Abstract
Escherichia coli has closely related amino acid chemoreceptors with distinct ligand specificity, Tar for l-aspartate and Tsr for l-serine. Crystallography of the ligand-binding domain of Tar identified the residues interacting with aspartate, most of which are conserved in Tsr. However, swapping of the nonconserved residues between Tsr and Tar did not change ligand specificity. Analyses with chimeric receptors led us to hypothesize that distinct three-dimensional arrangements of the conserved ligand-binding residues are responsible for ligand specificity. To test this hypothesis, the structures of the apo- and serine-binding forms of the ligand-binding domain of Tsr were determined at 1.95 and 2.5 Å resolutions, respectively. Some of the Tsr residues are arranged differently from the corresponding aspartate-binding residues of Tar to form a high affinity serine-binding pocket. The ligand-binding pocket of Tsr was surrounded by negatively charged residues, which presumably exclude negatively charged aspartate molecules. We propose that all these Tsr- and Tar-specific features contribute to specific recognition of serine and aspartate with the arrangement of the side chain of residue 68 (Asn in Tsr and Ser in Tar) being the most critical.
Keywords: Bacterial Signal Transduction, Chemotaxis, Crystal Structure, Protein Structure, Receptors
Introduction
Specific molecular recognition is of vital importance for virtually any function of biological macromolecules, including proteins involved in signal transduction. Understanding the mechanisms underlying such specific interactions is not only a challenge in basic biology but also provides opportunities for wide applications. Here, we focused on the closely related transmembrane chemoreceptors Tar and Tsr of Escherichia coli that mediate attractant responses to amino acids (for reviews see Refs. 1–4). Their ligand recognition is precise, Tar for l-aspartate and Tsr for l-serine (5–7). For example, Tar can mediate an attractant response to aspartate at concentrations as low as 100 nm but not to serine even at 10 mm.
The chemoreceptors have a common membrane topology as follows: from the amino terminus, the first transmembrane domain (TM1), the periplasmic ligand-binding domain, the second transmembrane domain (TM2), the HAMP domain, and the kinase control module. Regardless of the ligand occupancy state, the chemoreceptor exists as a homodimer (8) that forms, through its kinase control module, a ternary complex with the adaptor protein CheW and the histidine kinase CheA (9). An unliganded receptor activates CheA, which phosphorylates itself at His-48, and transfers the phosphoryl group to Asp-57 of CheY. Phospho-CheY binds to the flagellar motor, which otherwise rotates counterclockwise to induce its clockwise rotation. Attractant binding to the receptor inhibits the CheA activity, thereby reducing the cellular concentration of phospho-CheY and hence augmenting the counterclockwise flagellar rotation.
The high resolution structure of the ligand-binding domain of Tar has been determined both in the presence and absence of aspartate (10–15). The ligand-binding domain of each subunit consists of two long and two short α-helices to form a four-helix bundle. In the dimer, the first (α1 and α1′) and the fourth (α4 and α4′) helices form a quasi four-helix bundle contiguous to the transmembrane helices, and the α4-TM2 helix is thought to be a mobile element that plays a critical role in transmembrane signaling (16). The Tar dimer has two nonoverlapping ligand-binding pockets at the subunit interface that are located near the apex of the molecule about 80 Å away from the membrane plane and placed symmetrically around the 2-fold symmetry axis. Genetic analyses of Tar showed that Arg-64, Arg-69, Arg-73, Tyr-149, and Thr-154 are responsible for aspartate sensing but not the other general receptor functions (17–20). These residues indeed turned out to interact with the ligand aspartate in the crystal structure of the aspartate-bound ligand-binding domain of Tar. Structural similarities of the ligand-binding pocket between Tar and the trp repressor of E. coli that binds tryptophan have been reported (21). Yeh et al. (13) refined the three-dimensional structure and revealed that Ser-68 is also in contact with the β-carboxyl group of the ligand aspartate through a water molecule. Similarly, substitutions at Arg-64 and Thr-156 of Tsr (corresponding to Arg-64 and Thr-154 of Tar) specifically affect serine sensing (6, 22). The three-dimensional structure of the ligand-binding domain of Tsr, however, has not been determined.
Tar and Tsr are highly homologous even in their relatively diverse periplasmic domains (32% identity) (Fig. 1). In particular, the Arg triplet (positions 64, 69, and 73) of Tar is perfectly conserved in Tsr, which does not bind aspartate. Therefore, it is reasonable to assume that Tar and Tsr recognize their amino acid ligands essentially by a common mechanism. What is the nature of the specificity in their molecular recognition that enables discrimination between a carboxyl and a hydroxyl group at the β-position? Based on the homology modeling of the ligand-binding domain of Tsr, Jeffery and Koshland (23) suggested that a pair of corresponding residues (Phe-151 of Tsr and Tyr-149 of Tar) play important roles in ligand specificity. Yeh et al. (13) carried out energy calculations and speculated that residues Tyr-149, Ser-68, and Ile-65 of Tar (and corresponding Tsr residues, Phe-151, Asn-68, and Asn-65) are responsible for ligand specificity. The former two residues participate in the binding to the β-carboxyl group of aspartate through water molecules. However, no experimental study has been carried out to examine this hypothesis, and therefore the mechanism for ligand specificity in these amino acid chemoreceptors remains to be elucidated.
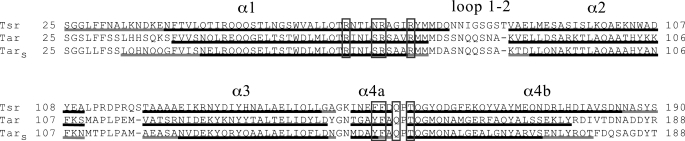
FIGURE 1.
Amino acid sequence alignment of the ligand-binding domains of E. coli Tsr (Tsr), E. coli Tar (Tar), and Salmonella enterica Tar (TarS). The sequences are aligned using ClustalW. The regions of α-helix are underlined, and the conserved α-helical regions in all known structures are highlighted in black. The ligand-binding residues are in gray boxes.
In this study, we first examined the roles of these nonconserved residues in ligand specificity by exchanging them between Tar and Tsr individually or together. Because this attempt failed to alter ligand specificity, we then constructed various chimeric receptors from Tar and Tsr to determine the region responsible for ligand specificity. This was further tested by random swapping experiments, in which the residues of Tsr were replaced randomly by the corresponding Tar-derived residues, and the mutant library was screened for aspartate-sensing receptors. Based on these results, we hypothesized that the arrangement of the nonconserved ligand-binding residues are important for ligand specificity. To test this hypothesis, we crystallized the periplasmic fragment of Tsr and determined the arrangement of the serine-binding residues of Tsr. The interpretation of the crystal structure was also verified by the ligand binding assay with isothermal titration calorimetry (ITC).4
EXPERIMENTAL PROCEDURES
Construction of Plasmids for Mutational Analyses
The plasmids encoding Tsr (pFH5) and Tar (pFH2), derived from pSU18 (24), are the parents for all other plasmids encoding mutant receptors. In these plasmids, unique PstI (corresponding to Leu-61 and Gln-62) and SacI (Glu-89 and Leu-90 of Tsr and Glu-88 and Leu-89 of Tar) sites have been introduced into the tsr and tar genes without changing the amino acid sequences. An Eco105I site was introduced into each of the tsr and tar genes with a single substitution of Tsr at position 73–74, Arg-Tyr to Leu-Asn. These restriction sites were used to construct the chimeric receptor genes, tasr, tsar, tasar, and tsasr, from pFH2 and pFH5.
