Background: Steatosis is a frequent complication in HCV infection and has been linked to the viral core protein.
Results: Core requires DGAT1 to localize to lipid droplets and to decrease their turnover.
Conclusion: The viral core protein stabilizes lipid droplets, which eventually causes lipid accumulation.
Significance: DGAT1 inhibitors may be useful in the treatment of HCV-associated steatosis.
Keywords: Hepatitis C Virus, Lipid Droplet(s), Lipid Synthesis, Lipolysis, Liver Metabolism, Triglyceride, DGAT Enzymes
Abstract
Steatosis is a frequent complication of hepatitis C virus infection. In mice, this condition is recapitulated by the expression of a single viral protein, the nucleocapsid core. Core localizes to the surface of lipid droplets (LDs) in infected liver cells through a process dependent on host diacylglycerol acyltransferase 1 (DGAT1), an enzyme that synthesizes triglycerides in the endoplasmic reticulum. Whether DGAT1 also plays a role in core-induced steatosis is uncertain. Here, we show that mouse embryonic fibroblasts isolated from DGAT1−/− mice are protected from core-induced steatosis, as are livers of DGAT1−/− mice expressing core, demonstrating that the steatosis is DGAT1-dependent. Surprisingly, core expression did not increase DGAT1 activity or triglyceride synthesis, thus excluding the possibility that core activates DGAT1 to cause steatosis. Instead, we find that DGAT1-dependent localization of core to LDs is a prerequisite for the steatogenic properties of the core. Using biochemical and immunofluorescence microscopy techniques, we show that the turnover of lipids in core-coated droplets is decreased, providing a physiological mechanism for core-induced steatosis. Our results support a bipartite model in which core first requires DGAT1 to gain access to LDs, and then LD-localized core interferes with triglyceride turnover, thus stabilizing lipid droplets and leading to steatosis.
Introduction
Abnormal accumulation of liver lipids (steatosis) occurs in more than 50% of individuals chronically infected with HCV4 (1). Liver steatosis is the only independent factor associated with hepatic fibrosis progression (2) and is significantly associated with the incidence of hepatocellular carcinoma in HCV-infected patients (3). The correlation between the degree of steatosis and viral replication, as assessed by viral load, points to an important role of steatosis in HCV infection with genotype 3a strains (4, 5). In genotype 3a-infected patients, steatosis also frequently resolves after successful clearance of HCV after antiviral therapy, providing further evidence for a direct association of steatosis with HCV infection (6, 7). Furthermore, in patients infected with HCV genotype 1, the most frequent genotype in the United States and Europe, the degree of steatosis is inversely related to the response to antiviral therapy (8). Interestingly, transgenic mice expressing the viral nucleocapsid protein core in their livers also develop steatosis, implying that viral infection itself may contribute to this process. In fact, some core-transgenic mouse strains progress from steatosis to hepatocellular carcinoma, recapitulating what is seen in some HCV-infected patients (9, 10).
Hepatic steatosis is characterized by the accumulation of intracellular lipid droplets (LDs), which serve as central organelles in the HCV life cycle. The viral core protein localizes to LDs and initiates production of viral particles at LD-associated membranes of the ER (11–13). Core is produced in the ER and consists of the first 191 amino acids of the large viral polyprotein precursor that is translated from the positive-strand HCV RNA genome. The polyprotein is co- and post-translationally processed to structural and nonstructural proteins by host and viral proteases (14). Core anchors the polyprotein at the ER membrane through a C-terminal signal peptide that translocates subsequent E1 and E2 envelope proteins into the ER lumen (14). After two sequential cleavages at the C terminus by host peptidases, processed core remains attached to the outer leaflet of the ER membrane and gains access to the surface of LDs (15). The localization of core to intracellular LDs provides a platform for viral assembly and therefore appears to be a key step in viral replication.
How the core protein induces LD accumulation and hepatic steatosis remains incompletely understood. One possibility that was previously reported was core-induced inhibition of the activity of microsomal triglyceride transfer protein (MTP), a key protein in VLDL assembly and lipid export from hepatocytes (16). More recent studies demonstrated that MTP function and lipoprotein secretion may be vital for functional HCV release (17–19). Core also alters the expression of genes involved in fatty acid biosynthesis by activating transcription factors such as sterol regulatory element-binding proteins and peroxisome proliferator-activated receptors α and γ in cultured cells and in core-transgenic mice (20–23). It is unclear whether these transcriptional changes are a direct consequence of core expression or reflect secondary changes because of altered metabolic homeostasis. In support of a model where core plays a direct role in altering transcriptional events, core was shown to bind and activate the DNA-binding domain of the retinoid X receptor α, a transcriptional regulator involved in the regulation of cellular lipid synthesis (24). It was recently shown that genotype 3a core protein induces the formation of large lipid droplets in hepatoma cells by decreasing the expression of phosphatase and tensin homolog and insulin receptor substrate 1 (IRS-1) (25). IRS-1 has also been linked to the development of insulin resistance in HCV-infected patients in a genotype-specific manner; genotype 3a core causes IRS-1 degradation through down-regulation of peroxisome proliferator-activated receptor γ and up-regulation of suppressor of cytokine signal 7 (SOCS7), whereas genotype 1b core instead activates the mammalian target of rapamycin (mTOR) pathway (26).
Ultimately, steatosis reflects the accumulation of lipid droplets, which reflects the production and clearance of cellular triacylglycerols (TGs). We showed recently that core specifically interacts with DGAT1 (27), one of the two host diacylglycerol acyltransferase (DGAT) enzymes that produce TGs in the ER (28). DGAT1 and DGAT2 account for nearly all TG synthesis in mammalian cells (29), and both enzymes reside in the ER, although they are found in distinct ER microdomains (30). Both DGATs have been implicated in the development of diet-induced hepatic steatosis (31–33). However, although DGAT2 deficiency in mice is lethal early after birth, DGAT1 deficiency protects against hepatic steatosis in both diet-induced obesity and fasting paradigms, but it has no effect on steatosis in situations where de novo lipogenesis is increased (32). DGAT1 activity is critical to the localization of core to the surface of LDs, and interference with DGAT1 activity restricts core localization to the ER and blocks the assembly and release of infectious HCV progeny virions (27). The link of core to DGAT1 suggested that the mechanism of steatosis might relate to TG synthesis.
