Background: The mechanisms of homologous recombination and recombinational repair of double-stranded DNA breaks in mitochondria are poorly understood.
Results: The yeast mitochondrial nucleoid protein, Mgm101, shares biochemical, structural, and functional similarities with the Rad52 family proteins.
Conclusion: Mgm101 is a Rad52-related molecular component involved in mtDNA recombination
Significance: This finding helps in understanding the mechanism of homologous recombination in mitochondria.
Keywords: Genetics, Mitochondria, Mitochondrial DNA (mtDNA), Yeast, Yeast Genetics
Abstract
Homologous recombination is a conserved molecular process that has primarily evolved for the repair of double-stranded DNA breaks and stalled replication forks. However, the recombination machinery in mitochondria is poorly understood. Here, we show that the yeast mitochondrial nucleoid protein, Mgm101, is related to the Rad52-type recombination proteins that are widespread in organisms from bacteriophage to humans. Mgm101 is required for repeat-mediated recombination and suppression of mtDNA fragmentation in vivo. It preferentially binds to single-stranded DNA and catalyzes the annealing of ssDNA precomplexed with the mitochondrial ssDNA-binding protein, Rim1. Transmission electron microscopy showed that Mgm101 forms large oligomeric rings of ∼14-fold symmetry and highly compressed helical filaments. Specific mutations affecting ring formation reduce protein stability in vitro. The data suggest that the ring structure may provide a scaffold for stabilization of Mgm101 by preventing the aggregation of the otherwise unstable monomeric conformation. Upon binding to ssDNA, Mgm101 is remobilized from the rings to form distinct nucleoprotein filaments. These studies reveal a recombination protein of likely bacteriophage origin in mitochondria and support the notion that recombination is indispensable for mtDNA integrity.
Introduction
Mitochondrial DNA (mtDNA)3 encodes integral components of the energy-transducing oxidative phosphorylation pathway. It is organized in vivo as protein-DNA complexes, known as mitochondrial nucleoids, with each nucleoid containing ∼2–10 copies of the genome (1–4). mtDNA is subject to constant attack by reactive oxygen species that are produced through metabolism in mitochondria. This is generally believed to be responsible for the higher mutational load in mtDNA compared with the nuclear genome (5, 6). In addition to oxidative damage, it has been suggested that replicative errors also contribute significantly to the overaccumulation of mutations in mtDNA during the lifetime of the cell (7). If unrepaired, the overaccumulated mutations in mtDNA can directly affect mitochondrial energy production and lead to severe physiological consequences, which are manifested by many human degenerative diseases (8, 9). Unfortunately, in contrast to what we have learned from the nuclear genome, our understanding of how damaged DNA is repaired in mitochondria is rather limited. So far, only a few enzymes involved in base excision repair have been documented inside the organelle (10, 11).
Homologous recombination (HR) is a DNA repair mechanism conserved from bacteriophage to humans that plays a critical role in the error-free repair of double strand breaks (DSBs) and stalled or collapsed replication forks. It is catalyzed by the Rad52 epistasis group proteins (12, 13). In conventional HR, DSBs are first processed by a nuclease into 3′ ssDNA tails. The 3′ ssDNA tails are subsequently coated by single strand DNA-binding proteins to prevent the formation of secondary structures. A recombination mediator, as exemplified by the bacterial RecO and yeast Rad52, displaces the single strand DNA-binding protein and recruits the core ATP-dependent recombinase (such as RecA in prokaryotes and its eukaryotic orthologs, Rad51) to form helical nucleoprotein filaments. The filaments then engage in homology search, strand invasion, and homologous pairing within duplex DNA templates. As a result, the invading 3′-end is used to prime DNA replication to copy the genetic information missing from the dsDNA breaks. The genome integrity is consequently restored following the resolution of recombination intermediates by different molecular strategies with or without involving DNA strand crossover (13–15). In canonical DSB repair, Rad52 has dual functions. In addition to its role as a recombination mediator for Rad51 recruitment, Rad52 also participates in ssDNA annealing, an activity critical for the capture of the second DNA end at the recombination site that generates Holliday junctions (16, 17).
Does HR, an almost universal DNA repair mechanism in all domains of life, also occur in mitochondria? Recombination between DNA markers has been well documented in yeast mitochondria (18). Shibata and co-workers (19–21) have identified the MHR1 gene that encodes a mitochondrial protein affecting gene conversion and the repair of oxidatively damaged mtDNA. Based on its ability in promoting homologous pairing between ssDNA and dsDNA duplex in vitro, Mhr1 was proposed to be an ATP-independent recombinase (22, 23). Because mutations in MHR1 have only a slight effect on the crossing over of unlinked mitochondrial genetic loci, it has been suggested that there is another pathway for homologous recombination in mitochondria (19).
We have previously identified the MGM101 gene in a genetic screen for temperature-sensitive (ts) mutants affecting mtDNA maintenance in Saccharomyces cerevisiae (24). Mature Mgm101 is a positively charged protein of 247 amino acids (25). Nunnari and co-workers (26, 27) have shown that Mgm101 is associated with actively replicating mitochondrial nucleoids and that mtDNA in mgm101 mutants is hypersensitive to several DNA-damaging agents, including ultraviolet, γ-ray irradiation, and hydrogen peroxide. These observations strongly suggested a role for Mgm101 in mtDNA repair. We have also found that the mgm101 mutant rapidly loses the wild-type (ρ+) but not the hypersuppressive ρ− mtDNA (28). These ρ− genomes contain highly recombinogenic GC-rich repeats, which may spare the requirement for Mgm101. This finding further suggested that Mgm101 may play a critical role in mtDNA repair via a recombination-based process. In this report, we show that Mgm101 is a Rad52-related protein of bacteriophage origin. The data suggest the presence of an evolutionarily conserved HR component in mitochondria that is essential for the maintenance of mtDNA integrity.
EXPERIMENTAL PROCEDURES
Growth Media, Yeast Strains, and Plasmid Construction
Complete (YP) and minimal medium (YNB) were prepared with 2% dextrose (D) or 2% glycerol plus 2% ethanol (GE). Yeast strains used in this study included M2915-7C (MATa, ade2, leu2, his4, ura3, mgm101-1ts), EAS748 (MATa, ura3-52, leu2-3,112, lys2, his3, arg8::hisG [cox2::ARG8m-Rep96], kindly provided by E. Sia, University of Rochester), CS1631 (as EAS748, but trp1Δ::mgm101N150A-kan), and CS1640 (as EAS748, but trp1Δ::mgm101N150A-kan, mgm101::URA3). The N150A, F153A, and F235A alleles of MGM101 were generated by site-directed in vitro mutagenesis using the plasmid pCXJ22-MGM101 as template. For chromosomal integration, the mutant alleles were cloned into the integrative vector, pUC-URA3/4. The resulting plasmids were linearized by digestion with StuI within the URA3 gene and integrated into the ura3 locus by selection for Ura+ transformants. To construct CS1631, the mgm101N150A allele was cloned adjacent to the kan gene in a plasmid. The mgm101N150A-kan cassette was then amplified by PCR using a pair of bipartite primers, which allowed the replacement of the chromosomal TRP1 gene in EAS748 by selecting for Trp− and G418R transformants. The wild-type MGM101 gene in CS1631 was then disrupted using an mgm101::URA3 cassette (24), which yielded CS1640.