A part of the tsr gene encoding residues 26–190 of Tsr was amplified by PCR using a set of primers, 5′-CGCGGATCCGGCGGTCTGTTCTTTAATGCC-3′(forward) and 5′-CCGGAATTCTCAGCTGTAGGAGGCATTGTTATCGC-3′ (reverse), introducing a BamHI site to the 5′-end and an EcoRI site to the 3′-end (hereafter the restriction sites are underlined in the primer sequences). The resulting fragment was cut with these restriction enzymes and cloned into the vector pGEX6p-2 (GE Healthcare) to express the ligand-binding fragment of Tsr fused to the carboxyl terminus of glutathione S-transferase (GST). The plasmid encoding the GST-fused Tar fragment (residues 25–188) was constructed similarly using the DNA fragment amplified with another set of primers, 5′-CGCGGATCCGGCAGTCTGTTTTTTTCTTC-3′ (forward) and 5′-CCGGAATTCTCATCGGTAATCATCTGCGTTGTC-3′ (reverse). The Cys-36 mutants of Tsr and Tar were constructed by site-directed mutagenesis. For the His6-tagged fragments, modified reverse primers with six ATG sequences inserted after the EcoRI site were used (note that CAU is a codon for histidine).
Random Swapping Experiments
We designed a set of degenerate oligonucleotides, 5′-GAGGCTGCAGACGCGTAWCAMCCTCARTCGCKCGGSTRTCCGCWWSATGATGGAT-3′ (forward) and 5′-GGAGGAGCTCAGCAACTKTGGMGYYRCTTYSTWKATTGYTCKRATCCATCAT-3′ (reverse), to create all possible combinations of amino acid residues in the 65–89 region that are not conserved between Tsr and Tar (illegitimate nucleotide codes used are as follows: K (keto), G or T; M (amino), A or C; R (purine), A or G; S (strong), C or G; W (weak), A or T; Y (pyrimidine), C or T). Without adding any DNA template, PCRs were carried out with the set of primers, and the resulting double-stranded DNA fragment was cloned between the PstI and SacI sites of pFH5. The library of the mutant genes was introduced into HCB339 cells (25), which lack all four chemoreceptors, and the resulting transformants were screened for taxis toward aspartate using aspartate-minimal semisolid agar.
Temporal Stimulation Assays
The ligand sensing ability of a particular receptor was examined by temporal stimulation assay essentially as described previously (26). Cells expressing the relevant receptor were grown at 30 °C in tryptone/glycerol broth (1% tryptone, 0.5% NaCl, 0.5% (w/v) glycerol) supplemented with 25 μg/ml chloramphenicol. Cells were harvested at the late exponential phase, washed with and resuspended in MLM medium (10 mm potassium phosphate buffer (pH 7.0), 0.1 mm EDTA, 10 mm l-lactate, 0.1 mm l-methionine), and incubated at room temperature. Swimming patterns of the cells were observed under a dark field optical microscope immediately after the addition of a ligand. Smoothly swimming fractions of cells were measured by using an image analyzer (Hamamatsu Photonics, Argus-10).
Expression and Purification of Periplasmic Fragment of Tsr or Tar
An overnight culture of receptor-less HCB436 cells (thr(Am)-1 leuB6 thi-1 LacY1 ara-14 xy-15 tsx-78 tonA31 rpsL136 his-4 mtl-1 metF159(Am) Δ(tar-cheB)2234 Δtsr-7021 trg-100 zdb::Tn5) expressing each fusion protein was diluted 1:100 with fresh LB medium supplemented with 50 μg/ml ampicillin and 0.1 mm isopropyl β-d-1-thiogalactopyranoside. Cells were grown with vigorous shaking at 37 °C for 6 h (for Tsr) or 25 °C for 8 h (for Tar). Cells were harvested, washed twice with phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4 (pH 7.3)), and disrupted by using French pressure cell press (SLM Aminco) at 600 × g. Cell debris and membrane vesicles were removed by ultracentrifugation at 120,000 × g. The supernatant was applied to a glutathione column, which was then washed with 15 column volumes of cleavage buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA). Then PreScission protease (GE Healthcare) was applied to the column to cleave off the GST tag. The eluate was concentrated up to 15 mg of protein/ml by using ultrafiltration. Nonreduced SDS-PAGE showed that the Tsr-D36C and Tar-S36C fragments form disulfide cross-linked homodimers (data not shown). Gel filtration profiles of the wild-type Tsr and Tsr-D36C fragments were similar (data not shown), suggesting that the former fragment also forms a homodimer in solution (27).
ITC Measurement
For ITC measurements, concentrations of the purified fragments were determined by using Micro BCA protein assay kit (Pierce). Titrations of Tsr or Tar fragments with l-serine or l-aspartate were carried out on a VP-ITC microcalorimeter (MicroCal Inc., Northampton, MA). During titration experiments, the fragment sample was thermostated in a stirred (307 rpm) reaction cell (1.4 ml) at 25 °C. An injection series (1.5 μl for the first injection and 3 μl each for later injections) was carried out using a 250-μl syringe filled with a serine or aspartate solution (30 injections with 240-s intervals). Data points were averaged and stored at 2-s intervals. Kd values were calculated using an Origin-ITC software package (MicroCal Inc.) based on the assumption that the Tsr or Tar dimer binds a single serine or aspartate molecule (i.e. n = 0.5).
Crystallization
Initial crystallization screening of the Tsr-D36C fragment with or without serine was performed by the sitting-drop vapor-diffusion method with the following screening kits: Crystal Screen and Crystal Screen2 (Hampton Research), and Wizard I and II (Emerald BioSystems). Crystallization drops were prepared by mixing 0.5 μl of protein solution (10 mg/ml) with 0.5 μl of reservoir solution and equilibrated to 100 μl of reservoir solution. Small needle crystals of the apo-Tsr-D36C fragment were grown from solutions containing LiSO4 or high molecular weight polyethylene glycol (PEG10000 to PEG20000) as a precipitant at neutral pH. We optimized the conditions by varying additives and the concentration of precipitant and finally obtained rod shape crystals with a typical size of 0.1 × 0.1 × 0.4 mm from a solution containing 0.1 m HEPES (pH 7.5), and 18–21% (v/v) PEG10000 at 20 °C. Crystals of the selenomethionine derivative of the Tsr-D36C fragment were obtained using the same conditions as used for the native crystals.