Here, we show that core-induced steatosis is indeed DGAT1-dependent. However, we found no effect of core on increasing TG synthesis, but we instead uncover a novel mechanism in which LD-bound core blocks TG turnover. Our studies suggest an unexpected bipartite mechanism in which the viral core protein utilizes the host DGAT1 enzyme to gain access to LDs, and once localized to this compartment, it interferes with LD TG turnover, thereby stabilizing LDs necessary for viral assembly.
EXPERIMENTAL PROCEDURES
Plasmids
Lentiviral expression constructs of core were as described previously (27). To generate the adenoviral core expression construct, the 191-amino acid core coding sequence (genotype 1b, NC1) was cloned into pAdEasy via the shuttle vector pAdTrack-CMV (34). This construct ensures co-expression of core with the marker EGFP.
Cell Lines and Culture Conditions
NIH/3T3, Huh7, HEK293, and HEK293T cells were obtained from the ATCC. All cells were grown under standard cell culture conditions and were transfected with FuGENE 6 (Roche Applied Science), according to the manufacturer's protocol. Calcium phosphate-mediated transfection of HEK293T cells was used for the production of lentiviral particles. Mouse embryonic fibroblasts were established from DGAT1−/− or DGAT2−/− embryos or their control littermates as described previously (29).
Animal Studies
DGAT1−/− mice have been described previously (35). Eight to 10-week-old male DGAT1−/− mice and control C57BL6 mice were administered 4.5 × 1010 pfu of adenovirus-expressing EGFP or core/EGFP in 300 μl of normal saline by tail vein injection. Four days later, mice were sacrificed and livers harvested. All animal experiments were approved by the University of California, San Francisco, IACUC.
Antibodies and Reagents
The following antibodies were obtained commercially: anti-core (clone C7-50; Affinity BioReagents), anti-tubulin (T6074, Sigma), anti-ADRP (AP125, Progen), anti-calreticulin (StressGen), anti-mTOR (22C2, Calbiochem), anti-mTOR (Ser(P)-2448) (Calbiochem), anti-mouse Alexa 647 (Invitrogen), anti-mouse Alexa 594 (Invitrogen), anti-rabbit Alexa 488 (Invitrogen), anti-rabbit Cy3 (Jackson ImmunoResearch), and anti-mouse Cy5 (Jackson ImmunoResearch). The DGAT1 inhibitor was as described previously (27). Enzymes for molecular cloning were purchased from New England Biolabs; cell culture reagents were from Invitrogen, and fine chemicals, if not noted otherwise, were from Sigma.
Immunofluorescence, Oil-Red-O (ORO) Staining, Epifluorescence Microscopy, and Quantification of Images
Immunofluorescence and ORO staining of cultured cells were done as described previously (27). For loading of cells with oleate, cells were incubated in the presence of 300 μm BSA-bound oleate (Sigma) for the indicated times. For the histological analysis of liver sections, livers were fixed in paraformaldehyde for 16 h, sucrose-infiltrated, and frozen in OCT using dry ice and 2-methylbutane. Sections were cut at 8 μm and allowed to dry overnight. OCT was removed by running water for 10 min and equilibrated in 50% and then 100% propylene glycol (Sigma) for 3 min each. The slides were stained in 1% ORO (Sigma) in propylene glycol for 18 h. Slides were then differentiated in three changes of 85% propylene glycol followed by a 10-min running water wash. Slides were counterstained in Mayers hematoxylin for 5 s, washed, and blued in running water and mounted using an aqueous mountant (CC-mount, American Mastertech). Cells and liver sections were analyzed with an Axio Observer Z1 microscope (Zeiss) equipped with EC Plan NeoFluar 10×/0.3 PHM27, EC Plan NeoFluar 20×/0.5 PHM27, EC Plan NeoFluar 40×/0.75 PH, and Plan Apo 63×/1.4 Oil DIC M27 objectives, filter sets 38HE, 43HE, 45, and 50, Optivar 1.25 and ×1.6 magnification, and an Axiocam MRM REV 3 and an Axiocam IC3. For quantification of LD content, we counted the ORO-positive area per cell with the automatic measurement program of the Zeiss Axiovision software. The ORO-positive area in EGFP-positive cells was quantified and divided by the total number of cells.
Sucrose Gradient Centrifugation and Western Blotting
For the sucrose gradient, liver samples were lysed in hypotonic buffer (50 mm HEPES, 1 mm EDTA, and 2 mm MgCl2, pH 7.4), supplemented with protease inhibitors, with 40 strokes in a tight-fitting Dounce homogenizer. After spinning for 5 min at 1500 rpm, post-nuclear fractions were mixed with equal volumes of 2 m sucrose in isotonic buffer (50 mm HEPES, 100 mm KCl, 2 mm MgCl2) and placed above a 2 m sucrose cushion in SW41 (Beckman) centrifuge tubes, overlaid with isotonic buffer containing decreasing concentrations of sucrose (0.75, 0.5, 0.25, and 0 m in isotonic buffer with 1 mm PMSF), and centrifuged for 16 h at 100,000 × g. Proteins from the fractions were precipitated with 15% trichloroacetic acid and 30% acetone, washed once with acetone, and resuspended in urea loading dye (200 mm Tris/HCl, pH 6.8, 8 m urea, 5% SDS, 1 mm EDTA, 0.1% bromphenol blue, 15 mm DTT). LDs were isolated as described previously (11, 27).
For Western blot analysis, tissues were homogenized in 2× sample buffer (20% glycerol, 4% SDS, 125 mm Tris/HCl, pH 6.8) with 20 strokes in a tight-fitting Dounce homogenizer. Before loading, the samples were supplemented with bromphenol blue and β-mercaptoethanol. Cultured cells were lysed in RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS) supplemented with protease inhibitor mixture (Sigma) for 30 min, followed by SDS-PAGE. For chemiluminescent detection, we used ECL and ECL Hyperfilm (Amersham Biosciences).
Lentivirus and Adenovirus Production and Transduction
Lentiviral particles were produced as described previously (27, 36). High titer adenoviral stocks were produced by the Vector Development Lab at the Baylor College of Medicine. Colony-forming units (cfu) were determined by infecting HEK293 cells with serial dilutions of the viral stocks and counting EGFP-positive foci 2 days post-infection.
DGAT1 Activity Assay and Measurement of TG Synthesis
DGAT assays were performed as described previously (27, 35) using microsomes prepared by differential centrifugation of liver homogenates. 100 mm MgCl2 was used for DGAT assays to selectively assay DGAT1 (37).
