. J., Nagy A., Greenspan D. S. (2011) J. Clin. Invest. 121, 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Linsenmayer T. F., Gibney E., Igoe F., Gordon M. K., Fitch J. M., Fessler L. I., Birk D. E. (1993) J. Cell Biol. 121, 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bruckner-Tuderman L., Nilssen O., Zimmermann D. R., Dours-Zimmermann M. T., Kalinke D. U., Gedde-Dahl T., Jr., Winberg J. O. (1995) J. Cell Biol. 131, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rattenholl A., Pappano W. N., Koch M., Keene D. R., Kadler K. E., Sasaki T., Timpl R., Burgeson R. E., Greenspan D. S., Bruckner-Tuderman L. (2002) J. Biol. Chem. 277, 26372–26378 [DOI] [PubMed] [Google Scholar]
- 58. Maertens B., Hopkins D., Franzke C. W., Keene D. R., Bruckner-Tuderman L., Greenspan D. S., Koch M. (2007) J. Biol. Chem. 282, 10647–10659 [DOI] [PubMed] [Google Scholar]
- 59. Borel A., Eichenberger D., Farjanel J., Kessler E., Gleyzal C., Hulmes D. J., Sommer P., Font B. (2001) J. Biol. Chem. 276, 48944–48949 [DOI] [PubMed] [Google Scholar]
- 60. Uzel M. I., Scott I. C., Babakhanlou-Chase H., Palamakumbura A. H., Pappano W. N., Hong H. H., Greenspan D. S., Trackman P. C. (2001) J. Biol. Chem. 276, 22537–22543 [DOI] [PubMed] [Google Scholar]
- 61. von Marschall Z., Fisher L. W. (2010) Matrix Biol. 29, 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steiglitz B. M., Ayala M., Narayanan K., George A., Greenspan D. S. (2004) J. Biol. Chem. 279, 980–986 [DOI] [PubMed] [Google Scholar]
- 63. Teixeira C. C., Xiang J., Roy R., Kudrashov V., Binderman I., Mayer-Kuckuk P., Boskey A. L. (2011) J. Cell. Biochem. 112, 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. von Marschall Z., Fisher L. W. (2010) Biochem. Biophys. Res. Commun. 391, 1374–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Scott I. C., Imamura Y., Pappano W. N., Troedel J. M., Recklies A. D., Roughley P. J., Greenspan D. S. (2000) J. Biol. Chem. 275, 30504–30511 [DOI] [PubMed] [Google Scholar]
- 66. Ge G., Seo N. S., Liang X., Hopkins D. R., Höök M., Greenspan D. S. (2004) J. Biol. Chem. 279, 41626–41633 [DOI] [PubMed] [Google Scholar]
- 67. Gonzalez E. M., Reed C. C., Bix G., Fu J., Zhang Y., Gopalakrishnan B., Greenspan D. S., Iozzo R. V. (2005) J. Biol. Chem. 280, 7080–7087 [DOI] [PubMed] [Google Scholar]
- 68. Veitch D. P., Nokelainen P., McGowan K. A., Nguyen T. T., Nguyen N. E., Stephenson R., Pappano W. N., Keene D. R., Spong S. M., Greenspan D. S., Findell P. R., Marinkovich M. P. (2003) J. Biol. Chem. 278, 15661–15668 [DOI] [PubMed] [Google Scholar]
- 69. Lesiak M., Auguciak-Duma A., Szydło A., Sieroń A. L. (2009) Pharmacol. Rep. 61, 468–476 [DOI] [PubMed] [Google Scholar]
- 70. Little S. C., Mullins M. C. (2006) Birth Defects Res. C Embryo Today 78, 224–242 [DOI] [PubMed] [Google Scholar]
- 71. Peluso C. E., Umulis D., Kim Y. J., O'Connor M. B., Serpe M. (2011) Dev. Cell 21, 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scott I. C., Blitz I. L., Pappano W. N., Maas S. A., Cho K. W., Greenspan D. S. (2001) Nature 410, 475–478 [DOI] [PubMed] [Google Scholar]
- 73. Branam A. M., Hoffman G. G., Pelegri F., Greenspan D. S. (2010) Dev. Biol. 341, 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Annes J. P., Munger J. S., Rifkin D. B. (2003) J. Cell Sci. 116, 217–224 [DOI] [PubMed] [Google Scholar]