Because no crystal was obtained for the Tsr-D36C fragment in the presence of serine, we tried to soak the native crystals into a reservoir solution containing 1–10 mm serine. However, all the crystals were dissolved within an hour. We then screened crystallization conditions for the wild-type Tsr fragment using the same method as used for the Tsr-D36C fragment. After optimizing the condition, diffraction quality crystals with prolate shape were grown from drops prepared by mixing 1 μl of protein solution (28 mg/ml) containing 2 mm serine with the equal quantity of reservoir solution containing 0.1 m acetate buffer (pH 5.0), 80–100 mm ammonium acetate, and 12–14% (v/v) PEG1000 at 20 °C.
Structure Determination
X-ray diffraction data were collected at SPring-8 (Harima, Japan) beamline BL41XU. Crystals were soaked in a solution containing 90% (v/v) reservoir solution and 10% (v/v) 2-methyl-2,4-pentanediol for a few seconds, immediately frozen in liquid nitrogen, and mounted in helium gas flow (Rigaku cryo-cooling device). The diffraction data were collected at 35 K on an ADSC Quantum 315 CCD detector (Area Detector Systems Corp.). The native Tsr-D36C fragment crystals belong to the monoclinic space group P21, with unit cell dimensions a = 42.5 Å, b = 55.1 Å, c = 73.3 Å, and β = 93.7°, and its selenomethionine derivative crystals showed almost same cell dimensions. The wild-type Tsr fragment serine complex crystals belong to the orthorhombic space group P21212, with unit cell dimensions a = 52.0 Å, b = 133.7 Å, c = 39.6 Å. The data were indexed, integrated, and scaled using the programs MOSFLM (28) and SCALA from the CCP4 program suite (29). The statistics of data collection are summarized in Table 1.
TABLE 1.
Summary of data collection and refinement statistics
Diffraction data were collected at SPring-8 BL41XU.
| Tsr-apo | Tsr-Ser | |||
|---|---|---|---|---|
| Data collectiona | ||||
| Space group | P21 | P21212 | ||
| Unit cell dimensions | a = 42.5 Å, b = 55.1 Å, c = 73.3 Å, β = 93.7° | a = 52.0 Å, b = 133.7 Å, c = 39.6 Å | ||
| Wavelength | 0.972 Å | 1.000 Å | ||
| Resolution range | 43.85 to 1.95 | (2.06 to 1.95) | 48.5 to 2.50 | (2.64 to 2.50) |
| Observations | 99,025 | (14,583) | 54,588 | (8,249) |
| Unique reflections | 24,467 | (3,520) | 9,819 | (1,447) |
| Completeness | 98.8% | (98.2%) | 98.4% | (100%) |
| Redundancy | 4.0 | (4.1) | 5.6 | (5.7) |
| I/σ(I) | 19.0 | (5.9) | 16.7 | (4.5) |
| Rsym | 5.6% | (21.9%) | 7.1% | (33.1%) |
| Refinement | ||||
| Resolution range | 43.85 to 1.95 Å | (2.00 to 1.95 Å) | 39.57 to 2.50 Å | (2.66 to 2.50 Å) |
| Reflections | ||||
| Working | 23,211 | (1,699) | 9,805 | (1,470) |
| Test | 1,242 | (71) | 981 | (160) |
| Rw | 21.6% | (27.0%) | 25.8% | (38.5%) |
| Rfreeb | 25.9% | (39.4%) | 28.0% | (37.2%) |
| Root mean square deviation | ||||
| Bond length | 0.013 Å | 0.010 Å | ||
| Bond angle | 1.3° | 1.7° | ||
| Ramachandran plot | ||||
| Most favored | 95.2% | 93.1% | ||
| Additionally allowed | 4.5% | 6.9% | ||
| Generously allowed | 0.3% | 0% | ||
| Disallowed | 0% | 0% | ||
| No. of atoms | ||||
| Protein | 2,544 | 2,401 | ||
| Solvent | 424 | 59 | ||
| Ligand | 14 | |||
| B-factors | ||||
| Protein | 35.7 | 60.8 | ||
| Solvent | 48.9 | 62.9 | ||
| Ligand | 64.6 | |||
a Values in parentheses are for the highest resolution shell.
b Rfree was calculated using 5% of the data set that was not used in the structural refinement.
The initial MAD phase for the apo-D36C mutant fragment crystal was calculated using SOLVE (30) and improved by density modification technique with DM (29). The initial model was built with O (31) and was refined to the native data at 1.95 Å using the program REFMAC5 (29). Then a series of “omit map” was calculated, and manual modification of the model was carried out. After several iterations of the refinement and manual modification cycles, an R factor and a free R factor were converged to 21.6 and 25.9%, respectively. The Ramachandran plot showed that 95.2 and 4.8% of the residues were located in the most favorable and allowed regions, respectively.
The structure of the wild-type Tsr fragment-bound serine was solved by molecular replacement method using program MOLREP (29). We tried the apo-Tsr D36C mutant and various Tar structures (Protein Data Bank codes 1LIH, 2LIG, 1VLS, 1VLT, and 2ASR) as search models, and the best result was obtained when the apo-N36C mutant fragment dimer was used. The model was refined using program CNS (32) and modified and completed with COOT (33). After several refinement and modification cycles, the refinement converged to an R factor of 25.8% and a free R factor of 28.0%. The Ramachandran plot indicated that 93.1 and 6.9% of the residues were located in the most favorable and allowed regions, respectively. Structural refinement statistics are summarized in Table 1.
RESULTS AND DISCUSSION
Exchange of Proposed Ligand Determinant Residues Does Not Alter Ligand Specificity
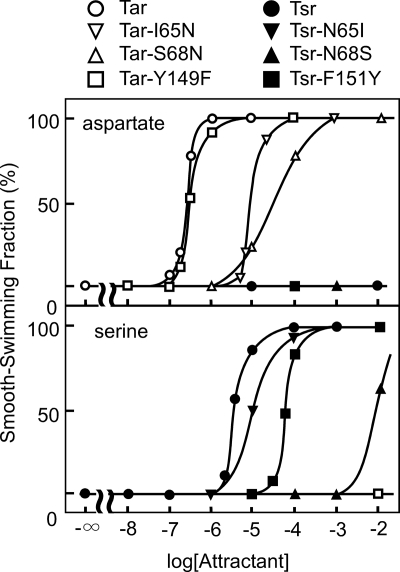
Based on the structure of the ligand-binding domain of Tar as well as the amino acid sequence comparison between Tar and Tsr, Ile-65, Ser-68, and Tyr-149 of Tar are proposed to determine ligand specificity (13). We exchanged these Tar residues with the corresponding Tsr residues individually or in combination. The resulting mutant receptors were expressed in HCB339 cells, which lack all chemoreceptors (Tsr, Tar, Trg, and Tap) (Δtsr Δ(tar-tap) trg::Tn10), and we examined them for their sensing abilities with the temporal stimulation assay. All the mutant Tsr or Tar receptors mediated repellent responses to 10% glycerol, suggesting that they retain general receptor functions. All of them, except Tar-Y149F, required more-or-less higher concentrations of serine or aspartate for responses other than the wild-type Tsr or Tar (Fig. 2). Among the single mutants, Tsr-N68S mediated a very weak response to serine, implying that Asn-68 is critical for serine sensing. The double and triple mutants, Tar-I65N/S68N, Tsr-N65I/N68S, Tar-I65N/S68N/Y149F, and Tsr-N65I/N68S/F151Y, showed severely impaired ligand sensing ability (supplemental Fig. S1). However, none of them showed altered ligand specificity, suggesting that residues that are not directly in contact with the ligands may contribute to discrimination of the ligands.