For the measurement of TG synthesis rates, cells were incubated in the presence of 0.125 μCi/ml [1-14C]oleic acid (GE Healthcare, 58 mCi/mmol) for 4 h. [1-14C]Oleic acid was dried under a nitrogen stream and then complexed to 10% bovine serum albumin before addition to the cells. Lipids were extracted with hexane/isopropyl alcohol (3:2), dried, loaded onto thin layer chromatography plates, and quantified as above with a Bioscan AR-2000 instrument.
MTP Activity Assay
Liver samples were homogenized in homogenization buffer (10 mm Tris, 150 mm NaCl, 1 mm EDTA, 0.5 mm PMSF, 2 μg/ml leupeptin, pH 7.4) by 40 strokes in a tight-fitting Dounce homogenizer. MTP activity assays (Roar Biomedical Inc.) were performed using 10 μg of total protein according to the manufacturer's protocol.
mTOR Activity Assay
Liver samples were homogenized in lysis buffer (50 mm Tris, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Tween 20, pH 7.4, supplemented with protease (Sigma) and phosphatase inhibitor mixture (Roche Applied Science)) by 40 strokes in a tight-fitting Dounce homogenizer. Endogenous mTOR was immunoprecipitated from clarified lysates with mTOR antibodies and protein G-agarose (Sigma). Equal amount of beads were used in mTOR activity assays according to the manufacturer's protocol (K-LISA, Calbiochem).
Measurement of Lipolysis
The measurement of lipolysis was performed as described previously (38). Cells were incubated with 400 μm BSA-bound oleate containing 0.125 μCi/ml [1-14C]oleic acid (GE Healthcare, 58 mCi/mmol) for 16 h to stimulate storage of TGs. Cells were then washed and incubated in fresh medium (containing 6 μm triacsin C for indicated times), and the amount of remaining labeled cellular TG was determined as above. For microscopic analysis, cells were loaded with 400 μm BSA-bound oleate for 16 h and incubated in fresh medium in the presence of 6 μm triacsin C. They were then fixed in paraformaldehyde and stained with ORO and Hoechst.
Statistical Analysis
When one variable differed between experimental groups, unpaired two-tailed Student's t tests were used. When experimental groups differed in two variables, two-way ANOVA was used with appropriate post-testing.
RESULTS
HCV Core Induces Accumulation of LDs in Cultured Cells in a DGAT1-dependent Manner
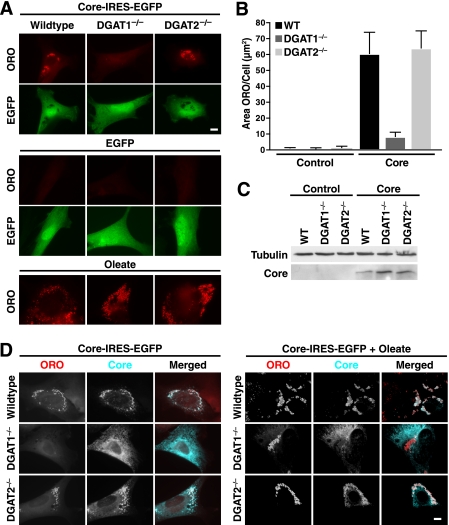
To examine whether core-induced lipid accumulation involves a specific DGAT enzyme, we introduced core into mouse embryonic fibroblasts from DGAT1- or DGAT2-deficient mice (29) and examined LD accumulation by ORO staining and fluorescent microscopy. Core expression caused an ∼60-fold increase in cellular LD content in wild-type (WT) and DGAT2−/− fibroblasts but not DGAT1−/− fibroblasts, pointing to a unique relationship between core and DGAT1-generated LDs (Fig. 1, A and B). DGAT1−/−, DGAT2−/−, and WT fibroblasts expressed core at similar levels (Fig. 1C), and mouse embryonic fibroblasts of each genotype formed LDs normally after incubation with monounsaturated fatty acids (oleate, Fig. 1A), indicating that either DGAT enzyme was competent for generating LDs from this stimulus.
FIGURE 1.
HCV core protein requires DGAT1 both for induction of LD and for LD localization. WT, DGAT1−/−, and DGAT2−/− mouse embryonic fibroblasts were transduced with lentiviral vectors expressing EGFP (control) or core-IRES-EGFP (core) and/or loaded with 300 μm oleate for 16 h and analyzed by immunostaining or Western blotting. A, epifluorescence microscopy after staining with ORO to visualize LDs and EGFP. B, quantification of A, mean of 50 cells ± S.E. Significance was determined by two-way ANOVA as a function of genotype (p < 0.05), core expression (p < 0.001), and interaction between the two variables (p < 0.05). C, analysis of core expression by Western blotting. D, indirect immunofluorescence of core and ORO staining (scale bars, 10 μm). Single channels are shown in black and white; the merged images are pseudocolored with ORO in red and core in turquoise.
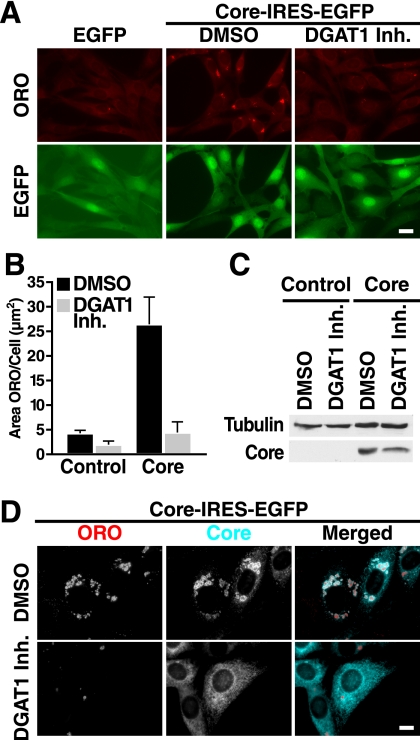
Core localized to LDs in WT and DGAT2−/− fibroblasts but not in DGAT1−/− cells where it showed a distinct reticular staining, consistent with its localization at the ER (Fig. 1D). Remarkably, core localization did not change in DGAT1−/− cells treated with oleate containing numerous LDs formed via DGAT2 (Fig. 1D). These results underscore a connection between the ability of the core to bind LDs and to induce lipid accumulation, and they indicate that DGAT1 is important in both processes. We confirmed these results with a DGAT1-specific small molecule inhibitor, 2-((1S,4S)-4-(4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl)cyclohexyl)acetic acid, described previously (27). Treatment with the inhibitor efficiently suppressed core-induced LD accumulation in NIH/3T3 fibroblasts and prevented core association with LDs (Fig. 2).
