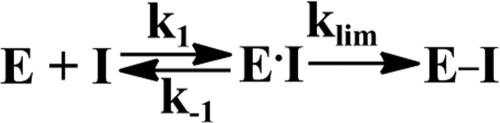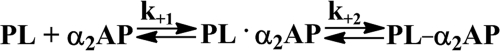Background: Elevated levels of extracellular DNA and aberrant fibrinolysis occur in a range of severe diseases.
Results: DNA competes with fibrin for fibrinolytic enzymes. DNA stimulates fibrin-independent plasminogen activation and increases enzyme susceptibility to serpins.
Conclusion: DNA is a macromolecular template that both potentiates and inhibits fibrinolysis.
Significance: Understanding the interaction of DNA with the fibrinolytic system could improve the outcomes of fibrinolytic therapy.
Keywords: DNA, Fibrinolysis, Plasmin, Plasminogen, Serpin, Fibrinolytic Enzymes, Plasminogen Activation, Template
Abstract
The increased levels of extracellular DNA found in a number of disorders involving dysregulation of the fibrinolytic system may affect interactions between fibrinolytic enzymes and inhibitors. Double-stranded (ds) DNA and oligonucleotides bind tissue-(tPA) and urokinase (uPA)-type plasminogen activators, plasmin, and plasminogen with submicromolar affinity. The binding of enzymes to DNA was detected by EMSA, steady-state, and stopped-flow fluorimetry. The interaction of dsDNA/oligonucleotides with tPA and uPA includes a fast bimolecular step, followed by two monomolecular steps, likely indicating slow conformational changes in the enzyme. DNA (0.1–5.0 μg/ml), but not RNA, potentiates the activation of Glu- and Lys-plasminogen by tPA and uPA by 480- and 70-fold and 10.7- and 17-fold, respectively, via a template mechanism similar to that known for fibrin. However, unlike fibrin, dsDNA/oligonucleotides moderately affect the reaction between plasmin and α2-antiplasmin and accelerate the inactivation of tPA and two chain uPA by plasminogen activator inhibitor-1 (PAI-1), which is potentiated by vitronectin. dsDNA (0.1–1.0 μg/ml) does not affect the rate of fibrinolysis by plasmin but increases by 4–5-fold the rate of fibrinolysis by Glu-plasminogen/plasminogen activator. The presence of α2-antiplasmin abolishes the potentiation of fibrinolysis by dsDNA. At higher concentrations (1.0–20 μg/ml), dsDNA competes for plasmin with fibrin and decreases the rate of fibrinolysis. dsDNA/oligonucleotides incorporated into a fibrin film also inhibit fibrinolysis. Thus, extracellular DNA at physiological concentrations may potentiate fibrinolysis by stimulating fibrin-independent plasminogen activation. Conversely, DNA could inhibit fibrinolysis by increasing the susceptibility of fibrinolytic enzymes to serpins.
Introduction
Growing interest in the clinical effects of extracellular (EC)3 DNA and a report that EC DNA in the circulatory system affects the fibrinolytic system (1) provide a strong rationale to elucidate the mechanisms of interaction of DNA with components of the fibrinolytic system. Fibrinolysins, including urokinase plasminogen activator (uPA) and tissue-type plasminogen activator (tPA), have long been used to treat a variety of thrombotic conditions as follows: acute myocardial infarction (2, 3), ischemic stroke (4), pulmonary emboli (5–7), organizing pleural effusions (8, 9), and acute respiratory distress syndrome (10, 11). An increase in the levels of EC DNA in plasma accompanies a number of pathological states (12), including cancer (13), myocardial infarction (14), stroke (15, 16), and trauma (17). Increased levels of EC DNA also serve as a predictor of the severity and clinical outcome in sepsis (18, 19), pulmonary emboli (20, 21), and exudative pleural injury (22, 23) as well as the mortality of critically ill patients in intensive care units (24, 25).