FIGURE 2.
Attractant responses mediated by the single mutants of Tar and Tsr receptors. HCB339 cells expressing wild-type or mutant receptor were pretreated with 1 m glycerol and then stimulated with various concentrations of l-aspartate (upper panel) or l-serine (lower panel). Fractions of smoothly swimming cells were measured at 30 s after the addition of an attractant.
Chimeric Receptors between Tsr and Tar Indicate the Region Essential for Ligand Specificity
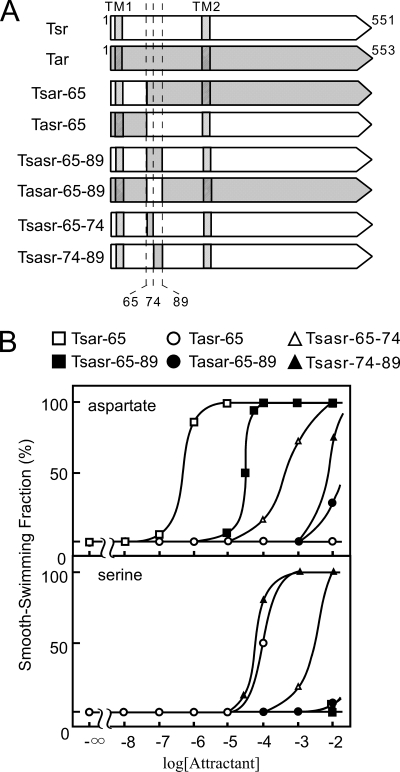
To find the region responsible for the ligand specificity, we constructed and characterized chimeric receptors of Tar and Tsr. Lee (34) showed that the chimeric receptor, in which the amino-terminal 89 residues of Tar are fused to the carboxyl-terminal residues of Tsr, named Tasr-89, recognizes aspartate, and the other one with the opposite combination, Tsar-89, recognizes serine. (Hereafter residue numbers of Tar are used for the names of chimeras to avoid confusion.) This observation suggests that the amino-terminal 89 residues determine the ligand specificity. We constructed two chimeric proteins, Tsar-65 and Tasr-65 (Fig. 3). Tsar-65 sensed aspartate with almost the same level as that of wild-type Tar but not serine. Tasr-65 sensed serine with less sensitivity compared with wild-type Tsr and not aspartate. These results suggest that the region 65–89 determines the ligand specificity. We therefore constructed the “sandwich-type” chimeric receptors (Tasar-65–89 and Tsasr-65–89), in which residues 65–89 of one receptor are replaced by the corresponding residues of the other. Cells expressing Tsasr-65–89 showed an attractant response to aspartate, although the concentration of aspartate required for a 50% response of cells expressing the chimera was approximately 2 orders higher than that for cells expressing wild-type Tar. No response to serine was observed. These results indicate that the 65–89 region of Tar is essential for aspartate sensing. Conversely, cells expressing Tasar-65–89 showed neither attractant response to aspartate nor serine when they were applied at concentrations less than 1 mm. It should be noted that this chimera mediated a repellent response to a high concentration of glycerol (data not shown), suggesting that it retains signaling function.
FIGURE 3.
Characterization of chimeric receptors between Tsr and Tar. A, schematic representation of the chimeras. Open and hatched portions indicate regions derived from Tsr and Tar, respectively. Diagonal portions represent transmembrane regions. B, aspartate- and serine-sensing abilities of the chimeric receptors. Responses of HCB339 cells expressing Tasr-65, Tsar-65, Tasar-65–89, Tsasr-65–89, Tsasr-65–74, or Tsasr-74–89 to l-aspartate (upper panel) or l-serine (lower panel) were examined as in the legend to Fig. 2.
To further narrow down the ligand specificity determinant, we constructed two more chimeric receptors (Tsasr-65–74 and Tsasr-74–89); the 65–74 and 74–89 regions correspond to the part of helix α1 and loop 1–2, respectively. Cells expressing Tsasr-65–74 responded weakly to both aspartate and serine, but aspartate gave stronger responses; the concentration of aspartate required for a 50% response was 1 order of magnitude lower than that of serine. In contrast, cells expressing Tsasr-74–89 responded weakly to serine but not to aspartate. These results suggest that the helical region of Tar may be more critical for ligand specificity than the loop.
Random Swapping of the Residues in 65–89 Region between Tsr and Tar Identify Critical Residues for Ligand Specificity
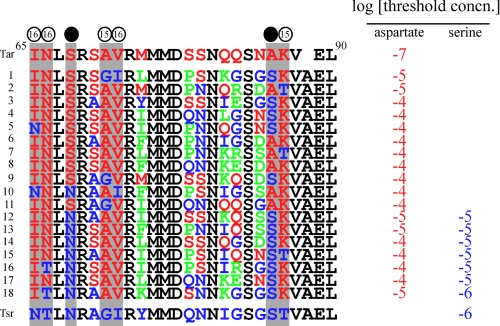
To identify residues responsible for the ligand specificity, we carried out “random swapping”; residues 65–89 of Tsr were replaced randomly by the corresponding Tar-derived residues, and the mutant Tsr library was screened for those mediating responses to aspartate. Eighteen aspartate-sensing mutants were isolated, and their ligand-sensing abilities were examined by temporal stimulation assay. Eleven mutants sensed only aspartate, but the rest responded to both aspartate and serine (Fig. 4). The nucleotide sequencing of the mutant gene revealed that the five Tar-derived residues, Ile-65, Asn-66, Ala-71, Val-72, and Lys-86 (the positions are marked with open circles in Fig. 4), are highly conserved among the mutants, but none of them was perfectly conserved. Residues at positions 68 and 85 (marked with filled circles in Fig. 4) appeared more characteristic; all of the mutants sensing both aspartate and serine have Asn and Ser at positions 68 and 85, whereas most (10 of 11) of the mutants sensing only aspartate have Ser at position 68, but they have Ala or Ser at position 85 with almost equal probability (6 and 5 of 11). Ser-68 may be critical for discrimination of aspartate from serine, whereas Asn-68 allows both aspartate and serine. In the Tar crystal, Ser-68 binds to the ligand aspartate through a water molecule, whereas Ile-65, Asn-66, Ala-71, Val-72, Ala-85, and Lys-86 do not interact with the ligand. These results suggest that residues that are not in contact with the ligand play critical roles in ligand specificity presumably by affecting the three-dimensional arrangement of the ligand-binding residues.
FIGURE 4.
Sequence alignment of the 65–90 region of the Tsr-based aspartate-sensing mutant receptors. Tar-derived, Tsr-derived, and conserved residues are shown in red, blue, and black. Non-Tsr and non-Tar residues that were introduced due to nucleotide swapping within a codon are shown in green. Positions highly conserved in the aspartate-sensing mutants are marked with open circles, and positions involved in discrimination between serine and aspartate are marked with filled circles. The number in the open circles shows the number of mutant receptors that have the Tar-derived residue at the position. Threshold concentrations of aspartate (red) or serine (blue) for attractant responses mediated by the mutant receptors are shown in the table.