In Vivo Repeat-mediated Deletion/Recombination Assay
The fluctuation assay for determining the rate of repeat-mediated deletions in mtDNA has been described by Sia and co-workers (29). Cells were preselected on minimal medium for Arg+ for 3 days and plated for colony formation on YPD medium for 3 days. Cells from 20 independent Arg+ colonies were then resuspended in water, and appropriate dilutions were plated on YPGE and YPD. Respiratory competent colonies (or recombinants) on YPGE were scored after growth at 30 °C for 7 days. The number of viable colonies on the YPD plates was used to deduce the total cell number plated on YPGE. The rate of recombination per cell division was estimated using the p0 method (30). For mtDNA instability and recombination rate, a two-sample t test (two-tailed) was used for statistical evaluation between the control and the mutant cells.
Expression and Purification of Mgm101, Rim1, and Mhr1 from Escherichia coli
The MGM101 ORF lacking the first 22 codons was amplified by PCR using the primers MGM101P1 (5′-CGGGGATCCGTGGTGAGTACCGGCACTAG-3′) and MGM101P2 (5′-AAAACTGCAGCTATTTATAAGGATATTCAACTTG-3′). The BamHI and PstI sites incorporated in the primers (underlined) allowed the cloning of the gene downstream of the malE sequence encoding the maltose-binding protein (MBP) in the expression vector pMal-c2e (New England Biolabs). This created the MBP-Mgm101 fusion with a linker containing a cleavage site for the PrescissionTM protease (GE Healthcare). The fusion plasmid pMalc2e-MGM101 was introduced into the E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene). Expression of fusion protein was induced with isopropyl β-d-1-thiogalactopyranoside at 0.3 mm in LB medium supplemented with 0.2% glucose, ampicillin (100 μg/ml), and chloramphenicol (50 μg/ml). Cells were cultured at 30 °C for 5 h with an A600 of ∼0.5. After centrifugation, the cell pellet was resuspended in the column buffer (20 mm Tris-HCl, pH 7.4, 50 mm glucose, and 1 mm EDTA) containing protease inhibitors (25 μm leupeptin, 10 μm pepstatin, and 0.5 mm phenylmethanesulfonyl fluoride). The cell suspension was sonicated for 30 s five times, and DNase I (50 μg/ml) was added to cell lysate and incubated at 4 °C for 2 h. The final concentration of NaCl in the crude lysate was adjusted to 500 mm, and cell debris was removed by centrifugation at 10,000 × g at 4 °C. The cell lysate was then incubated with amylose resin at 4 °C overnight. After several washes with column buffer, MBP-Mgm101 was eluted with 10 mm maltose in column buffer. The fusion protein was cleaved with the PrescissionTM protease (0.33 units/100 μg) overnight to release Mgm101. The recombinant Mgm101 thus has the sequence GPKVPEFGS added to the N terminus of the native protein. After dialysis against 10 mm sodium phosphate (pH 7.2, 4 °C, 100 mm NaCl), Mgm101 was separated from MBP and purified to homogeneity by cation exchange chromatography using a Bio-ScaleTM Mini Prep UNOsphereTM cartridge (Bio-Rad). Mgm101 was further purified by size exclusion chromatography on a calibrated Superose 6 column equilibrated with 20 mm MOPS, pH 7.0, 150 mm NaCl, 5 mm β-mercaptoethanol, 1 mm EDTA, 0.2 mm phenylmethanesulfonyl fluoride. Peak fractions were analyzed by SDS-PAGE, concentrated, and stored at −80 °C.
Rim1 was expressed in an MBP-fused form. After proteolytic cleavage by Prescission Protease (GE Healthcare), Rim1 was released from MBP. Rim1 was then bound to single strand DNA cellulose. After washing six times with 20 Tris, pH 7.4, 500 mm NaCl, 1 mm EDTA, 10% glycerol, 2.5 mm β-mercaptoethanol, and 1 mm PMSF, Rim1 was eluted with the same buffer but containing 1.5 m NaCl. Mhr1 was expressed in a His6-tagged form as described by Ling and Shibata (22). After the removal of His6 with Prescission Protease (GE Healthcare), Mhr1 was further purified to homogeneity by size exclusion chromatography on a calibrated S75 column equilibrated in 20 mm Tris, pH 7.0, 150 mm NaCl, 10 mm MgCl2, 5 mm β-mercaptoethanol, 1 mm EDTA, and 0.2 mm PMSF.
Gel Retardation and Single Strand Annealing Assay
The GC-containing B and A box probe used for DNA binding is HS40BA-F (5′-GAAGATATCCGGGTCCCAATAATAATTATTATTGAAAATAATAATTGGGACCCCCACAATAAA-3′, with the B and A boxes underlined). The AT-rich probe is HS40AT-F (5′-TTAATATTTAATAATATAATCAATAAATAATATTATAATAATATAATATAA-3′). The probes were used in both single-stranded and double-stranded forms. Both sequences are derived from the HS40 ρ− genome. For electrophoretic mobility shift assays, oligonucleotide probes were end-labeled with [γ-32P]ATP (6000 Ci/mmol; PerkinElmer Life Sciences) using T4 polynucleotide kinase (3′-phosphatase-free; Roche Applied Science). Recombinant proteins were mixed with 2.5 × 10−3 μm [γ-32P]ATP end-labeled oligonucleotide probes in the binding buffer (20 mm Tris-HCl, pH 8.0, 100 mm KCl, 4 mm MgCl2, 1 mm EDTA, 10% glycerol, 1 mm DTT, 50 μg/ml BSA, 1 mm PMSF) and incubated at room temperature for 30 min. The samples were then loaded onto a 6% native acrylamide gel in 0.5× TBE buffer at 4 °C.
For the single strand annealing assay, the 51-base HS40AT-F (see above) oligonucleotide was end-labeled with [32P]ATP. A 200 nm concentration of the labeled HS40AT-F and 1.75 μm Mgm101 were added to the annealing buffer containing 50 mm triethanolamine-HCl, pH 7.5, 0.5 mm Mg(OAc)2, 1 mm DTT, and BSA at 100 μg/ml. After incubation for 5 min at 25 °C, the reactions were initiated by adding a 200 nm concentration of the unlabeled complementary oligonucleotide HS40AT-R and incubated at 25 °C for the designated time. The reactions were stopped by the addition of 2 μm cold HS40AT-F, 0.5% SDS, and proteinase K at 1 mg/ml. The deproteination was continued at 25 °C for 15 min, and the samples were loaded on a 10% native PAGE, which was run at 4 °C in Tris-acetate-EDTA buffer.
Native Agarose Gel Electrophoresis and Dithiobis(Succinimidyl Propionate) (DSP) Cross-linking
Native agarose gel electrophoresis was performed using a 0.8% agarose gel. The gel was run in TA buffer (40 mm Tris-base, 20 mm acetic acid, pH 8.25) for ∼1 h on ice using an applied voltage of 100 V. The gel was then fixed in fixing solution (25% isopropyl alcohol, 10% acetic acid) for 30 min, washed in 95% ethanol two times for 15 min and once overnight, blot-dried, and dried in a vacuum slab drier for 1 h at 80 °C. The gel was subsequently stained in Coomassie Blue and destained in fixing solution to visualize bands. For DSP-mediated cross-linking, 73 μg of purified Mgm101 at a concentration of 2.02 mg/ml in sodium phosphate buffer (10 mm sodium phosphate, pH 7.2, 500 mm NaCl) was diluted in water to a final volume of 200 μl. DSP was added from a 1.0 mm stock to obtain a final concentration of 0.5 mm. Prior to the addition of DSP, a 10.7-μl aliquot of Mgm101 was removed as uncross-linked control and treated with glycine at a final concentration of 100 mm. Once cross-linker was added, the protein was incubated on ice for 1 h 20 min, and aliquots of 10.7 μl were removed at distinct time points. The cross-linking reaction in each aliquot was quenched with glycine (100 mm) at the point of removal and analyzed on a 12% non-reducing SDS-polyacrylamide gel. The cross-linked products were visualized by Western blot analysis using an anti-Mgm101 antibody.