FIGURE 2.
DGAT1 inhibition blocks trafficking of core to LDs and their accumulation. NIH/3T3 cells were transduced with lentiviral vectors expressing EGFP (control) or Core-IRES-EGFP (core), treated with DMSO or 20 μm DGAT1 inhibitor (Inh.) (day 1), fixed, and stained or lysed for SDS-PAGE (day 3). A, epifluorescence microscopy after staining with ORO (scale bar, 20 μm). B, quantification of A (mean of 500 cells ± S.E.). C, Western blot analysis with core and tubulin antibodies. D, indirect immunofluorescence with core antibodies and ORO staining (scale bar, 10 μm). Single channels are shown in black and white; the merged images are pseudocolored with ORO in red and core in turquoise.
DGAT1 Is Required for Core-induced Lipid Accumulation in Vivo
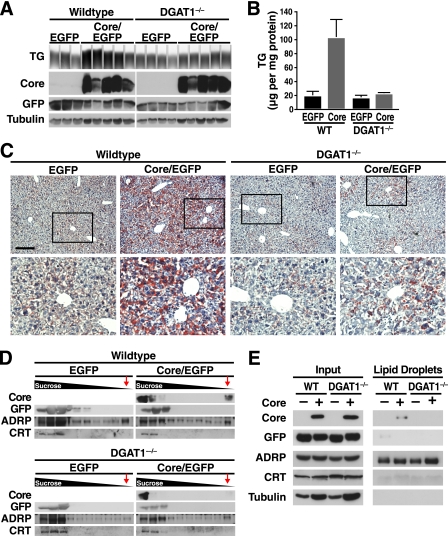
To determine whether DGAT1 is also important for core-induced steatosis in vivo, we expressed core in the livers of DGAT1−/− mice. We generated adenoviral vectors expressing both core and EGFP or EGFP alone, and these were injected into tail veins of WT and DGAT1-deficient mice. Four days after injection, livers were harvested, and lysates were subjected to Western blotting. Similar levels of core expression were detected in WT and DGAT1−/− mouse livers (Fig. 3A). In addition, regardless of the DGAT1 genotype, EGFP was similarly expressed in mice after injections with control or core-expressing viral stocks. Core or EGFP expression was not detected in organs other than the liver in injected mice (data not shown).
FIGURE 3.
DGAT1−/− mice are protected from HCV core-induced steatosis. WT and DGAT1−/− mice were injected with core/EGFP- or EGFP-expressing adenovirus. Livers were harvested 4 days after infection. A, extracted lipids were loaded on a thin layer chromatography plate. Protein extracts were analyzed by Western blotting with core, EGFP, and tubulin antibodies. B, quantification of TG in A, mean ± S.E., significance was determined by two-way ANOVA as a function of DGAT1 genotype (p < 0.05), core expression (p < 0.05), and interaction between the two variables (p < 0.05). C, ORO staining of liver sections (scale bar, 20 μm). Enlarged sections are marked by a box. D, sucrose gradient centrifugation of liver lysates. The fractions were analyzed by Western blotting with core, EGFP, ADRP, and calreticulin (CRT) antibodies. LDs float on the gradient; a red arrow marks the LD fractions. E, LDs were isolated by two subsequent centrifugations. LD fractions were normalized to the levels of ADRP.
Core expression induced an ∼5-fold increase in TG levels (compared with EGFP control-injected mice) in WT but not in DGAT1−/− mouse livers (Fig. 3, A and B). Therefore, WT mice were more susceptible to core-induced steatosis than DGAT1−/− mice (p < 0.05, two-way ANOVA). In fact, TG levels in DGAT1−/− mice injected with core-expressing viral stocks were similar to those of WT or DGAT1−/− mice infected with control virus (Fig. 3, A and B). The same was observed in histological sections after staining with ORO; accumulation of ORO-positive LDs occurred only in livers of core-expressing WT mice, indicating that DGAT1 is required for core-induced steatosis in vivo (Fig. 3C).
As DGAT1 apparently directs core onto the surface of LDs (27), we isolated LDs by sucrose gradient centrifugation from livers of core-expressing WT and DGAT1−/− mice. Core was readily detected by Western blot analysis in the LD fraction in liver extracts from WT, but not DGAT1−/−, mice (Fig. 3D, LD fraction marked by red arrows). This fraction was also enriched in ADRP, another LD-associated protein, but it lacked EGFP and calreticulin. Calreticulin is an ER-anchored protein, which, as expected, was only present in higher density fractions (Fig. 3D). A similar result was observed when two sequential sucrose gradient centrifugations were performed, and the amount of LD protein loaded was normalized to ADRP levels in both genotypes (Fig. 3E). These results show that DGAT1 is required for core to access LDs in vivo and confirm the requirement for DGAT1 in mediating core-induced steatosis.
Core Does Not Cause Steatosis by Increasing DGAT1 Activity or TG Synthesis
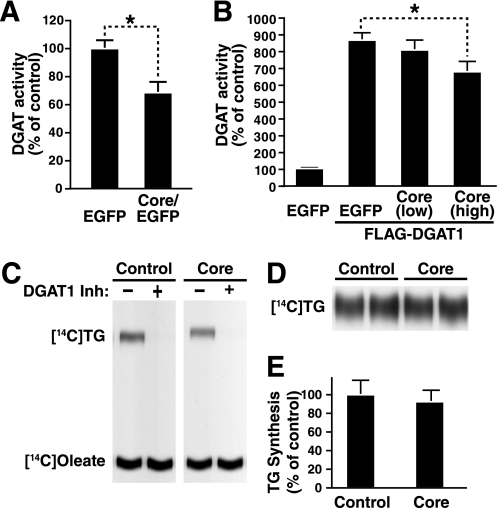
Given that DGAT1 was required for core-induced hepatic steatosis and because core and DGAT1 physically interact (27), we tested whether DGAT1 activity was increased in core-expressing livers as a possible mechanism contributing to steatosis. Microsomes from the livers of WT mice that had been injected with core-expressing or control EGFP-expressing virus were assayed for DGAT1 activity. Surprisingly, there was no increase in DGAT1 activity in core-expressing livers, but rather less DGAT1 activity than in controls (Fig. 4A).
FIGURE 4.