Structure of the Periplasmic Domain of Tsr
To understand the precise mechanism of ligand specificity, we tried to crystallize a couple of versions of the periplasmic ligand-binding fragment of Tsr (residues 26–190) as follows: with or without a His6 tag at the carboxyl terminus or the Cys substitution for Asp-36 at the subunit interface, which allows a functional cross-linked dimer (8). A monoclinic P21 crystal of the apo-form of the His6-tagged fragment with the Cys substitution (Tsr-D36C-His6) and an orthorhombic P21212 crystal of the serine-bound form of the wild-type fragment (Tsr-WT) were obtained, and their structures were determined at 1.95 and 2.5 Å resolution, respectively. The asymmetric units of both crystals contain two polypeptide chains, which form a functional dimer related by a local pseudo-2-fold axis. Unfortunately, no crystals were grown for the apo-form of the wild-type and the serine-bound form of the His6-tagged D36C mutant.
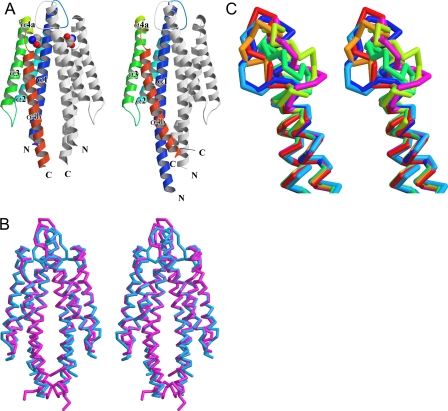
The overall structure of the Tsr fragment is basically identical to those of the Tar fragment (Fig. 5, A and B). The Tsr fragment structure consists of four long helices, α1, α2, α3, and α4b, and a short helix α4a (Fig. 5A). The interface of the dimer is constructed by α1 and α4b, which form an anti-parallel four-helix bundle structure with α1′ and α4b′ of its partner subunit (prime denotes the other subunit). The Tsr dimer has two nonoverlapping ligand-binding pockets at the subunit interface. The long helix α1 and the short helix α4a with the following loops contribute to the formation of the ligand-binding pocket. These structural features are conserved between Tsr and Tar. The corresponding Cα atoms of each Tsr subunit can be superimposed to those of Tar with root mean square deviations of 1.0–1.3 Å. The most remarkable difference between Tsr and Tar is the conformation of loop 1–2 (i.e. the loop between helices α1 and α2).
FIGURE 5.
Three-dimensional structures of the periplasmic domain of Tsr. A, ribbon models of the crystal structures of the Tsr fragments in the presence (left) and absence (right) of l-serine. The α-helices (α1, α2, α3, α4a, and α4b) are colored in blue, light blue, green, yellow, and red, respectively. B, superposition of Cα traces of Tsr (cyan) and Tar (magenta) (Protein Data Bank code 2LIG) are shown in stereo diagram. C, stereo diagram of loop 1–2. Cα traces of the apo- and serine-bound Tsr (this study) and five Tar (2LIG (B subunit), 2ASR, 1WAS, 1VLS, and 1LIH) structures are superimposed. The apo- and serine-bound Tsr are shown in blue and cyan, respectively. 2LIG, 2ASR, 1WAS, 1VLS, and 1LIH are shown in red, orange, yellow, green, and purple, respectively.
Serine-binding Sites
The Tsr dimer, like that of Tar, contains two ligand-binding pockets in the subunit interface. However, unlike the aspartate-binding sites of Tar, only one of which is occupied, both of the serine-binding pockets of Tsr bind single serine molecules, but their conformations are different. One site is filled with a clear electron density of serine, indicating tight binding of the ligand, but the other site has a rather disordered density although it can be recognized as serine. It is possible that the serine molecule at the second site may represent an intermediate state in serine binding or dissociation. This could also be related to the proposed high and low affinity serine-sensing systems of Tsr (35).
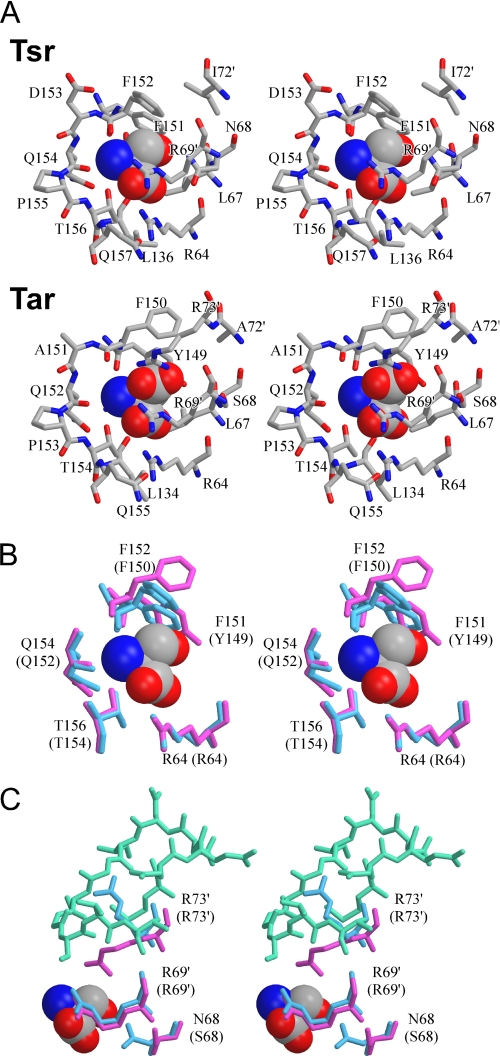
Each ligand-binding pocket is formed by Arg-64, Leu-67, Asn-68, Leu-136, Leu-139, and residues 151–157 from one subunit, and Arg-69′ and Ile-72′ from the other (Fig. 6A). The empty pocket is open wide enough to accommodate the ligand serine, but the binding of serine is supposed to trigger an induced fit. The hydrophobic residues, Leu-67, Leu-136, and Leu-139, form a back-end wall of the pocket. In the high affinity site, the α-carboxyl oxygen atoms of serine form hydrogen bonds with the ω- and the ω′-nitrogen atoms of the side chain of Arg-64 in the same plane. The extended conformation of the side chain of Arg-64 seems to be stabilized via a hydrogen bond with Gln-157, thereby providing a cavity for α-carboxyl group of the ligand. The α-amino group of serine directly interacts with Phe-151, Phe-152, Gln-154, and Thr-156 (Fig. 6B). These interactions are conserved in the Tar-aspartate complex structure (10). It should be noted, however, that the aromatic rings of Phe-151 and Phe-152 are placed in different ways from those of the counterparts in Tar (Tyr-149 and Phe-150), presumably contributing in part to restrict the pocket volume (see below for detail).
FIGURE 6.