Sedimentation Velocity Analytical Ultracentrifugation
We used a Beckman Coulter ProteomeLabTM XL-A analytical ultracentrifuge equipped with absorbance optics and an 8-hole An-50 Ti analytical rotor. Sedimentation velocity experiments were conducted at 40,000 rpm (128,000 × g) at 10 °C using 3-mm two-sector charcoal-filled Epon centerpieces. Protein samples were run in 10 mm sodium phosphate, pH 7.2, 100 mm NaCl, and 0.1% sodium azide. 300 scans were collected with the time interval between scans set to zero. Sedimentation boundaries were analyzed by the continuous distribution (c(s)) method using the program SEDFIT (31). The program SEDNTERP version 1.09 (32) was used to correct the experimental S value (s*) to standard conditions at 20 °C in water (s20,w) and to calculate the partial specific volume of each protein.
Transmission Electron Microscopy and Image Analysis
For negative stain electron microscopy, Mgm101 (with or without DNA) was diluted to between 20 and 100 μg/ml and spread on carbon-coated copper grids. For visualizing Mgm101-ssDNA interaction, 2 μm Mgm101 was incubated with ssDNA oligonucleotide (8 μm) or M13mp18 ssDNA (5 μm) at 37 °C for 10 min before being applied to the grids. The buffer contained 20 mm Tris, pH 7.0, 10 mm MgCl2, 1 mm EDTA, and 0.2 mm PMSF. The grids were washed once with water and stained with 1% uranyl acetate. For electron cryomicroscopy, protein at 0.2–1 mg/ml was applied to holey carbon grids (C-flat), blotted with filter paper and flash-frozen by plunging into liquid ethane. Negative stained and frozen hydrated samples were visualized in a JEM-2100 transmission electron microscope (JEOL) operating at 200 kV.
Electron micrographs were recorded on a 4096 × 4096 pixels CCD camera (TVIPS F415-MP) in minimum dose mode at an electron optical magnification of 40,000× and a defocus of −1.5 μm. Micrographs were displayed using the boxer program of the EMAN software package (33), and data sets of 4,500 top and 930 side view images were collected using the “autobox” function (for the top views) or manually (for the side views). All subsequent image analyses were done with the IMAGIC 5 package of programs (34). Images were normalized and were band pass-filtered to remove low (<0.05 Å−1) and high (>0.16 Å−1) spatial frequencies, and a circular mask was applied. Data sets were treated by multivariate statistical analysis and sorted into 16–36 initial classes by hierarchical ascendant classification. The best averages of the classes as determined by IMAGIC were then used as references for multireference or single reference alignment, and the multivariate statistical analysis/classification was repeated until stable results were obtained.
RESULTS
Mgm101 Shares Functionally Conserved Sequence Signatures with Rad52
Rad52 is a central recombination protein best known as a mediator of the Rad51 recombinase in S. cerevisiae. It is evolutionarily related to a highly diverged superfamily of recombination proteins that are widely present in temperate bacteriophages (35–37). These viral proteins, as exemplified by Redβ, Erf, and Sak, have single strand annealing (SSA) activity. The eukaryotic Rad52 has retained the SSA activity on its N terminus, but it has also acquired a large C-terminal domain that recruits Rad51 onto ssDNA (38, 39). Yeast cells also express Rad59, a Rad52 paralogue that lacks the C-terminal Rad51-interacting domain (40). Rad59 functionally overlaps with Rad52 in DSB repair, but it also appears to have unique functions, including stimulation of Rad52-mediated single strand annealing (41–43).
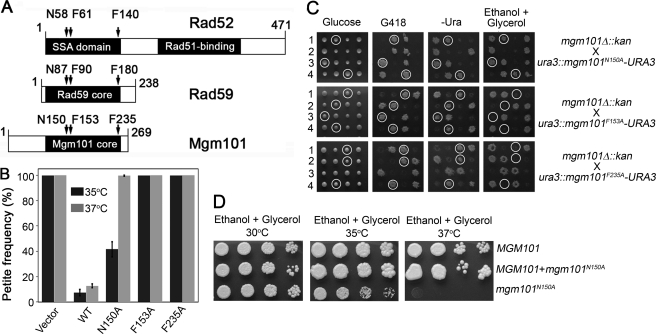
The central domain of Mgm101 between residues 108 and 241 shares 17 and 15% sequence identities with the N-terminal ssDNA-annealing domain of yeast Rad52 (25) (supplemental Fig. S1) and with Rad59, respectively (Fig. 1A). Sequence conservation is hardly recognizable between Mgm101 and the Rad52 orthologs in bacteriophage, such as Redβ, Erf, and Sak. To test whether Mgm101 is a Rad52-related protein, we first asked whether highly conserved amino acids critical for Rad52 activity are required for Mgm101 function in vivo. Previous studies have shown that the nuclear genome of yeast cells expressing the N58A, F61A, and F140A alleles of Rad52 are hypersensitive to γ-ray irradiation (44). These residues are also conserved in Rad59, which correspond to Asn-87, Phe-90, and Phe-180. Therefore, we mutated the equivalent amino acids in Mgm101 (Asn-150, Phe-153, and Phe-235) to alanine. The mutant alleles were examined for their activity in supporting mtDNA maintenance in two series of experiments. As shown in Fig. 1B, the mgm101-1ts mutant harboring the control vector is completely converted into petite colonies lacking mtDNA at both 35 and 37 °C. Petite formation was largely suppressed by the wild type MGM101. At 37 °C, the mgm101N150A, mgm101F153A, and mgm101F235A alleles expressed from a centromeric vector failed to suppress petite formation, suggesting that the mutant proteins are functionally defective in vivo. The N150A allele was found to be relatively milder compared with F153A and F235A. It partially suppressed the petite phenotype when cells were incubated at 35 °C.
FIGURE 1.
Mgm101 and Rad52 share functionally conserved sequence signatures. A, schematic diagram showing the molecular organization of Mgm101, Rad52, and Rad59. Asn-58 (N58), Phe-61 (F61), and Phe-140 (F140) of Rad52, which are required for Rad52 function in vivo (44), are conserved in Mgm101 (corresponding to Asn-150, Phe-153, and Phe-235) and Rad59 (corresponding to Asn-87, Phe-90, and Phe-180). B, stability of the ρ+ genome in the mgm101-1ts mutant transformed with mutant alleles of MGM101. The mgm101–1ts mutant was transformed with a centromeric vector expressing mgm101N150A, mgm101F153A, and mgm101F235A. Ura+ transformants were plated on YPD medium and incubated at 35 and 37 °C. Petites were scored as colonies unable to grow on non-fermentable carbon sources after replica plating. The data are the averaged petite frequency of at least three independent transformants. Error bars, average deviations. C, meiotic analysis showing that mgm101N150A, but not mgm101F153A and mgm101F235A, complements the null mgm101Δ::kan allele for respiratory growth on ethanol plus glycerol medium at 30 °C. Four complete tetrads independently segregating the null mgm101Δ::kan and the mgm101N150A, mgm101F153A, or mgm101F235A allele, were dissected on complete glucose medium. The meiotic segregants were then replica-plated onto G418, −Ura, and ethanol plus glycerol medium. G418R marks the presence of the null mgm101Δ::kan allele. Ura+ marks the mgm101N150A, mgm101F153A, or mgm101F235A allele, as a wild-type URA3 gene was introduced together with mutant alleles into the ura3 locus after chromosomal integration. Circled are the meiotic spores that cosegregate mgm101Δ::kan with mgm101N150A, mgm101F153A, or mgm101F235A. D, the respiratory growth of cells expressing only the mgm101N150A allele is completely inhibited at 37 °C but partially inhibited at 35 °C.