The major endogenous inhibitor of tPA and uPA, plasminogen activator inhibitor 1 (PAI-1), is markedly increased in pleural loculation (26–28), myocardial infarction (29–34), acute respiratory distress syndrome, and sepsis (35–37). High levels of PAI-1 inhibit the fibrinolytic system and promote thrombosis and uncontrolled fibrin deposition (38, 39). The level of PAI-1 strongly correlates with the size of myocardial infarction (40) and unfavorable outcomes in severe multiple organ failure in sepsis, disseminated intravascular coagulation (41–44), and pleural injury (26). It has been unclear as to whether there is a molecular mechanism that connects physiological effects of high levels of EC DNA and PAI-1 to the fibrinolytic system.
A better understanding of the molecular mechanisms of fibrin-independent activation of plasminogen (Plg) by tPA and uPA and the effects of DNA on the reactions of fibrinolytic enzymes with serpins is of potential clinical importance. Although the activation of tPA by proteins, protein aggregates, and macromolecular templates other than fibrin have been described (45–49), to the best of our knowledge there are no prior studies of the effects of DNA on fibrinolytic enzymes.
In this study, we investigated the effects of nucleic acids on the activation of Glu- and Lys-Plg by tPA and uPA and on the reactions between the serpins PAI-1 and α2-antiplasmin (α2AP) and their target proteinases. DNA markedly accelerates Plg activation by tPA in a manner similar to that previously described for fibrin (50–55). Higher concentrations of DNA in solution or DNA incorporated into the FITC-labeled fibrin film (FITC-fibrin) inhibit fibrinolysis. Although dsDNA slightly decreases the rate of reaction between plasmin (PL) and α2AP, it affects the kinetics of the reactive center loop (RCL) insertion because of the reaction of two-chain (tc) and single chain (sc) tPA and tcuPA with S338C PAI-1 selectively labeled with N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-3-diazole (NBD P9 PAI-1) and its complex with vitronectin (Vn). dsDNA caused no effect on the stoichiometry of inhibition (SI) of fibrinolytic proteinases by serpins.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
Human recombinant WT sctPA (Activase®) was obtained from Genentech (San Francisco); sctPA was converted to tc form by treatment with immobilized PL (Molecular Innovations, Novi, MI), as described previously (56). Human recombinant tcuPA was a gift from Abbott; a tcuPA activity standard (100,000 IU/mg) was purchased from American Diagnostica (Stamford, CT). Human Glu- and Lys-Plg, PL, α2AP, thrombin, and fluorogenic PL substrate were obtained from Hematologic Technologies Inc. (Essex Junction, VT). FITC-fibrinogen (FITC-Fbg; 3 mol of fluorescein per mol of Fbg), FITC-tPA, FITC-uPA, and FITC-Plg were purchased through Molecular Innovations (Novi, MI). Fluorogenic uPA, tPA, and chromogenic PL substrates were acquired from Centerchem Inc. (Norwalk, CT). Human Vn (2 mg/ml), purified from plasma, was from Promega (Madison, WI). WT and S338C (S338C mutation at position P9 of RCL) (P9 Cys) PAI-1 were purified and characterized as described elsewhere (56, 57). P9 Cys PAI-1 was labeled with N-((2-(iodoacetoxy)ethyl)-N-methyl) amino-7-nitrobenz-2-oxa-3-diazole as described previously (57, 58). The concentrations of tPA, uPA, or PAI-1 were calculated from absorbance values at 280 nm, using Mr values of 63,500, 54,000 and 43,000, and extinction coefficients of ϵ280 of 1.90, 1.36, and 0.93 ml mg−1 cm−1, respectively. Protein concentration was also determined using a BCA protein assay kit (Pierce). Human DNA, salmon DNA, RNA (Saccharomyces cerevisiae), and bovine serum albumin (BSA) were purchased from Sigma. Custom oligonucleotides (oligo(dT)20; oligo(dT)65, oligo(dAT)10 (a 20-mer containing 10 AT repeats); oligo(dAT)33 (a 66-mer containing 33 AT repeats); and TEX615-oligo(dAT)33, (an oligonucleotide labeled with red wavelength dye TEX615 at the 5′ end) were synthesized by Integrated DNA Technologies Inc. (Iowa City, IA). All experiments were carried out in 0.05 m Hepes/NaOH buffer (pH 7.4) with or without NaCl (20–200 mm) and/or BSA (1 mg/ml).