Stereo diagrams of the ligand-binding pockets. A, close up view of the ligand-binding pockets of Tsr (upper panel) and Tar (lower panel). The ligand molecules (serine and aspartate) are shown in CPK representation. B, superposition of the Tsr and Tar residues interacting with the α-amino and α-carboxyl groups of the ligand. C, superposition of the Tsr and Tar residues interacting with the β-hydroxyl group of serine or β-carboxyl group of aspartate. B and C, a serine molecule is shown in CPK representation, and residues of Tsr and Tar are shown in blue and magenta, respectively. Residues that constitute loop1–2 of Tsr are shown in turquoise.
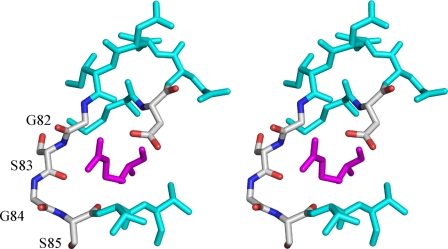
In contrast to the amino acid backbone, the groups at the β-position appear to be recognized by Tsr and Tar in a different way. The β-hydroxyl group of serine forms a hydrogen bond with the side chain of Asn-68. Arg-73′, unlike its counterpart in Tar that interacts with the β-carboxyl group of aspartate, is oriented away from the ligand serine. Indeed the side chain δ-guanidino group of Arg-73′ of Tsr is trapped by a quadruple residue sequence (Gly-82–Ser-83–Gly-84–Ser-85) in loop 1–2 and interacts with the β-carboxyl group of Asp-77 (Fig. 7), sequestering it from the ligand serine (Fig. 6C). The space where Arg-73′ is located in the Tar structure is now occupied by Phe-152, which forms a hydrophobic wall with Phe-151 and Ile-72′ just above the ligand side chain. These residues may contribute to restricting the pocket volume with Asn-68, whose side chain protrudes into the pocket and directly interacts with the ligand.
FIGURE 7.
Stereo view of the arrangement of Arg-73 and loop 1–2 in apo-Tsr. Arg-73 is highlighted in magenta, and Asp-77 and Gly-82 to Ser-85 are shown in CPK color. Others are in cyan.
Arg-69′ of Tsr, a member of the well conserved triplet, does not bind to the ligand serine, whereas the side chain of the corresponding residue Arg-69′ of Tar interacts with the β-carboxyl group of the ligand aspartate. Nevertheless, the side chain of Arg-69′ of Tsr is located around the entrance of the pocket. The apo-ligand-binding site of Tsr is wide open. The arrangements of Phe-151 and Phe-152 in the apo-structure of Tsr are similar to those of Tyr-149 and Phe-150 in both the apo- and the aspartate-bound structures of Tar. In the serine-bound form of Tsr, however, these residues slid into the ligand-binding pocket, indicating that serine binding may trigger a shrinkage of the pocket. Such a conformational change would enable the side chain of Arg-69′ to interact with the oxygen atom of the main chain carbonyl of Phe-152 through a water molecule. As a result, the entrance of the pocket is closed and the bound serine is surrounded by the Tsr molecule. This sort of induced fit might also contribute to ligand specificity.
Mechanism of Ligand Specificity
Comparison of the crystal structures of Tsr and Tar revealed how these receptors recognize and distinguish their ligands. Tsr and Tar share a common recognition mechanism for the amino acid backbone of their ligands. The α-carboxyl group is bound to the extended Arg residue, and the α-amino group interacts with the oxygen atoms of residues in helix α4a and the following short loop. The hydroxyl and carboxyl groups at β-positions of serine and aspartate are directly in contact with the side chains of Asn-68 of Tsr and Arg-73′ of Tar, respectively. These residues can thus be regarded as the primary ligand specificity determinants. In fact, substitutions of these residues impaired sensing abilities. Arg-73′, however, is conserved between Tsr and Tar. In the Tsr structure, the side chain of Arg-73′ points away from the binding pocket, by interacting with loop 1–2 as described above (Fig. 6C). Phe-152, which is also conserved, fills the space that Arg-73′ would have occupied if it adopted a Tar-like conformation and forms the compact binding pocket together with Ile-72′ and Asn-68 to accommodate the small side chain of serine. In other words, the altered conformation of loop 1–2 seems to play a key role in ligand specificity, by hanging over the pocket and trapping Arg-73′, thereby reducing the pocket volume and preventing larger molecules from entering. Significance of this conformation is demonstrated by comparing the conformations of loop 1–2 in all the available crystal structures of Tsr and Tar (Fig. 5C). Although it seems more flexible than the helices as expected, loop 1–2 of Tsr adopts significantly distinct conformations of those of the Tar counterpart. These observations can account for the results of the mutational experiments.
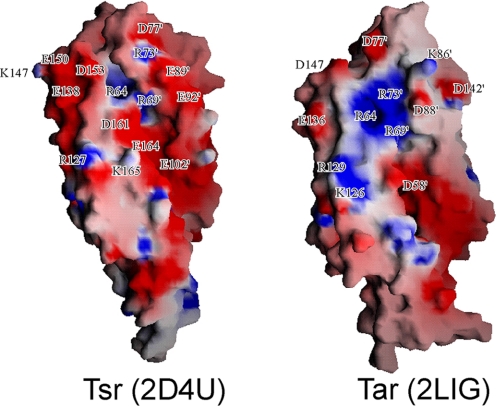
The electrostatic surface potential analysis of the periplasmic domains of Tsr and Tar suggests another mechanism for selecting ligands (Fig. 8). Tar has a positively charged surface in and around the ligand-binding pocket (13), to which negatively charged molecules have easier access. In contrast, the positively charged ligand-binding pocket of Tsr is surrounded by strong negative potentials. The positively charged pocket is partly hidden by loop 1–2. Following the loop, two residues, Glu-89′ and Glu-92′, provide strong negative charges just next to the pocket. Below the pocket entrance, Asp-161 and Glu-164 contribute to form a negative surface, whereas Lys-126 and Arg-129 of Tar form a positive surface. Around helix α4, Glu-138, Glu-150, and Asp-153 form a negatively charged patch. These “negative potential barriers” of Tsr may prevent aspartate molecules from accessing the entrance of the serine-binding pocket.
FIGURE 8.
Surface potential maps of the periplasmic domains of Tsr (left) and Tar (2LIG, right). Positive and negative potentials are shown in blue and red, respectively. 2LIG is viewed from the binding pocket that is not binding aspartate.
Why does Tar not mediate a response to serine? The ligand-binding pocket of Tsr is smaller than that of Tar as mentioned above. The latter probably has too wide an opening to hold serine stably, whereas the former is compact and well fit to serine. The smaller size of the binding pocket of Tsr was supported by the chemical modification analysis (data not shown). We replaced Thr-156 in Tsr and Thr-154 in Tar by Cys as described previously (36–38), and the resulting mutant proteins were modified with a fluorescent thiol-reacting reagent, iodoacetoamide fluorescein. Tsr-T156C was modified with much less efficiency than Tar-T154C, suggesting that Cys-156 in the ligand-binding pocket of Tsr is less accessible to aqueous solution than the corresponding residue of Tar. Thus, Tsr is supposed to favorably accommodate amino acid ligands with small side chains. In fact, Tsr can bind small amino acids, such as d-serine, glycine, alanine, and cysteine, although with lower affinities than serine (39, 40).