To exclude possible interallelic complementation and the synthetic defects that might occur with mgm101-1ts, we generated meiotic segregants expressing only mgm101N150A, mgm101F153A, or mgm101F235A (Fig. 1C). The mutant alleles were introduced into a diploid strain in which one copy of the wild type MGM101 is disrupted by the insertion of the kan (G418R) marker. The mutant alleles were placed adjacent to the URA3 gene, which facilitated the integration into the ura3 locus by selecting for Ura+ transformants. After sporulation and tetrad dissection, G418R and Ura+ segregants were examined for respiratory competency. We found that the mgm101F153A and mgm101F235A alleles are unable to support respiratory growth on ethanol plus glycerol medium. The meiotic segregants expressing only mgm101N150A are respiratory competent at 30 °C. However, these cells exhibited partial and complete loss of respiratory growth at 35 and 37 °C, respectively (Fig. 1D). Taken together, the data demonstrated that Asn-150, Phe-153, and Phe-235 in Mgm101 are functionally critical for in vivo function, supporting the idea that Rad52, Rad59, and Mgm101 may share similar molecular functions despite a weak similarity in their primary sequences.
Mgm101 Is Required for mtDNA Recombination and Suppression of mtDNA Fragmentation in Vivo
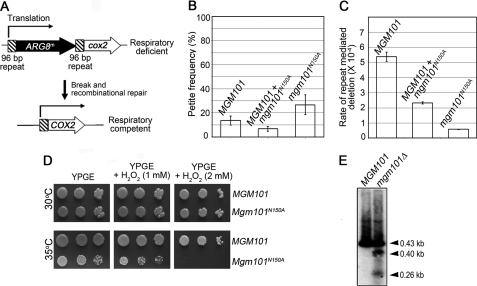
We used a reporter system developed by Sia and co-workers (29) to determine whether Mgm101 affects mtDNA recombination in vivo (Fig. 2A). In this system, expression of the mitochondrial COX2 gene, encoding subunit 2 of cytochrome c oxidase, is interrupted by the insertion of ARG8m, which has been recoded for expression in mitochondria. ARG8m is expressed via an in-frame translational fusion with COX2, which complements the nuclear arg8 mutation (45). ARG8m is flanked by 96-bp direct repeats. Recombination events that occur between the repeats following dsDNA breaks result in the deletion of ARG8m and the restoration of COX2 expression. Thus, the number of respiratory competent and Arg− colonies that arise on a non-fermentable carbon source can be used as an indicator for the frequency of recombination-based ARG8m deletions.
FIGURE 2.
Defect in Mgm101 function reduces repeat-mediated recombination, decreases tolerance to oxidative damage, and induces mtDNA fragmentation in vivo. A, schematic representation of the cox2::ARG8m-Rep96 allele in mtDNA used as a recombination reporter. B, petite frequency of the haploid strains EAS748 (MGM101), CS1631 (MGM101, trp1Δ::mgm101N150A), and CS1640 (mgm101::URA3, trp1Δ::mgm101N150A). C, repeat-mediated recombination rate as expressed by the rate of respiratory competent colonies per cell division during colony formation on YPD at 30 °C. The haploid strains EAS748 (MGM101), CS1631 (MGM101, trp1Δ::mgm101N150A), and CS1640 (mgm101::URA3, trp1Δ::mgm101N150A) were used. Error bars, average deviations. The statistical significance levels (P) were all <0.0001 in B and C between the mutant and control strains. D, hypersensitivity of the mgm101N150A mutant to H2O2 on complete ethanol plus glycerol (YPGE) medium. E, Southern blot analysis of the 430-bp ρ− N1 genome showing mtDNA fragmentation in mgm101Δ cells. Disruption of MGM101 reduces the copy number of N1 by ∼5-fold. Total DNA was digested with NspI that has a single cut to N1. Five times more DNA was used for the mgm101Δ mutant compared with MGM101 cells in order to have approximately equal amounts of mtDNA loaded. Purified N1 mtDNA was used as a probe after labeling with 32P.
We took advantage of the relatively mild N150A allele of MGM101 (see Fig. 1) to evaluate its effect on repeat-mediated mtDNA deletion/recombination. The choice of such a mild mutant is critical to avoiding possible nonspecific effects on recombination in severely affected mutants having a highly destabilized mitochondrial genome and reduced mtDNA copy number. The N150A allele was introduced into the chromosome, and the endogenous wild-type MGM101 was subsequently disrupted. At 30 °C, haploid cells expressing mgm101N150A had only a mildly increased ρ− and ρ° frequency, which was 26.7% compared with 13.6% in the MGM101 control strain (Fig. 2B). However, we found that the rate of repeat-mediated mtDNA deletions in the mutant is reduced by 8.1-fold compared with the control (Fig. 2C). Haploid cells co-expressing MGM101 and mgm101N150A also had a reduced recombination rate, suggesting a partially dominant effect of the mutant allele despite its slight reduction in the petite formation. We speculate that a mild decrease in recombination may partially stabilize instead of destabilizing the mitochondrial genome under unstressed conditions by lowering ectopic recombinations, which give rise to spontaneous petites. In summary, these data demonstrate that defects in Mgm101 function affect recombination between non-tandem repeats in the mitochondrial genome.
To learn whether reduced recombination in the mgm101N150A mutant impairs mtDNA repair, we directly examined the sensitivity of the mutant cells to hydrogen peroxide on non-fermentable carbon sources. As shown in Fig. 2D, the growth of the mgm101N150A mutant is partially reduced on ethanol plus glycerol medium (YPGE) at the semipermissive temperature (35 °C), but cell growth is inhibited with an increasing level of hydrogen peroxide. Consistent with previous observations (26), these results further support the idea that Mgm101 is required for the repair of oxidatively damaged mtDNA. Furthermore, we found that mtDNA fragmentations can be readily detected in mgm101 mutants even under normal growth conditions. We have previously shown that in a nascent null mutant of MGM101, the copy number of the 430-bp N1 ρ− genome is reduced by ∼4–5-fold compared with that in cells expressing the wild type (28). The partial retention of mtDNA permitted us to determine possible molecular changes to mtDNA brought by MGM101 inactivation. We performed Southern blot analysis of the NspI-digested DNA samples, which generates a single cut in the N1 ρ− genome. It was found that in addition to the expected 430-bp N1 repeats, several fragments of different subgenomic sizes were frequently detected (see Fig. 2E). These subgenomic fragments, which probably arise from replication of broken DNA, were not seen in cells expressing the wild-type MGM101. The capture of these subgenomic mtDNA species, together with a role of Mgm101 in repeat-mediated recombination and the hypersensitivity of the mgm101N150A mutant to hydrogen peroxide, strongly supports the idea that Mgm101 may be involved in the repair of dsDNA breaks, probably via a recombination-based mechanism.