- 75. Ge G., Greenspan D. S. (2006) J. Cell Biol. 175, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wolfman N. M., McPherron A. C., Pappano W. N., Davies M. V., Song K., Tomkinson K. N., Wright J. F., Zhao L., Sebald S. M., Greenspan D. S., Lee S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15842–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ge G., Hopkins D. R., Ho W. B., Greenspan D. S. (2005) Mol. Cell. Biol. 25, 5846–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ge G., Fernández C. A., Moses M. A., Greenspan D. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. St Croix B., Rago C., Velculescu V., Traverso G., Romans K. E., Montgomery E., Lal A., Riggins G. J., Lengauer C., Vogelstein B., Kinzler K. W. (2000) Science 289, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 80. Chau P., Fielding P. E., Fielding C. J. (2007) Biochemistry 46, 8445–8450 [DOI] [PubMed] [Google Scholar]
- 81. Steiglitz B. M., Keene D. R., Greenspan D. S. (2002) J. Biol. Chem. 277, 49820–49830 [DOI] [PubMed] [Google Scholar]
- 82. Takahara K., Kessler E., Biniaminov L., Brusel M., Eddy R. L., Jani-Sait S., Shows T. B., Greenspan D. S. (1994) J. Biol. Chem. 269, 26280–26285 [PubMed] [Google Scholar]
- 83. Kronenberg D., Vadon-Le Goff S., Bourhis J. M., Font B., Eichenberger D., Hulmes D. J., Moali C. (2009) J. Biol. Chem. 284, 33437–33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ge G., Zhang Y., Steiglitz B. M., Greenspan D. S. (2006) J. Biol. Chem. 281, 10786–10798 [DOI] [PubMed] [Google Scholar]
- 85. Bekhouche M., Kronenberg D., Vadon-Le Goff S., Bijakowski C., Lim N. H., Font B., Kessler E., Colige A., Nagase H., Murphy G., Hulmes D. J., Moali C. (2010) J. Biol. Chem. 285, 15950–15959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Weiss T., Ricard-Blum S., Moschcovich L., Wineman E., Mesilaty S., Kessler E. (2010) J. Biol. Chem. 285, 33867–33874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moali C., Font B., Ruggiero F., Eichenberger D., Rousselle P., François V., Oldberg A., Bruckner-Tuderman L., Hulmes D. J. (2005) J. Biol. Chem. 280, 24188–24194 [DOI] [PubMed] [Google Scholar]
- 88. Zhu J., Gardner J., Pullinger C. R., Kane J. P., Thompson J. F., Francone O. L. (2009) J. Lipid Res. 50, 1330–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Francone O. L., Ishida B. Y., de la Llera-Moya M., Royer L., Happe C., Zhu J., Chalkey R. J., Schaefer P., Cox C., Burlingame A., Kane J. P., Rothblat G. H. (2011) J. Lipid Res. 52, 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mesilaty-Gross S., Anikster Y., Vilensky B., Wolf I., Phillip M., Gat-Yablonski G. (2009) Clin. Chim. Acta 403, 76–80 [DOI] [PubMed] [Google Scholar]
- 91. Morimoto H., Wada J., Font B., Mott J. D., Hulmes D. J., Ookoshi T., Naiki H., Yasuhara A., Nakatsuka A., Fukuoka K., Takatori Y., Ichikawa H., Akagi S., Nakao K., Makino H. (2008) Matrix Biol. 27, 211–219 [DOI] [PubMed] [Google Scholar]
- 92. Inomata H., Haraguchi T., Sasai Y. (2008) Cell 134, 854–865 [DOI] [PubMed] [Google Scholar]
- 93. Huang G., Zhang Y., Kim B., Ge G., Annis D. S., Mosher D. F., Greenspan D. S. (2009) J. Biol. Chem. 284, 25879–25888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fogelgren B., Polgár N., Szauter K. M., Ujfaludi Z., Laczkó R., Fong K. S., Csiszar K. (2005) J. Biol. Chem. 280, 24690–24697 [DOI] [PubMed] [Google Scholar]
- 95. Maruhashi T., Kii I., Saito M., Kudo A. (2010) J. Biol. Chem. 285, 13294–13303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee H. X., Ambrosio A. L., Reversade B., De Robertis E. M. (2006) Cell 124, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kobayashi K., Luo M., Zhang Y., Wilkes D. C., Ge G., Grieskamp T., Yamada C., Liu T. C., Huang G., Basson C. T., Kispert A., Greenspan D. S., Sato T. N. (2009) Nat. Cell Biol. 11, 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. He W., Zhang L., Ni A., Zhang Z., Mirotsou M., Mao L., Pratt R. E., Dzau V. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21110–21115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang Y., Ge G., Greenspan D. S. (2006) J. Biol. Chem. 281, 39096–39104 [DOI] [PubMed] [Google Scholar]