What Determines the Conformation of Loop 1–2?
Which residues determine the loop 1–2 conformation? The structures of this loop are almost identical between the apo- and the serine-bound forms, indicating that serine binding does not induce a significant conformational change in this region. We argued above that the carboxyl-terminal part of helix α1 may be more critical for ligand specificity than the loop because the Tsr-based chimera Tsasr-65–74, in which the helical region is replaced, mediated stronger responses to aspartate than to serine, and the other chimera Tsasr-74–89, in which the loop region is replaced, retained the ability to sense serine exclusively. Moreover, a deletion of Ala-89 from Tsr or an insertion of Ala at the corresponding position of Tar did not change ligand specificity (data not shown). These facts imply that the conformation of the loop is imposed by the constraint from the other parts of the molecule, especially helix α1.
Ligand Binding Affinities of Periplasmic Fragments of Tsr or Tar with Mutation at Position 68 or 73
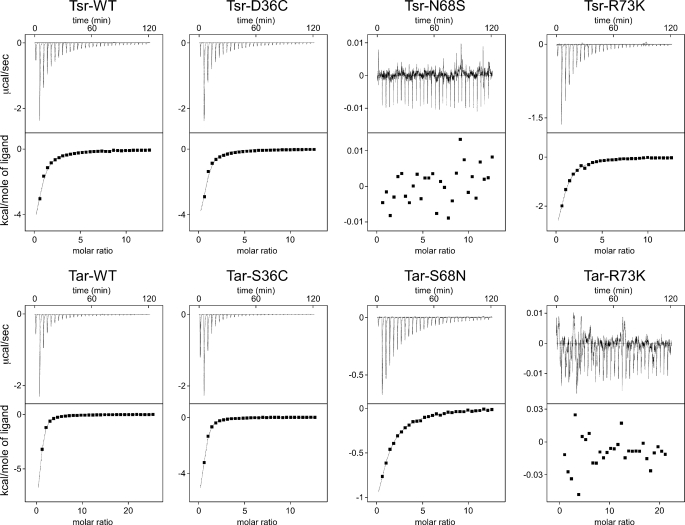
To test the proposed roles of Asn/Ser-68 and Arg-73 of Tsr and Tar, we performed ITC measurements of ligand binding. Titration curves of the wild-type and mutant Tsr and Tar fragments with their ligands are shown in Fig. 9. All of the receptor constructs, except those of wild-type Tsr and Tar, have a Cys residue at position 36 (D36C in Tsr and S36C in Tar) to facilitate dimerization and a His6 tag at the carboxyl terminus. The Tsr and Tar fragments, regardless of the Cys mutation and the His6 tag, did not bind aspartate and serine, respectively (Table 2), indicating that the original ligand specificity was retained. The dissociation constants (Kd) for the serine-Tsr (both wild-type and the D36C-His6 mutant) interaction were 20–40 μm (Table 2), indicating that the D36C mutation of Tsr scarcely affects the serine binding affinity. This Kd value is consistent with the reported value of that of full-length Tsr (5–30 μm) (6, 41). The R73K mutation of Tsr affected the Kd value and ΔH only slightly (about 50 μm), whereas aspartate binding to the Tar-R73K fragment was not detected, a result that is consistent with the prediction that the conserved residue plays different roles in ligand binding between Tsr and Tar. Moreover, the N68S mutation in Tsr severely impaired serine binding; the addition of serine to the mutant fragment caused neither release nor uptake of heat. By contrast, the S68N mutation in Tar mildly affected the Kd value. These mutations did not change the ligand specificity; Tsr-N68S and Tar-S68N did not bind aspartate and serine, respectively (Table 2), excluding the possibility that they can bind the wrong ligand without eliciting signals. These results are consistent with our speculation on the ligand specificities of Tsr and Tar; Asn-68 plays an essential role in serine binding of Tsr, whereas Arg-73 of Tar is important for aspartate binding of Tar. On the contrary, Arg-73 of Tsr and Ser-68 of Tar are insignificant for their ligand recognition.
FIGURE 9.
Isothermal titration calorimetry of the wild-type and mutant receptor fragments. The upper and lower sets show the titration data of the fragments of Tsr or its derivatives with serine and those of the fragments of Tar or its derivatives with aspartate, respectively. In each set of panels, the upper traces represent raw data, and the lower plots represent enthalpy changes per mol plotted as a function of the molar ratio of the ligand to the receptor fragment. The calculated binding parameters are listed in Table 2.
TABLE 2.
ITC measurements of the receptor fragments
| Receptor | Mutation | Titrated ligand | Kda |
|---|---|---|---|
| μm | |||
| Tsr | WT | Serine | 35.6 |
| D36C-His6 | Serine | 21.0 | |
| D36C-His6 | Aspartate | NDb | |
| D36C,N68S-His6 | Serine | ND | |
| D36C,N68S-His6 | Aspartate | ND | |
| D36C,R73K-His6 | Serine | 51.7 | |
| Tar | WT* | Aspartate | 12.8 |
| S36C-His6 | Aspartate | 6.8 | |
| S36C-His6 | Serine | ND | |
| S36C,S68N-His6 | Aspartate | 99.4 | |
| S36C,S68N-His6 | Serine | ND | |
| S36C,R73K-His6* | Aspartate | ND |
a For ITC measurements, we titrated an amino acid solution (10 mm) to a receptor fragment solution (50 μm) except for Tar-WT (25 μm) and Tar-S36C,R73K-His6 (30 μm), both of which are marked with asterisks in the table.
b ND means not determined.
Concluding Remarks
Taken together, we propose a plausible model of the ligand specificities of the two closely related amino acid receptors. First, the two key features of the serine-binding pocket of Tsr serve as initial filters. The more acidic electrostatic surface potentials around it and its smaller size prevent amino acids with acidic and larger side chains from entering into the pocket. Second, unlike their counterparts in Tar, which interact with the β-carboxyl group of the ligand aspartate, the conserved residues Arg-69 and Arg-73 in Tsr do not directly interact with the ligand serine and therefore are not directly involved in ligand recognition. Rather, the former contributes to induced fit of the serine-binding pocket, and the latter is trapped by loop 1–2, thereby sequestered from the ligand serine. Third, Asn-68, whose counterpart in Tar (Ser-68) is not in contact with the ligand aspartate, interacts directly with the β-hydroxyl group of the ligand serine, playing the principal role in precise recognition of serine. As such, distinct three-dimensional arrangements of the conserved and nonconserved ligand-binding residues are primarily responsible for the ligand specificities of these receptors.