Mgm101 Preferentially Binds to Single-stranded DNA and Promotes Annealing of Rim1-coated Single-stranded DNA
To characterize Mgm101 biochemically, we attempted to produce and purify the recombinant yeast protein from E. coli. Direct expression of Mgm101 in bacteria has been unsuccessful, probably due to its cellular toxicity and/or poor solubility in bacteria. However, we succeeded in producing Mgm101 at high yield in an MBP-tagged form. Cell lysates were treated with DNase I to minimize contamination by bacterial DNA before amylose affinity chromatography. After proteolytic cleavage by the PrescissionTM protease, Mgm101 was released from MBP and purified to homogeneity in a soluble form by cation exchange and size exclusion chromatography (Fig. 3A). We compared the quality of Mgm101 with regard to DNA contamination in preparations with or without DNase I treatment. We found that this treatment improves the A280/A260 ratio from ∼1.1 to 1.55–1.63, suggesting that the final preparations have minimal DNA contamination.
FIGURE 3.
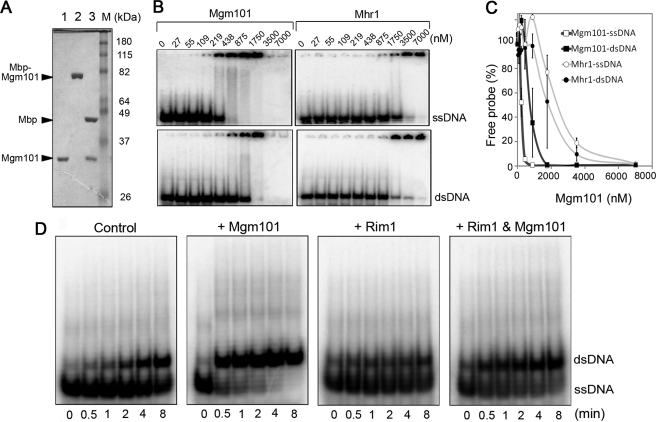
Purification, substrate specificity, and ssDNA annealing activity of Mgm101. A, SDS-PAGE showing the purified Mgm101 (lane 1), MBP-Mgm101 fusion (lane 2), and MBP-Mgm101 cleaved by PrescissionTM protease (lane 3). Mgm101 was separated from MBP by cation exchange chromatography. B, electrophoretic mobility shift assays showing the binding of Mgm101 and Mhr1 to the single-stranded (top) and double-stranded (bottom) HS40AT mtDNA probes. C, graphic representation of the results from B. Error bars, average deviations of three and two independent experiments for Mgm101 and Mhr1, respectively. D, single strand annealing in the absence and presence of the mitochondrial single strand binding protein, Rim1. The annealing reactions were performed at 25 °C for 0–8 min with or without Mgm101 (1.75 μm) and Rim1 (3.0 μm). Rim1 was preincubated with the probe for 2 min at room temperature before the addition of Mgm101.
Using electrophoretic mobility shift assays, we found that Mgm101 binds to ssDNA with an affinity 5.5 times higher than to dsDNA (Fig. 3B), with a Kd value of 192 nm for ssDNA versus 1.068 μm for dsDNA (p = 0.005). This property is reminiscent of Rad52 family proteins (46, 47). The binding affinity of Mgm101 to ssDNA is roughly comparable with that of Rad52. For comparison, we purified and analyzed the DNA-binding affinity of Mhr1, a protein previously known to be required for mtDNA recombination (22). We found that Mgm101 binds to ssDNA with an affinity ∼8-fold higher than Mhr1 (Fig. 3, B and C).
To know whether Mgm101 is functionally analogous to Rad52, we determined whether Mgm101 catalyzes ssDNA annealing. In this assay, a 32P-labeled 51-base oligonucleotide (HS40AT-F) was incubated with Mgm101. The annealing reactions were initiated by the addition of the complementary strand (HS40AT-R) and stopped by a 10-fold excess of the unlabeled HS40AT-F. After deproteination, the reaction products were resolved on a native polyacrylamide gel. As shown in Fig. 3D, spontaneous annealing between the two oligonucleotides occurred slowly in a time-dependent manner. The annealing process was drastically accelerated by Mgm101. The oligonucleotides were almost completely annealed to form the slowly migrating dsDNA after only 30 s of incubation in the presence of Mgm101.
The in vivo substrates for Rad52 are ssDNA that are coated by their cognate single strand binding proteins to prevent the formation of secondary structures (48, 49). The yeast mitochondrial single strand binding protein is encoded by the RIM1 gene, which contains an intron on its 5′ moiety (50). We completely synthesized the intronless open reading frame of RIM1. After expression in E. coli, the protein was purified to homogeneity (supplemental Fig. S2). To assess whether Mgm101 anneals Rim1-complexed ssDNA, the 32P-labeled oligonucleotides were preincubated with Rim1. Annealing reactions were then initiated with or without the addition of Mgm101. As predicted, Rim1 alone inhibited the self-annealing of the complementary oligonucleotides (Fig. 3D). However, Mgm101 considerably stimulated strand annealing in the presence of Rim1, although the rate of annealing is significantly lower compared with that in the reaction lacking Rim1. The data provide strong support for a role of Mgm101 in promoting single strand annealing under physiologically relevant conditions.
Mgm101 Forms Homo-oligomers
Several lines of evidence suggest that Mgm101 forms homo-oligomers. First, recombinant Mgm101 migrates poorly to the cathode in a native agarose gel at pH 5.5, 7.4, and 8.25 despite an estimated isoelectric point of 9.09 (Fig. 4A). Second, treatment of purified Mgm101 with DSP generated oligomeric species with a molecular mass above 180 kDa (Fig. 4B). Third, size exclusion chromatography clearly showed that purified Mgm101 was eluted as a single peak with an estimated molecular mass of ∼400 kDa (Fig. 4C). The tagging by MBP does not appear to affect the oligomerization of Mgm101, because the MBP-Mgm101 fusion was eluted as an expected molecular species of ∼940 kDa. Note that no protein fractions corresponding to the monomeric form were detected with Mgm101 or MBP-Mgm101.
FIGURE 4.
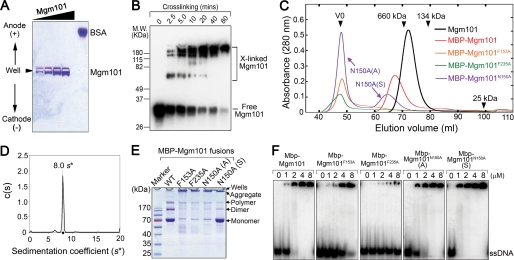
Oligomerization is required for the functionality of Mgm101. A, native agarose gel electrophoresis showing that the positively charged Mgm101 migrates poorly toward the cathode. 5, 10, 20, and 30 μg of purified Mgm101 were loaded, respectively. 10 μg of bovine serum albumin (BSA) was included as control. B, Western blot analysis showing DSP-assisted cross-linking of recombinant Mgm101 using anti-Mgm101 antibody. C, size exclusion chromatography of purified Mgm101 and MBP-fused Mgm101 variants on a Superose 6 column. V0, void volume; N150A(A), aggregated form of MBP-Mgm101N150A; N150A(S), soluble form of MBP-Mgm101N150A. D, diffusion free sedimentation coefficient distribution c(s) derived from fitting sedimentation velocity data (see supplemental Fig. S3) of Mgm101 at a concentration of 0.46 mg/ml. E, SDS-PAGE analysis of MBP-Mgm101 (WT), MBP-Mgm101F153A (F153A), MBP-Mgm101F235A (F235A), and the aggregated (N150A (A)) and soluble (N150A (S)) forms of MBP-Mgm101N150A. F, electrophoretic mobility shift assays showing the binding of MBP-Mgm101, MBP-Mgm101F153A, MBP-Mgm101F235A, MBP-Mgm101N150A (A), and MBP-Mgm101N150A (S) to the single-stranded HS40AT mtDNA probes.