Measurement of uPA, tPA, and PL Amidolytic Activity
Amidolytic uPA and tPA activities were determined from time traces of the change in fluorescence emission at 440 nm (excitation 344 nm) of fluorogenic uPA- and tPA-based substrates (Pefafluor uPA benzyl-β-Ala-Gly-Arg-7-amino-4-methylcoumarin AcOH and tPA CH3-SO2-d-Phe-Gly-Arg-7-amino-4-methylcoumarin AcOH, respectively; Centerchem Inc. (Norwalk, CT)) in 0.05 m Hepes/NaOH (pH 7.4) as described previously (59). PL activity was measured using either fluorogenic (6-amino-1-naphthalenesulfonamide-based) d-Ala-Phe-Lys-ANS-NH-iC4H9·2HBr (HTI, Essex Junction, VT) or chromogenic (p-nitroanilide (pNA) based) H-d-Ala-CHA-Lys-pNA 2AcOH (Centerchem Inc., Norwalk, CT) substrates (0.5 mm). Activity was measured in either white 96-well flat bottom plates from Costar (Corning Inc.) or Pro-Bind 353915 (BD Biosciences). PL activity was calculated from an increase in either fluorescence emission at 470 nm (F470) (excitation at 352 nm), using a Varian Cary Eclipse fluorescence spectrophotometer (Varian Inc.), or in absorbance at 405 nm (A405), using a SpectraMax 96-well optical absorbance plate reader (Molecular Devices, Sunnyvale, CA). Enzymes with a known specific activity were used as standards.
Effect of DNA on the Rate of Plg Activation
Glu- or Lys-Plg (0–20 μm) was incubated in 50 μl of 0.05 m Hepes/NaOH buffer (pH 7.4) (1 mg/ml BSA) with 1 mm chromogenic (Sc) (37 °C) or fluorogenic (Sf) (room temperature) PL substrate in 96-well plates. To study the effects of DNA, RNA, or oligonucleotides on Plg activation, increasing amounts of the ligand were added to the reaction mixture 10–20 min prior to PL activity measurements. The reaction was started by adding 50 μl of 0.05 m Hepes/NaOH (pH 7.4) (1 mg/ml BSA), containing 0.01–2.0 nm tPA or uPA. Changes in PL activity in the reaction mixtures were monitored using either a SpectraMax or Varian Cary Eclipse. Initial increases in A405 and F470 were proportional to the square of time (t2) as follows: F470 = A1 B1 kcat [E] [Plg] t2/2(Km + [Plg]) + C1; and A405 = ϵ405 B2 kcat [E] [Plg] t2/2(Km + [Plg]) + C2; respectively, where E is tPA or uPA. For fluorescence measurements, A1 represents changes in F470 (8.3 × 106 AU m−1 determined experimentally for the linear part of the dependence of F470 on [6-amino-1-naphthalenesulfonamide], which corresponds to the complete digestion of 1 m of Sf), B1 = kS1 [Sf]/(KS1m + [Sf]), where kS1 = 220 min−1, and KS1m corresponds to the hydrolysis of Sf by PL. For A405 measurements, ϵ405 is the extinction coefficient for pNA generated from Sc (determined experimentally for the 96-well plate assay) and B2 = kS2 [Sc]/(KS2m + [Sc]), where kS2 = 680 min−1, KS2m = 440 μm, and ϵ405 = 6.16 × 103 m−1 correspond to the hydrolysis of Sc by PL. C1 and C2 correspond to the nonspecific degradation of Sf or Sc, respectively, and their hydrolysis by PL impurities in the Plg sample. Therefore, the slopes of the linear dependences of the concentration of the products of the hydrolysis of the fluorogenic (6-amino-1-naphthalenesulfonamide) and chromogenic (pNA) substrates (determined from F470 and A405, respectively) with t2 are equal to 0.5 B v (where v is the rate of PL generation (v = kcat [E] [Plg]/(Km + [Plg]) and B corresponds to B1 and B2 for Sf or Sc, respectively). kcat and Km were calculated from double-reciprocal plots of the rate of PL formation v by tPA or uPA and Plg concentration (0.025–1.0 μm). A linear equation was fit to the data, and the x and y axis intercepts were −1/Km and 1/Vm, respectively. The values of kcat were calculated as Vm/[E].