HCV core decreases DGAT1 activity in mice and in cultured cells. A, WT mice were injected with core/EGFP- or EGFP-expressing adenovirus. Livers were harvested at 4 days after infection, and in vitro DGAT activity assays were performed with isolated microsomal fractions (mean ± S.E.; nEGFP = 6, ncore/EGFP = 11; *, p < 0.05, Student's t test). B, in vitro DGAT activity assay of cell lysates from HEK293T cells transfected with constructs expressing FLAG-DGAT1 and core. Co-transfections were performed with low (DGAT1/core = 3:1) or high (DGAT1/core = 1:1) concentrations of core plasmids. Extracted lipids were loaded on a thin layer chromatography plate, analyzed by autoradiography, and quantified using the Bioscan AR-2000 instrument (mean ± S.D.; n = 3; *, p < 0.05, Student's t test). C, in vitro DGAT activity assays of cell lysates prepared from Huh7 cells transduced with lentiviral vectors expressing EGFP (control) or core-IRES-EGFP (core). Assays were performed in the presence or absence of the DGAT1 inhibitor. Extracted lipids were loaded on a thin layer chromatography plate and analyzed by autoradiography. D, Huh7 cells transduced with lentiviral vectors expressing EGFP (control) or core-IRES-EGFP (core) were incubated with radiolabeled oleate to quantify cellular TG synthesis. Extracted lipids were loaded on a thin layer chromatography plate and analyzed by autoradiography. E, quantification of a TG synthesis experiment performed as in D (mean ± S.D.; n = 6).
Core expression also moderately decreased the activity of co-expressed DGAT1 enzyme in co-transfection assays in HEK293T cells (Fig. 4B). In cultured hepatoma cells, no difference was observed between in vitro DGAT1 activity in core-expressing cells and DGAT1 activity in EGFP-expressing cells, although treatment with a DGAT1 inhibitor efficiently suppressed the activity in both cell lines (Fig. 4C). Because in vitro assays for DGAT1 activity are performed in the presence of fixed substrate concentrations to measure Vmax enzyme activity, these results may not represent the conditions in cells. We therefore analyzed TG synthesis in cells by measuring the incorporation of radiolabeled oleic acid into TG in core-expressing and EGFP-expressing cells. TG synthesis was unchanged with core expression (Fig. 4, D and E). Collectively, these data exclude enhanced TG synthesis as a cause of core-induced steatosis. They also uncover an unexpected core-induced decrease in DGAT1 activity under certain experimental conditions.
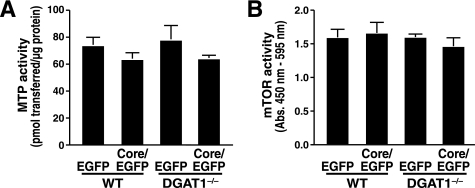
MTP inhibition is one of the mechanisms described for core-induced steatosis in core-transgenic mice, and it also represents a mechanism that is independent of TG synthesis (16). We therefore assessed MTP activity in liver homogenates of WT and DGAT1−/− mice expressing core. We observed a slight but not significant decrease in MTP activity in core-expressing compared with control mice in both genetic backgrounds (WT and DGAT1−/−, Fig. 5A). Activation of mTOR has been shown to cause core-induced down-regulation of IRS1–1 in hepatoma cells in a genotype-specific manner (26), and mTOR signaling in turn is involved in the regulation of lipolysis (39). Because our studies were performed with genotype 1b, we measured mTOR activity in the livers of WT and DGAT1−/− mice-expressing core. We could not detect any effect of core expression on mTOR activity in mouse liver (Fig. 5B).
FIGURE 5.
Effect of HCV core expression on MTP and mTOR activity. WT and DGAT1−/− mice were injected with core/EGFP- or EGFP-expressing adenovirus. Livers were harvested at 4 days after infection. A, in vitro MTP activity assays of liver homogenates (mean ± S.E.; n = 4). B, in vitro activity assays of immunoprecipitated mTOR (mean ± S.E.; n = 4). No significant changes were observed using two-way ANOVA.
Core Reduces Turnover of LD TGs
Because steady-state levels of cellular TGs result from the equilibrium of TG synthesis and breakdown, we focused next on the possibility that core expression in cells may reduce TG turnover. LD-binding proteins are known to affect TG breakdown. In adipocytes, coating of LDs with perilipin prevents access of hormone-sensitive lipase to these droplets during the fed state (40). Similarly, ectopic expression of another abundant LD-binding protein, ADRP in HEK293T cells, causes accumulation of TGs by slowing their turnover (41).
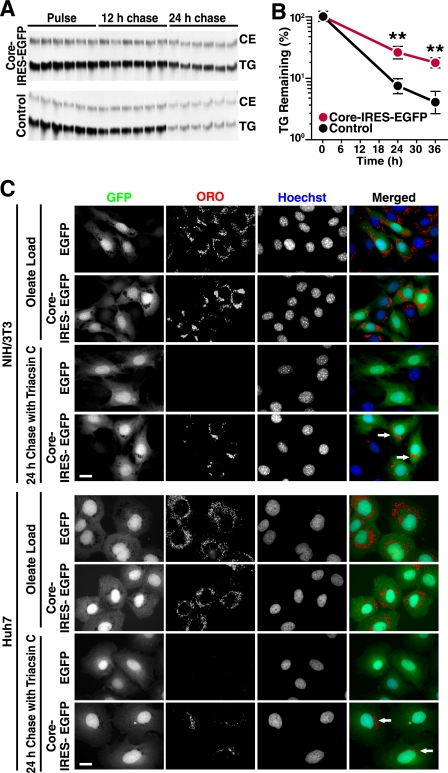
To test whether core expression has a stabilizing effect on LDs, we used an established lipolysis assay where NIH/3T3 fibroblasts are labeled with radiolabeled oleate and then chased in normal medium in the presence of triacsin C, a drug that prevents synthesis of TGs by inhibiting acyl-CoA synthetases (38). At the end of the labeling period, core-expressing and control NIH/3T3 cells had incorporated similar levels of 14C-radiolabeled oleic acid, confirming that in vivo TG synthesis rates were not affected by core (Fig. 6A). In contrast, after oleate removal and treatment with triacsin C, the decline in TG levels was significantly delayed in core-transduced as compared with control cells, indicating that core inhibits the rate of TG breakdown in cells (Fig. 6, A and B).
FIGURE 6.