To more precisely determine the molecular weight of Mgm101 oligomers, we characterized the recombinant protein by sedimentation velocity analytical ultracentrifugation. The majority of Mgm101 sedimented as a monodispersed species with an estimated sedimentation coefficient (s*) of 8.0 s (8.3 s for the standard sedimentation coefficient, s20,w, after correcting for water at 20 °C) (Fig. 4D and supplemental Fig. S3). The native molecular mass calculated from the s20,w value is ∼397 kDa, which is consistent with the presence of ∼14 subunits in the Mgm101 oligomers as estimated with size exclusion chromatography.
Mgm101 Oligomeric State Is Affected by the F153A, F235A, and N150A Mutations
Interestingly, we found that the oligomeric state of Mgm101 is dramatically altered by the functionally defective F153A, F235A, and N150A mutations. We prepared the mutant proteins by the MBP-tagging strategy. However, after the proteolytic release from MBP, no mutant Mgm101 was recovered due to poor solubility. Direct analysis of the fusion proteins revealed that the mutant proteins are aggregated. By size exclusion chromatography, we found that MBP-Mgm101F153A and MBP-Mgm101F235A eluted in the void volume fraction, in contrast to MBP-Mgm101, which formed a distinct peak of ∼940 kDa as expected (Fig. 4C). This suggests that Mgm101F153A and Mgm101F235A form large aggregates. Consistent with its mild phenotype, Mgm101N150A behaved differently. A large fraction of Mgm101N150A eluted in the void volume, suggesting aggregate formation (denoted as fraction A). In addition, Mgm101N150A forms a second peak with an estimated molecular mass of ∼1.4 MDa, which remains soluble (denoted as fraction S), suggesting the formation of oligomers significantly larger than MBP-Mgm101. The molecular species in the void volume appeared to result from protein aggregates rather than from the formation of higher order structures. On SDS-PAGE, MBP-Mgm101F153A, MBP-Mgm101F235A, and the aggregated fraction of MBP-Mgm101N150A (N150A (A)) formed SDS-resistant aggregates that mainly remained in the wells, whereas the soluble fraction of MBP-Mgm101N150A (N150A (S)) was readily resolved into monomers like MBP-Mgm101 (Fig. 4E).
Electrophoretic mobility shift assays showed that in comparison with MBP-Mgm101, the DNA binding activity of MBP-Mgm101F153A and MBP-Mgm101F235A is drastically reduced, providing an explanation for the loss of their function in vivo (see Fig. 1, B and C). Intriguingly, the aggregated fraction of MBP-Mgm101N150A retains a DNA binding activity comparable with that of MBP-Mgm101 at 25 °C, whereas the soluble fraction of MBP-Mgm101N150A has an increased DNA binding activity (Fig. 4F). It is important to note that despite its DNA binding activity in vitro, Mgm101N150A is mainly present in aggregates (see Fig. 4C). We speculate that this may be further accentuated when cells are grown at the non-permissive temperature (37 °C). The aggregated protein is either inactive in DNA transactions after ssDNA binding or unable to interact with other components in the recombination pathway, which ultimately results in mtDNA disintegration in the mgm101N150A mutant. In summary, the ensemble of the data demonstrates that the oligomerizing property of Mgm101 is altered by the F153A, F235A, and N150A mutations, thereby leading to protein aggregation and loss of in vivo function.
Higher Order Structural Organization of Mgm101 in the Absence and Presence of DNA
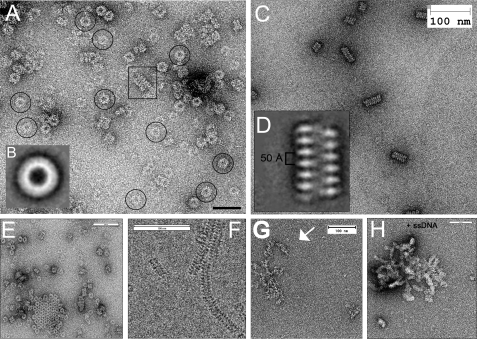
We used transmission electron microscopy to directly visualize the oligomeric structure of Mgm101. We found that Mgm101 shares some higher order structural features similar to the Rad52 family proteins, especially those from bateriophages. Analysis of the negatively stained specimen showed that Mgm101 forms homogeneous ring-shaped structures (Fig. 5A). Alignment and classification of a data set of ∼4,500 molecular images (without imposing a symmetry) revealed an estimated diameter of ∼200 Å for the rings and ∼60 Å for the central channel (Fig. 5B).
FIGURE 5.
Higher order structural organization of Mgm101 as revealed by transmission electron microscopy. Images were acquired either after negative staining of samples (A, C, E, G, and H) or directly by electron cryomicroscopy (F) in the absence (A–F) or presence of a 60-base oligonucleotide (G) or M13 ssDNA (H). Scale bar, 50 nm (A) and 100 nm (C, E–H).
Mgm101 also exhibited dynamic structural organization like several known recombination proteins. The Mgm101 rings interacted laterally to form honeycomb-like structures (Fig. 5E). Short filaments were frequently observed in freshly prepared samples (Fig. 5A, boxed). Interestingly, these filaments became the dominant molecular species in samples after storage for 4 weeks at 4 °C (Fig. 5C). We found that there is no significant difference between the ring- and the filament-enriched Mgm101 samples in binding to ssDNA (our unpublished observation). Further inspection of aged samples by electron cryomicroscopy revealed that some filaments can reach a length of several hundred nm (Fig. 5F).
To visualize in greater detail the structures of the Mgm101-based filaments, we analyzed 930 images from negatively stained samples by the IMAGIC 5 software. The data suggested that the filaments contain compressed helical structures with a pitch of ∼50 Å (Fig. 5D). The filaments are therefore formed by following a distinct structural pattern and not simply by the stacking of conventional oligomeric rings.
Finally, we used negative staining to visualize Mgm101-DNA interaction. Incubation with an oligonucleotide of only 66 bases gave rise to distinct nucleoprotein filaments (Fig. 5G). In the presence of the 7.25-kb M13mp18 ssDNA, the Mgm101-DNA complexes were mostly condensed into thick nucleoprotein filaments (Fig. 5H). Neither ring nor helical structures could be discerned in the Mgm101-based nucleoprotein filaments.
DISCUSSION
In this report, we show that the yeast mitochondrial nucleoid protein, Mgm101, shares similar biochemical, structural, and functional properties with several Rad52 family proteins involved in DNA recombination. The work reveals the presence of an evolutionarily conserved recombination protein in mitochondria that is essential for the maintenance of mtDNA integrity. The Rad52-type proteins are widespread among bacteriophages. Based on structure-based modeling, these viral recombination proteins are grouped into the Rad52-, RecA/Rad51-, and Gp2.5-type subfamilies (37). The Rad52 family proteins are structurally characterized by a typical β-β-β-α fold, despite very limited similarity in their primary sequences. The best characterized Rad52-type proteins include Redβ and Erf from the bacteriophages λ and P22, RecT from the prophage rac, and Sak from the lactococcal phage ul36. These proteins have a higher order structural organization similar to the ssDNA binding domain of Rad52, all forming homo-oligomeric rings of 10–14-fold symmetry (35, 47, 51–53). Mgm101 shares similar features with these proteins, by forming rings of ∼14-fold symmetry with a diameter of ∼200 Å, compared with ∼130, 145, and 155 Å for Rad52, Redβ, and Sak, respectively.