In this study, we could not obtain apo- and serine-bound structures from the same fragment under any condition tested. Unlike the ligand-binding residues, the α1, α1′, α4b, and α4b′ helices are arranged in a substantially different way between the apo-form of the Tsr-D36C-His6 fragment and the serine-bound form of the Tsr-WT fragment. This difference in helical packing could reflect the conformational change upon transmembrane signaling, because the α4b helix is directly contiguous to the second transmembrane helix, which connects the periplasmic domain to the cytoplasmic HAMP domain, and hence it is one of the key components of signaling. However, the more compact packing in the apo-form of the mutant fragment is supposed to result at least in part from the disulfide bond formed between the α1 and α1′ helices. We therefore should refrain from arguing about the signaling mechanism at this stage. To fully understand receptor signaling, the full-length structure will be required.
Supplementary Material
Acknowledgments
We express sincere gratitude, with deep sorrow, to the late Professor M. Kasahara of Teikyo University, who died unexpectedly on November 4, 2010. Professor Kasahara provided helpful advice on random-swapping experiments and had cordial relations with M. H. and I. K. We thank K. Shimono of Matsuyama University and A. Tsuneshige of Hosei University for helpful advice on ITC measurements; N. Shimizu, M. Kawamoto, K. Hasegawa, and M. Yamamoto at SPring-8 for technical help in the use of beam lines; and K. Namba of Osaka University, T. Yakushi and S. Kojima of Nagoya University, and S. Nishiyama, T. Inaba, and Y. Sowa of Hosei University for discussions and encouragement.
This work was supported in part by grants-in-aid for scientific research and the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K. I. and I. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 2D4U and 3ATP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Stock. J. B., Surette M. G. (1996) in Escherichia coli and Salmonella typhimurium (Neidhardt F. C., Curtiss R., 3rd, Ingraham J. L., Lin E. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. eds) 2nd Ed., pp. 1103–1129, American Society for Microbiology, Washington, D. C [Google Scholar]
- 2. Szurmant H., Ordal G. W. (2004) Microbiol. Mol. Biol. Rev. 68, 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wadhams G. H., Armitage J. P. (2004) Nat. Rev. Mol. Cell Biol. 5, 1024–1037 [DOI] [PubMed] [Google Scholar]
- 4. Baker M. D., Wolanin P. M., Stock J. B. (2006) BioEssays 28, 9–22 [DOI] [PubMed] [Google Scholar]
- 5. Melton T., Hartman P. E., Stratis J. P., Lee T. L., Davis A. T. (1978) J. Bacteriol. 133, 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clarke S., Koshland D. E., Jr. (1979) J. Biol. Chem. 254, 9695–9702 [PubMed] [Google Scholar]
- 7. Hedblom M. L., Adler J. (1983) J. Bacteriol. 155, 1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Milligan D. L., Koshland D. E., Jr. (1988) J. Biol. Chem. 263, 6268–6275 [PubMed] [Google Scholar]
- 9. Gegner J. A., Graham D. R., Roth A. F., Dahlquist F. W. (1992) Cell 70, 975–982 [DOI] [PubMed] [Google Scholar]
- 10. Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr., Kim S. H. (1991) Science 254, 1342–1347 [DOI] [PubMed] [Google Scholar]
- 11. Scott W. G., Milligan D. L., Milburn M. V., Privé G. G., Yeh J., Koshland D. E., Jr., Kim S. H. (1993) J. Mol. Biol. 232, 555–573 [DOI] [PubMed] [Google Scholar]
- 12. Yeh J. I., Biemann H. P., Pandit J., Koshland D. E., Kim S. H. (1993) J. Biol. Chem. 268, 9787–9792 [PubMed] [Google Scholar]
- 13. Yeh J. I., Biemann H. P., Privé G. G., Pandit J., Koshland D. E., Jr., Kim S. H. (1996) J. Mol. Biol. 262, 186–201 [DOI] [PubMed] [Google Scholar]
- 14. Bowie J. U., Pakula A. A., Simon M. I. (1995) Acta Crystallogr. D Biol. Crystallogr. 51, 145–154 [DOI] [PubMed] [Google Scholar]
- 15. Chi Y. I., Yokota H., Kim S. H. (1997) FEBS Lett. 414, 327–332 [DOI] [PubMed] [Google Scholar]
- 16. Falke J. J., Hazelbauer G. L. (2001) Trends Biochem. Sci. 26, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolff C., Parkinson J. S. (1988) J. Bacteriol. 170, 4509–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee L., Imae Y. (1990) J. Bacteriol. 172, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mowbray S. L., Koshland D. E., Jr. (1990) J. Biol. Chem. 265, 15638–15643 [PubMed] [Google Scholar]
- 20. Gardina P., Conway C., Kossman M., Manson M. (1992) J. Bacteriol. 174, 1528–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lynch B. A., Koshland D. E., Jr. (1992) FEBS Lett. 307, 3–9 [DOI] [PubMed] [Google Scholar]
- 22. Lee L., Mizuno T., Imae Y. (1988) J. Bacteiol. 170, 4769–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jeffery C. J., Koshland D. E., Jr. (1993) Protein Sci. 2, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bartolomé B., Jubete Y., Martínez E., de la Cruz F. (1991) Gene 102, 75–78 [DOI] [PubMed] [Google Scholar]
- 25. Wolfe A. J., Conley M. P., Kramer T. J., Berg H. C. (1987) J. Bacteriol. 169, 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tatsuno I., Lee L., Kawagishi I., Homma M., Imae Y. (1994) Mol. Microbiol. 14, 755–762 [DOI] [PubMed] [Google Scholar]
- 27. Milligan D. L., Koshland D. E., Jr. (1993) J. Biol. Chem. 268, 19991–19997 [PubMed] [Google Scholar]
- 28. Leslie A. G. (1991) in Crystallographic Computing (Moras D., Podjarny A. D., Thiery J. C. eds) Vol. 5, pp. 27–38, Oxford University Press, Oxford [Google Scholar]
- 29. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 32. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Lee L. (1990) Mechanism of the Ligand Recognition of Serine and Aspartate Chemoreceptors in Escherichia coli. Ph.D. thesis, Nagoya University, Japan [Google Scholar]
- 35. Hedblom M. L., Adler J. (1980) J. Bacteriol. 144, 1048–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gomi S., Lee L., Iwama T., Imae Y. (1993) J. Biochem. 113, 208–213 [DOI] [PubMed] [Google Scholar]
- 37. Gomi S., Lee L., Iwama T., Imae Y., Kawagishi I. (1994) in Olfaction and Taste (Kurihara K., Suzuki N., Ogawa H. eds) Vol. 9, pp. 210–214, Springer-Verlag, Tokyo [Google Scholar]
- 38. Iwama T., Kawagishi I., Gomi S., Homma M., Imae Y. (1995) J. Bacteriol. 177, 2218–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacAB R. M. (1987) in Escherichia coli and Salmonella typhimurium (Neidhardt F. C., Ingraham J. L., Low K. B., Magasanik B., Schaechter M., Umbarger H. E. eds) pp. 732–759, American Society for Microbiology, Washington, D. C [Google Scholar]
- 40. Iwama T., Homma M., Kawagishi I. (1997) J. Biol. Chem. 272, 13810–13815 [DOI] [PubMed] [Google Scholar]
- 41. Lin L. N., Li J., Brandts J. F., Weis R. M. (1994) Biochemistry 33, 6564–6570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.