HCV core expression delays TG breakdown. A, TG turnover assay in NIH/3T3 cells transduced with lentiviral vectors expressing EGFP (control) or core-IRES-EGFP (core). Cells were loaded with radiolabeled oleate and chased in the presence of triacsin C to inhibit esterification of fatty acids, including the potential re-esterification of fatty acids released during the chase. Extracted lipids were examined by thin layer chromatography. CE, cholesterol ester. B, quantification of a TG turnover assay performed as in A (mean ± S.D.; n = 6; **, p < 0.01, Student's t test). C, epifluorescence microscopy of NIH/3T3 or Huh7 cells transduced with lentiviral vectors expressing EGFP or core-IRES-EGFP, loaded with oleate, and chased in the presence of triacsin C for 24 h. Cells were stained with ORO (LDs) and Hoechst (Nuclei). Single channels are shown in black and white; the merged images are pseudocolored with GFP in green, ORO in red, and Hoechst in blue. (Scale bars, 20 μm.) Arrows point to core/EGFP-expressing cells retaining LDs at the end of the chase period.
To visualize this effect in cells, we performed a similar experiment with non-labeled oleate and monitored LD content by fluorescence microscopy using ORO staining. As expected, oleate treatment induced LD formation both in core-EGFP-expressing and EGFP-expressing control cells (Fig. 6C). At the end of the chase period in the presence of triacsin C (24 h), groups of LDs were still visible in core-expressing (EGFP-positive) cells, although no LDs could be detected in neighboring uninfected (EGFP-negative) or in control transduced cell lines, lacking core but expressing EGFP. Similar results were obtained in Huh7 hepatoma cells permissive for HCV infection (Fig. 6C). These data demonstrate that the HCV core protein stabilizes LDs by reducing TG turnover.
Ability of Core to Access LDs Is Linked to Its Ability to Inhibit LD Turnover
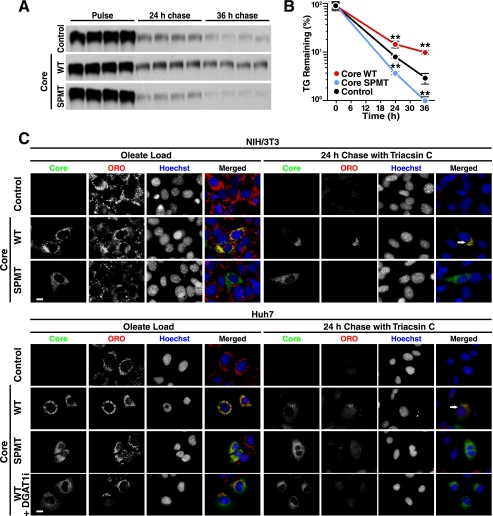
Because core requires DGAT1 to access LDs in cells (27), we examined whether the localization of core at LDs is required for the delay in LD turnover. To address this question, we analyzed a signal peptide mutant (SPMT) form of core that is not properly processed and cannot localize to LDs (15). The SPMT mutant did not delay TG breakdown in radiolabeled pulse-chase experiments as observed with WT core, confirming that the localization of core to LDs is critical for the anti-lipolytic effects of core (Fig. 7, A and B). In fact, for reasons that are not clear, the expression of the SPMT mutant in cells increased turnover of TGs when compared with control-transduced cells (Fig. 7, A and B). Importantly, by immunostaining, LDs remaining after the chase period were all coated with WT core protein, whereas in cells expressing the SPMT mutant, core was found in a reticular staining pattern reflecting ER, and no droplets remained after a 24-h chase period (Fig. 7C).
FIGURE 7.
Migration of HCV core to the LD surface is required for its ability to delay LD turnover. A and B, TG turnover assay in cells transduced with lentiviral vectors expressing WT core or the SPMT mutant of core. NIH/3T3 cells were loaded with radiolabeled oleate, washed, and chased in medium containing triacsin C. Extracted lipids were examined by thin layer chromatography (A) and quantified (B) (mean ± S.D.; n = 4; **, p < 0.01, Student's t test). C, cells were loaded with oleate in the absence or presence of 20 μm DGAT1 inhibitor and chased in the presence of triacsin C for 24 h. Epifluorescence microscopy after staining with anti-core antibodies, ORO (LDs) and Hoechst (nuclei). Single channels are shown in black and white; the merged images are pseudocolored with core in green, ORO in red, and Hoechst in blue. (Scale bar, 10 μm). Arrows point to core/EGFP expressing cells retaining LDs at the end of the chase period.
Similar results were found in Huh7 hepatoma cells expressing either core or the SPMT mutant (Fig. 7C). In addition to testing the core mutant, we also tested whether preventing core localization by using a DGAT1 inhibitor (27) would have similar effects on TG turnover in cells. In the presence of the inhibitor, ample LDs were present due to the remaining activity of DGAT2 (Fig. 7C). However, less co-localization of core and LDs was observed, which is consistent with the model that core requires DGAT1 activity to access LDs (Fig. 7C). Again, core was not able to protect LDs from lipid turnover. These data underscore the important role of DGAT1 and the localization of core at LDs for the steatogenic properties of core.
DISCUSSION
We previously showed in cultured hepatoma cells that core requires DGAT1 to access lipid droplets. Here, we report that the same is observed in livers of core-expressing mice and is linked to core-induced steatosis. Interestingly, core does not activate DGAT1 or TG synthesis to increase LD production. Instead, our findings indicate that once core is localized to LDs, it causes steatosis by inhibiting lipolysis and slowing turnover of TGs in LDs. Thus, our data support a model in which core, a single HCV protein, quite remarkably supports viral replication by at least two distinct mechanisms as follows: binding to DGAT1 for access to LDs, and subsequent inhibition of TG turnover in LDs, which causes LD accumulation to possibly promote viral assembly.
Surprisingly, microsomes from core-expressing cells had decreased, rather than increased, DGAT1 activity. The mechanism for this decreased activity is currently unknown. Given the strong interaction between core and DGAT1, core might interfere with the binding of DGAT1 substrates (e.g. diacylglycerol or acyl-CoA). Alternatively, the binding of core may impinge on the catalytic sites either directly or allosterically. We were not able to test these models by adding recombinant core to DGAT assays because of difficulties purifying soluble core protein.
How can core increase cellular TG while modestly decreasing DGAT1 activity? Using two complementary approaches, we found that core robustly stabilizes the intracellular TG pool of LDs. First, we performed a pulse-chase experiment of radiolabeled oleic acid, followed the intracellular TG pool, and found reduced turnover. Second, we found reduced catabolism of LDs by microscopy. These approaches also showed that the SPMT core mutant, which is not cleaved and therefore does not translocate to LDs, was unable to stabilize TG LDs as WT core did. In fact, cells expressing the SPMT mutant turned over LD TGs more rapidly than control-infected cells. Thus, our data collectively indicate that core causes LD accumulation by slowing TG turnover.