Biochemical data suggest that the ring structure is functionally important in vivo. The F153A, F235A, and N150A alleles affecting oligomerization are all defective in maintaining mtDNA. By using several techniques, including native gel electrophoresis, size exclusion chromatography, and sedimentation velocity analytical ultracentrifugation, we did not observe the presence of protein fractions corresponding to monomeric Mgm101. Even in the cases of the aggregation-prone MBP-Mgm101F153A, MBP-Mgm101F235A, and Mgm101N150A, the monomeric form of the proteins was undetectable. It is possible that the monomers are unstable in solution in the absence of DNA. The F153A, F235A, and N150A mutations probably induce aggregation by disassembling the ring structure into the unstable monomeric components. Thus, the ring structure may play a critical role in stabilizing Mgm101.
Although Mgm101 shares hardly recognizable sequence similarity with the bacteriophage enzymes, its structural organization appears to be more close to the phage proteins than to Rad52. For example, Mgm101 forms highly compressed helical filaments like Redβ (35). The functional implications of these structures are not well understood. We speculate that the higher order structures, including rings and filaments, may serve as storage forms of Mgm101 in the nucleoids. This is particularly relevant in view of the fact that monomers are unstable and prone to aggregation. In response to DNA damage, the protein can therefore be rapidly remobilized from the stores to bind and repair damaged mtDNA that is present in the vicinity. However, future in vivo experiments are required to validate this hypothesis.
Although Mgm101 and Redβ form similar helical filaments, they display structural differences when bound to DNA. In contrast to Redβ, which to some extent retains the helical organization (35), Mgm101 forms distinct nucleoprotein filaments on ssDNA. No ring structure is recognizable in the nucleoprotein filaments. This type of organization is rather close to that observed with the Sak protein in lactococcal phage ul36 (47). The unique DNA condensing activity may provide a mechanism for bringing the substrates into close proximity, thereby facilitating a homology search. Because neither the DNA-free helical structure nor the condensed nucleoprotein filaments have been seen with Rad52, Mgm101 would have most likely arisen from a viral protein. From an evolutionary perspective, this may not be surprising because several mtDNA-transacting enzymes have previously been known to have a viral origin (e.g. mitochondrial DNA and RNA polymerases) (54, 55).
We have demonstrated that defects in Mgm101 function lead to mtDNA fragmentation and reduce repeat-mediated mtDNA deletions. Together with the hypersensitivity of the mgm101N150A mutant to hydrogen peroxide and other DNA-damaging agents (26), the data strongly support a role of the protein in recombinational repair in vivo. Recombination between non-tandem direct repeats generally results from recombinational repair of dsDNA breaks, which are mediated either by non-allelic strand invasion between the repeats or by annealing of non-allelic repeated sequences after exonuclease resection of dsDNA breaks. Because Mgm101 lacks the large C-terminal domain that is present in Rad52 for the recruitment of Rad51 (see Fig. 1A), it is unlikely that Mgm101 operates in the classic strand invasion mode by acting as a mediator for a RecA/Rad51-type recombinase. Based on its single strand annealing activity in vitro, the simplest model would be that Mgm101 operates by the strand annealing mode. In this regard, Mgm101 shares similar molecular organization with Rad59. Rad59 has been reported to promote Rad51-independent inverted repeat recombination by a strand annealing mechanism (56).
It is important to note that, unlike the strand invasion mechanism, repair of dsDNA breaks by the single strand annealing mode is error-prone and generates deletions that could be detrimental for the functionality of the genome. Therefore, a more conciliatory model would predict that Mgm101 may promote strand invasion and error-free recombinational repair by an unconventional deployment of its strand annealing activity. mtDNA is rich in ssDNA stretches (57). Instead of invading dsDNA duplex, which is dependent on Rad51/RecA, the Mgm101-ssDNA nucleoprotein filaments may be directly annealed to these homologous single-stranded donor sequences. In this way, although Mgm101 primarily operates in the strand annealing mode, strand invasion could be one of the recombination products during mtDNA repair in vivo. The use of the annealing activity for strand invasion bypasses the requirements for ATP-dependent ssDNA binding and dsDNA remodeling in a conventional recombination reaction.
Shibata and co-workers (19, 22) have identified the MHR1 gene, which affects mitochondrial gene conversion after mating of yeast cells carrying different mitochondrial genotypes. Mhr1 is a DNA-binding protein of 27 kDa that lacks recognizable sequence homology to the ATP-dependent Rad51/RecA recombinases. Unlike the currently known DNA recombinases that all form oligomers, Mhr1 is clearly monomeric in solution (supplemental Fig. S4). Mhr1 has been shown to promote ATP-independent D-loop formation in vitro (22), but the implications for mtDNA recombination in vivo need to be further evaluated. Interestingly, Mhr1 has recently been shown to share sequence similarity with Rad54 (58), a dsDNA-dependent helicase that promotes dsDNA unwinding, stimulates homologous pairing, stabilizes the Rad51-ssDNA nucleoprotein filament, and contributes to the late phase of homologous recombination, especially the branch migration of Holliday junctions (13, 59). Further studies are required to determine the functional relationship between Mgm101 and Mhr1.
Although the Rad52-type recombination proteins are only conserved at the tertiary and quaternary structural levels rather than at primary sequences (37), close orthologs of Mgm101 are readily identified in lower animals (e.g., Nematostella venctensis) and in Dictyostelium discoideum (see supplemental Fig. S5). For a long time, animal mtDNA has been believed not to recombine. On the other hand, extensive evidence has also been provided that supports mtDNA recombination in species across the animal kingdom, including humans (e.g. see Refs. 60 and 61). Notably, recent electron microscopic studies have revealed the presence of abundant four-way and three-way junctions suggestive of recombination intermediates in the oxyradical-rich human adult heart and brain mitochondria (62–64). A low level of recombination, which is probably limited between intranucleoidal mtDNA molecules (65), may be in place in animal mitochondria from specific tissues as an effective mechanism for mutational clearance while minimizing the invasion by selfish DNAs (66). RecA orthologs have been identified in plant mitochondria (67). Because fungi, dictyostelids, and animals belong to the unikonts clade on the eukaryotic tree, the Mgm101 orthologs in these species may have arisen from a common ancestral gene during evolution. In light of the present work, it would be interesting to see whether the evolutionary lineage of the Rad52-type proteins, with or without primary sequence similarity to Mgm101, can be extended to the mitochondria of higher animals. More recently, a co-fractionation of the Rad51-type proteins with mitochondria has been reported in carcinoma cell lines (68). This would stipulate the need for the presence of a Rad52-type activity as a recombination mediator in mitochondria. In summary, the data reported in the current work could set the stage for studying Rad52-type proteins in mtDNA recombination in different species.
Supplementary Material
Acknowledgments
We thank E. A. Sia for the generous gift of yeast strains; Stewart Loh for the use of FPLC and help in data analysis; and Mark Schmitt, Des Clark-Walker, and David Mitchell for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AG023731, R01GM058600, and R01CA140522 (to X. J. C., S. W., and M. S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- mtDNA
- mitochondrial DNA
- DSB
- double strand break
- HR
- homologous recombination
- MBP
- maltose-binding protein
- SSA
- single strand annealing
- ts
- temperature-sensitive.