Pleural Fluids and Isolation of Total Nucleic Acids
Pleural fluids (PFs) were collected as described previously (26). Total DNA was purified from 100 μl of human PF (pneumonia (PN), lung cancer (LC), empyema (EMP), and congestive heart failure (CHF); n = 2–3 per group) using a DNeasy blood and tissue kit from Qiagen (Valencia, CA) as described in the manufacturer's protocol. The amounts and size distribution of DNA were estimated by A260 and agarose gel electrophoresis (EGel, 0.8%, Invitrogen) and visualized and analyzed using Molecular Imager supplemented with Quantity One (version 4.2.3) software (Bio-Rad).
Electrophoretic Gel Mobility Shift Assay
Ability of tPA, uPA, Plg, and PL to bind oligonucleotides and TEX615-oligo(dAT)33 was assessed by EMSA using 6% DNA-retardation PAGE (Invitrogen) and 4% agarose E-gels (Invitrogen). Reaction mixtures (10 μl) contained oligonucleotide (0.08–0.4 μm) and enzyme (0.25–10 μm) in a 10 mm Hepes/NaOH buffer (pH 7.4), with 0.5× Orange DNA loading dye (Fermentas, Glen Burnie, MD) or in 0.5× TBE (pH 8.0) buffer (Invitrogen) with 100 mm NaCl and 0.5× Orange DNA loading dye. The mixtures were incubated for 10 min at room temperature, loaded in 6% DNA-retardation PAGE, and resolved as described in the manufacturer's protocol (Invitrogen). At the end of the run, gels that contained unlabeled oligonucleotides were incubated in ethidium bromide for visualization. SimplyBlue SafeStain (Invitrogen) was employed to visualize protein bands, according to the manufacturer's protocol. In parallel experiments, mixtures (20 μl) containing oligonucleotides with a final concentration of 3 μm and various concentrations tPA or uPA (0–16 μm) in 10 mm Hepes/NaOH buffer (pH 7.4) were subjected to agarose electrophoresis. Mixtures were incubated at room temperature for 10 min and loaded on a 4% E-gel containing ethidium bromide and resolved as suggested by the manufacturer's protocol. The gels were then visualized and analyzed using a Molecular Imager supplemented with Quantity One (version 4.2.3) software (Bio-Rad).
Fluorescence Titration
The effects of dsDNA and oligo(dT)65 on the intrinsic tryptophan fluorescence emission of tctPA and tcuPA at 340 nm (excitation at 290 nm) were studied as described previously (60, 61). The effects of dsDNA and TEX615-oligo(dAT)33 on the fluorescence emission of FITC-tctPA and FITC-tcuPA at 520 nm were studied in a similar manner. Briefly, ligands (L) (dsDNA or oligonucleotides) were added to 0.6–1.0 ml of the enzyme solution (20–100 nm) in 0.05 Hepes/NaOH buffer (pH 7.4) with 20 or 200 mm NaCl in a quartz cuvette (wave pathway of 1 cm) at 25 °C. Fluorescence emission spectra (excitation at 290 (tryptophan) or 493 (FITC) nm) were recorded at 25 °C using a Varian Cary Eclipse fluorescence spectrophotometer equipped with a Peltier thermocontroller (Varian Inc.). The changes in fluorescence emission at 340 (tryptophan) or 520 (FITC) nm (ΔF) induced by ligands were calculated as 100·(F − Fmin)/(F0 − Fmin), where F0, Fmin, and F are fluorescence emission before, at saturation, and after the addition of the indicated amount of ligand, respectively. The changes in fluorescence emission were plotted against [oligonucleotides] or dsDNA concentrations (μg/ml). The values of the apparent dissociation constants (Kd) were calculated as the concentration of the oligonucleotide that induced half of the maximal quenching of the fluorescence emission by fitting hyperbolic Equation 1,
to the data using SigmaPlot 11.0.