The precise mechanism for lipolysis inhibition by core is unknown, but several models are attractive for further testing. One model involves core slowing TG turnover by interfering with the action of lipases. Although lipolysis in hepatocytes is not as well understood as in adipocytes, a mechanism involving blocking of adipose TG lipase is attractive because global (42) and hepatocyte-specific adipose TG lipase deficiency (43) result in hepatic steatosis, whereas deletion of other TG lipases, such as TG hydrolase (44) or adiponutrin/patatin-like phospholipase domain-containing protein 3 (PNPLA3) (45), does not increase hepatic TG content in mice. A second potential mechanism is that core interferes with macroautophagy of LDs, which has been linked to LD turnover in hepatocytes (46). However, we did not observe recruitment of activated LC3 to LDs in our experimental system, arguing against this possibility (data not shown). In addition, we note that core increased the total LD and TG content in a wide variety of cell types, suggesting that the DGAT1-dependent mechanism does not require hepatocyte-specific factors.
Our data support those of others, linking polymorphisms in the LD-binding domain of genotype 3 core to steatosis development in HCV-infected patients (47). Although the strongest correlation between core and steatosis is seen with genotype 3a (48), polymorphisms in genotype 1b, the genotype employed in our study, are also frequently associated with steatosis (49, 50). These studies, along with the present study, indicate that the mechanisms of core-induced steatosis in vitro are likely to be similar to that during infection.
Our studies were based on short term expression of core and therefore do not exclude that core or HCV utilizes mechanisms other than blocking TG turnover to promote steatosis during long term infection. In line with previous results (16), we have seen a slight decrease in MTP activity in core-expressing mice. This effect might be more pronounced in transgenic mice where core is expressed over longer periods of time. Consistent with several mechanisms contributing to promote hepatic steatosis, core induces expression of SREBP1, a transcription factor involved in fatty acid synthesis, as well as several SREBP1 targets (21). However, lipogenesis was not directly measured in these studies, and SREBP1C and target mRNAs are not increased in steatotic HCV patients (51), making this mechanism uncertain.
Our in vivo studies show that DGAT1 is required for the development of hepatic steatosis in mice expressing core in the liver via adenovirus. Furthermore, DGAT1 was required for core localization to LDs in mouse liver, as revealed by immunoblot analysis of subcellular fractions. We previously showed that some, but not all, causes of hepatic steatosis are DGAT1-dependent (32). The steatosis that accompanies lipodystrophy or treatment with the liver X receptor agonist T0901317 is DGAT1-independent, whereas fasting-induced steatosis and diet-induced obesity-associated steatosis are DGAT1-dependent (32). The molecular basis for how the different DGATs relate to specific causes of steatosis remains unclear.
DGAT1 inhibitors are currently being evaluated in clinical trials for metabolic disease. Our previous findings demonstrating inhibition of viral replication with DGAT1 inhibitors (27) and our current data demonstrating prevention of core-induced steatosis via both genetic and pharmacological inhibition of DGAT1 support the model that DGAT1 inhibitors may be useful in the treatment of HCV infection and its associated hepatic steatosis.
Acknowledgments
We thank the Gladstone Histology Core for the preparation of histological liver sections, Gary Howard and Jill Dunham for editorial assistance, Marina Vayner for assistance with animal husbandry, and members of the Farese and Ott laboratories and Tobias Walther for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R56 AI085056 (to M. O.) and R01-DK056084 (to R. V. F.), Grant DK081680 (Mentored Clinical Scientist Research Career Development Award) from NIDDK (to C. H.), and Grant CO6 RR-018928 from NCRR (to Gladstone animal facilities). This work was also supported by the Hellman Family Foundation (to M. O.), Pilot/Feasibility Grant P30 DK026743 from the University of California, San Francisco, Liver Center (to C. H. and E. H.), fellowship from the Human Frontiers Science Program (to E. H.), and the J. David Gladstone Institutes.
- HCV
- hepatitis C virus
- LD
- lipid droplet
- MTP
- microsomal triglyceride transfer protein
- mTOR
- mammalian target of rapamycin
- TG
- triacylglycerol
- ORO
- Oil-Red-O
- ADRP
- adipocyte differentiation-related protein
- ER
- endoplasmic reticulum
- ANOVA
- analysis of variance
- EGFP
- enhanced GFP
- DGAT
- diacylglycerol acyltransferase
- SPMT
- signal peptide mutant.
REFERENCES
- 1. Asselah T., Rubbia-Brandt L., Marcellin P., Negro F. (2006) Gut 55, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castéra L., Hézode C., Roudot-Thoraval F., Bastie A., Zafrani E. S., Pawlotsky J. M., Dhumeaux D. (2003) Gut 52, 288–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohata K., Hamasaki K., Toriyama K., Matsumoto K., Saeki A., Yanagi K., Abiru S., Nakagawa Y., Shigeno M., Miyazoe S., Ichikawa T., Ishikawa H., Nakao K., Eguchi K. (2003) Cancer 97, 3036–3043 [DOI] [PubMed] [Google Scholar]
- 4. Adinolfi L. E., Gambardella M., Andreana A., Tripodi M. F., Utili R., Ruggiero G. (2001) Hepatology 33, 1358–1364 [DOI] [PubMed] [Google Scholar]
- 5. Rubbia-Brandt L., Quadri R., Abid K., Giostra E., Malé P. J., Mentha G., Spahr L., Zarski J. P., Borisch B., Hadengue A., Negro F. (2000) J. Hepatol. 33, 106–115 [DOI] [PubMed] [Google Scholar]
- 6. Kumar D., Farrell G. C., Fung C., George J. (2002) Hepatology 36, 1266–1272 [DOI] [PubMed] [Google Scholar]
- 7. Poynard T., Ratziu V., McHutchison J., Manns M., Goodman Z., Zeuzem S., Younossi Z., Albrecht J. (2003) Hepatology 38, 75–85 [DOI] [PubMed] [Google Scholar]