REFERENCES
- 1. Chen X. J., Butow R. A. (2005) Nat. Rev. Genet. 6, 815–825 [DOI] [PubMed] [Google Scholar]
- 2. Holt I. J., He J., Mao C. C., Boyd-Kirkup J. D., Martinsson P., Sembongi H., Reyes A., Spelbrink J. N. (2007) Mitochondrion 7, 311–321 [DOI] [PubMed] [Google Scholar]
- 3. Bogenhagen D. F. (2010) Exp. Gerontol. 45, 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spelbrink J. N. (2010) IUBMB Life 62, 19–32 [DOI] [PubMed] [Google Scholar]
- 5. Brown W. M., George M., Jr., Wilson A. C. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howell N. (1996) Am. J. Hum. Genet. 59, 749–755 [PMC free article] [PubMed] [Google Scholar]
- 7. Larsson N. G. (2010) Annu. Rev. Biochem. 79, 683–706 [DOI] [PubMed] [Google Scholar]
- 8. Wallace D. C. (2005) Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Copeland W. C. (2008) Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bogenhagen D. F., Pinz K. G., Perez-Jannotti R. M. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 257–271 [DOI] [PubMed] [Google Scholar]
- 11. Weissman L., de Souza-Pinto N. C., Stevnsner T., Bohr V. A. (2007) Neuroscience 145, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 12. Pâques F., Haber J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Symington L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 16. Shi I., Hallwyl S. C., Seong C., Mortensen U., Rothstein R., Sung P. (2009) J. Biol. Chem. 284, 33275–33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sugiyama T., Kantake N., Wu Y., Kowalczykowski S. C. (2006) EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dujon B. (1981) in The Molecular Biology of the Yeast Saccharomyces (Strathern J. N., Jones E. W., Broach J. R. eds) pp. 505–635, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Ling F., Makishima F., Morishima N., Shibata T. (1995) EMBO J. 14, 4090–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ling F., Morioka H., Ohtsuka E., Shibata T. (2000) Nucleic Acids Res. 28, 4956–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shibata T., Ling F. (2007) Mitochondrion 7, 17–23 [DOI] [PubMed] [Google Scholar]
- 22. Ling F., Shibata T. (2002) EMBO J. 21, 4730–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling F., Yoshida M., Shibata T. (2009) J. Biol. Chem. 284, 9341–9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X. J., Guan M. X., Clark-Walker G. D. (1993) Nucleic Acids Res. 21, 3473–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuo X., Xue D., Li N., Clark-Walker G. D. (2007) FEMS Yeast Res. 7, 131–140 [DOI] [PubMed] [Google Scholar]
- 26. Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., Yates J. R., Nunnari J. (1999) J. Cell Biol. 145, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meeusen S., Nunnari J. (2003) J. Cell Biol. 163, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuo X. M., Clark-Walker G. D., Chen X. J. (2002) Genetics 160, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phadnis N., Sia R. A., Sia E. A. (2005) Genetics 171, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luria S. E., Delbrück M. (1943) Genetics 28, 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laue T. M., Shah B. D., Ridgeway T. M., Pelleteir S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S., Rowe A., Horton J. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 33. Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 34. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 35. Passy S. I., Yu X., Li Z., Radding C. M., Egelman E. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4279–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iyer L. M., Koonin E. V., Aravind L. (2002) BMC Genomics 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lopes A., Amarir-Bouhram J., Faure G., Petit M. A., Guerois R. (2010) Nucleic Acids Res. 38, 3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sung P. (1997) J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 39. New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. (1998) Nature 391, 407–410 [DOI] [PubMed] [Google Scholar]
- 40. Bai Y., Symington L. S. (1996) Genes Dev. 10, 2025–2037 [DOI] [PubMed] [Google Scholar]
- 41. Davis A. P., Symington L. S. (2001) Genetics 159, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng Q., Düring L., de Mayolo A. A., Lettier G., Lisby M., Erdeniz N., Mortensen U. H., Rothstein R. (2007) DNA Repair 6, 27–37 [DOI] [PubMed] [Google Scholar]
- 43. Wu Y., Sugiyama T., Kowalczykowski S. C. (2006) J. Biol. Chem. 281, 15441–15449 [DOI] [PubMed] [Google Scholar]
- 44. Mortensen U. H., Erdeniz N., Feng Q., Rothstein R. (2002) Genetics 161, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steele D. F., Butler C. A., Fox T. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ploquin M., Bransi A., Paquet E. R., Stasiak A. Z., Stasiak A., Yu X., Cieslinska A. M., Egelman E. H., Moineau S., Masson J. Y. (2008) Curr. Biol. 18, 1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kantake N., Madiraju M. V., Sugiyama T., Kowalczykowski S. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sugiyama T., Zaitseva E. M., Kowalczykowski S. C. (1997) J. Biol. Chem. 272, 7940–7945 [DOI] [PubMed] [Google Scholar]
- 50. Van Dyck E., Foury F., Stillman B., Brill S. J. (1992) EMBO J. 11, 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S. (2002) Mol. Cell 10, 359–371 [DOI] [PubMed] [Google Scholar]
- 52. Singleton M. R., Wentzell L. M., Liu Y., West S. C., Wigley D. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poteete A. R., Sauer R. T., Hendrix R. W. (1983) J. Mol. Biol. 171, 401–418 [DOI] [PubMed] [Google Scholar]
- 54. Kaguni L. S. (2004) Annu. Rev. Biochem. 73, 293–320 [DOI] [PubMed] [Google Scholar]
- 55. Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 56. Mott C., Symington L. S. (2011) DNA Repair 10, 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gerhold J. M., Aun A., Sedman T., Jõers P., Sedman J. (2010) Mol. Cell 39, 851–861 [DOI] [PubMed] [Google Scholar]
- 58. Masuda T., Ling F., Shibata T., Mikawa T. (2010) FEBS J. 277, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 59. Mazin A. V., Mazina O. M., Bugreev D. V., Rossi M. J. (2010) DNA Repair 9, 286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kraytsberg Y., Schwartz M., Brown T. A., Ebralidse K., Kunz W. S., Clayton D. A., Vissing J., Khrapko K. (2004) Science 304, 981. [DOI] [PubMed] [Google Scholar]
- 61. Bacman S. R., Williams S. L., Moraes C. T. (2009) Nucleic Acids Res. 37, 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pohjoismäki J. L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P. J., Griffith J. D., Holt I. J., Jacobs H. T. (2009) J. Biol. Chem. 284, 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pohjoismäki J. L., Goffart S., Taylor R. W., Turnbull D. M., Suomalainen A., Jacobs H. T., Karhunen P. J. (2010) PLoS One 5, e10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pohjoismäki J. L., Goffart S. (2011) BioEssays 33, 290–299 [DOI] [PubMed] [Google Scholar]
- 65. Gilkerson R. W., Schon E. A., Hernandez E., Davidson M. M. (2008) J. Cell Biol. 181, 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Neiman M., Taylor D. R. (2009) Proc. Biol. Sci. 276, 1201–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shedge V., Arrieta-Montiel M., Christensen A. C., Mackenzie S. A. (2007) Plant Cell 19, 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sage J. M., Gildemeister O. S., Knight K. L. (2010) J. Biol. Chem. 285, 18984–18990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.