- 8. Sanyal A. J. (2011) Liver Int. 31, Suppl. 1, 23–28 [DOI] [PubMed] [Google Scholar]
- 9. Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., Koike K. (1998) Nat. Med. 4, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 10. Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., Koike K. (1997) J. Gen. Virol. 78, 1527–1531 [DOI] [PubMed] [Google Scholar]
- 11. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 12. Boulant S., Targett-Adams P., McLauchlan J. (2007) J. Gen. Virol. 88, 2204–2213 [DOI] [PubMed] [Google Scholar]
- 13. Roingeard P., Hourioux C., Blanchard E., Prensier G. (2008) Histochem. Cell Biol. 130, 561–566 [DOI] [PubMed] [Google Scholar]
- 14. Selby M. J., Choo Q. L., Berger K., Kuo G., Glazer E., Eckart M., Lee C., Chien D., Kuo C., Houghton M. (1993) J. Gen. Virol. 74, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 15. McLauchlan J., Lemberg M. K., Hope G., Martoglio B. (2002) EMBO J. 21, 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perlemuter G., Sabile A., Letteron P., Vona G., Topilco A., Chrétien Y., Koike K., Pessayre D., Chapman J., Barba G., Bréchot C. (2002) FASEB J. 16, 185–194 [DOI] [PubMed] [Google Scholar]
- 17. Huang H., Sun F., Owen D. M., Li W., Chen Y., Gale M., Jr., Ye J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang K. S., Jiang J., Cai Z., Luo G. (2007) J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F. V. (2008) J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaguchi A., Tazuma S., Nishioka T., Ohishi W., Hyogo H., Nomura S., Chayama K. (2005) Dig. Dis. Sci. 50, 1361–1371 [DOI] [PubMed] [Google Scholar]
- 21. Kim K. H., Hong S. P., Kim K., Park M. J., Kim K. J., Cheong J. (2007) Biochem. Biophys. Res. Commun. 355, 883–888 [DOI] [PubMed] [Google Scholar]
- 22. Moriishi K., Mochizuki R., Moriya K., Miyamoto H., Mori Y., Abe T., Murata S., Tanaka K., Miyamura T., Suzuki T., Koike K., Matsuura Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka N., Moriya K., Kiyosawa K., Koike K., Gonzalez F. J., Aoyama T. (2008) J. Clin. Invest. 118, 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsutsumi T., Suzuki T., Shimoike T., Suzuki R., Moriya K., Shintani Y., Fujie H., Matsuura Y., Koike K., Miyamura T. (2002) Hepatology 35, 937–946 [DOI] [PubMed] [Google Scholar]
- 25. Clement S., Peyrou M., Sanchez-Pareja A., Bourgoin L., Ramadori P., Suter D., Vinciguerra M., Guilloux K., Pascarella S., Rubbia-Brandt L., Negro F., Foti M. (2011) Hepatology 54, 38–49 [DOI] [PubMed] [Google Scholar]
- 26. Pazienza V., Clément S., Pugnale P., Conzelman S., Foti M., Mangia A., Negro F. (2007) Hepatology 45, 1164–1171 [DOI] [PubMed] [Google Scholar]
- 27. Herker E., Harris C., Hernandez C., Carpentier A., Kaehlcke K., Rosenberg A. R., Farese R. V., Jr., Ott M. (2010) Nat. Med. 16, 1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr. (2008) J. Lipid Res. 49, 2283–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris C. A., Haas J. T., Streeper R. S., Stone S. J., Kumari M., Yang K., Han X., Brownell N., Gross R. W., Zechner R., Farese R. V., Jr. (2011) J. Lipid Res. 52, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., Rothstein S. J., Mullen R. T., Dyer J. M. (2006) Plant Cell 18, 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi C. S., Savage D. B., Kulkarni A., Yu X. X., Liu Z. X., Morino K., Kim S., Distefano A., Samuel V. T., Neschen S., Zhang D., Wang A., Zhang X. M., Kahn M., Cline G. W., Pandey S. K., Geisler J. G., Bhanot S., Monia B. P., Shulman G. I. (2007) J. Biol. Chem. 282, 22678–22688 [DOI] [PubMed] [Google Scholar]
- 32. Villanueva C. J., Monetti M., Shih M., Zhou P., Watkins S. M., Bhanot S., Farese R. V., Jr. (2009) Hepatology 50, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., Bhanot S. (2005) Hepatology 42, 362–371 [DOI] [PubMed] [Google Scholar]
- 34. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., Erickson S. K., Farese R. V., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 37. Cases S., Stone S. J., Zhou P., Yen E., Tow B., Lardizabal K. D., Voelker T., Farese R. V., Jr. (2001) J. Biol. Chem. 276, 38870–38876 [DOI] [PubMed] [Google Scholar]
- 38. Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., Londos C. (2000) J. Biol. Chem. 275, 38486–38493 [DOI] [PubMed] [Google Scholar]
- 39. Zhang C., Yoon M. S., Chen J. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E862–E868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 41. Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. (2007) J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 42. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 43. Wu J. W., Wang S. P., Alvarez F., Casavant S., Gauthier N., Abed L., Soni K. G., Yang G, Mitchell G. A. (2011) Hepatology 54, 122–132 [DOI] [PubMed] [Google Scholar]
- 44. Wei E., Ben Ali Y., Lyon J., Wang H., Nelson R., Dolinsky V. W., Dyck J. R., Mitchell G., Korbutt G. S., Lehner R. (2010) Cell Metab. 11, 183–193 [DOI] [PubMed] [Google Scholar]
- 45. Basantani M. K., Sitnick M. T., Cai L., Brenner D. S., Gardner N. P., Li J. Z., Schoiswohl G., Yang K., Kumari M., Gross R. W., Zechner R., Kershaw E. E. (2011) J. Lipid Res. 52, 318–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jhaveri R., McHutchison J., Patel K., Qiang G., Diehl A. M. (2008) J. Infect. Dis. 197, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abid K., Pazienza V., de Gottardi A., Rubbia-Brandt L., Conne B., Pugnale P., Rossi C., Mangia A., Negro F. (2005) J. Hepatol. 42, 744–751 [DOI] [PubMed] [Google Scholar]
- 49. Sumida Y., Kanemasa K., Hara T., Inada Y., Sakai K., Imai S., Yoshida N., Yasui K., Itoh Y., Okanoue T., Yoshikawa T. (2011) J. Gastroenterol. Hepatol. 26, 836–842 [DOI] [PubMed] [Google Scholar]
- 50. Tachi Y., Katano Y., Honda T., Hayashi K., Ishigami M., Itoh A., Hirooka Y., Nakano I., Samejima Y., Goto H. (2010) Liver Int. 30, 554–559 [DOI] [PubMed] [Google Scholar]
- 51. McPherson S., Jonsson J. R., Barrie H. D., O'Rourke P., Clouston A. D., Powell E. E. (2008) J. Hepatol. 49, 1046–1054 [DOI] [PubMed] [Google Scholar]