Stopped-flow Kinetics of Interaction between tPA, uPA, and Oligonucleotide
Changes in the fluorescence emission detected through a 650-nm cutoff filter (excitation at 493 nm) resulting from interaction between TEX615-oligo(dAT)33 (20–40 nm) and FITC-tctPA or FITC-tcuPA (0.12–0.70 μm) was employed to study the kinetics of the interaction of plasminogen activators with DNA. FITC proteins were mixed with TEX615-oligo(dAT)33 in a microvolume stopped-flow reaction analyzer (model SX-20, Applied Photophysics Ltd., Leatherhead, UK), equipped with a fluorescence detector and a thermostated (25 °C) cell. Pro-Data Viewer software (Applied Photophysics Ltd.) was employed to fit single, two-, and three-exponential equations to the data to find the best fit. The values of the observed first-order rate constants (kobs) were calculated from the traces by fitting three-exponential Equation 2,
where Ft is the fluorescence emission at time t, F∞ the final fluorescence (arbitrary units, AU); kobs and A the observed rate constants and amplitudes, respectively, to the changes in the fluorescence emission recorded at different [FITC-enzyme]. Dependences of kobs on [FITC-enzyme] were plotted using SigmaPlot 11.0. Linear (kobs = k−1 + k1·[FITC-enzyme]) and kobs = constant or hyperbolic (kobs = klim·[FITC-enzyme]/(K0.5 + [FITC-enzyme]) equations were fit to the data, where k1 and k−1 are rate constants for formation and dissociation of the initial enzyme·DNA complex; constants are the rate constant of the monomolecular step, which is independent [FITC-enzyme]; klim is kobs at infinite [FITC-enzyme], and K0.5 is the [FITC-enzyme] at kobs = klim/2).
Effect of DNA on Rate of RCL Insertion for the Reaction of NBD P9 PAI-1 with tPA and uPA
Time-dependent inactivation of tcuPA, sc, and tctPA was measured by incubating various amounts of the enzyme (0.02–2.5 μm) with 5–20 nm NBD P9 PAI-1 (or its complex with Vn) in the presence of DNA (2–20 μg/ml). SX-20 (Applied Photophysics Ltd.) was used to record time changes in NBD fluorescence emission through a 515-nm cutoff filter (excitation at 490 nm). kobs values were calculated from the traces by fitting a single exponential equation (Ft = F∞ + Ae−(kobs)t) using Pro-Data Viewer software (Applied Photophysics Ltd.) to the recorded changes in the NBD fluorescence. Ft is the fluorescence emission at time t; F∞ is the final fluorescence (AU); kobs and A are the observed rate constant and amplitude, respectively. The simplest mechanism of the serpin reaction (Scheme 1) includes the formation of the Michaelis complex (E·I) between a serpin (I) and proteinase (E), which transforms into the final inhibitory complex (E-I*). The values of klim and Km = (klim + k−1)/k1 were estimated by fitting a hyperbolic equation kobs = klim·[E]/(Km + [E]) to the dependence of kobs on enzyme concentration.
SCHEME 1.
Effect of DNA on Stoichiometry of Inhibition of uPA and tPA by WT PAI-1 (PAI-1·Vn Complex)
The number of moles of PAI-1 required for the inactivation of 1 mol of proteinase is called the SI. The SI for the reaction between WT PAI-1 and its complex with Vn and tPA or uPA in the presence of DNA was estimated from the results of the analysis of the products of the reaction by SDS-PAGE (4–12% gradient gel; Invitrogen) as described previously (58, 62). Briefly, 3–5 μm uPA or tPA was preincubated with dsDNA (10 μg/ml) for 10 min at room temperature in 0.05 m Hepes/NaOH (pH 7.4). A 1.5–2.0 molar excess of PAI-1 or its complex with Vn was added to the proteinase and incubated for 5 min. SDS loading buffer (Invitrogen) was added to each reaction mixture, heated at 100 °C for 2 min, and analyzed by SDS-PAGE. The amount of PAI-1 (active plus latent, cleaved, and complexed with proteinase) was estimated from the intensity of the corresponding bands on gel scans. The SI was calculated as an average of 3–5 measurements ± S.E.
Effect of DNA on Interaction of PL with α2AP
The effects of DNA on the fast step of the reaction between PL and α2AP were determined observing the change in the intrinsic tryptophan fluorescence resulting from the Michaelis complex formation (63). Briefly, equimolar amounts of PL and α2AP were mixed in an SX-20 thermostabilized at 25 °C; the change in fluorescence emission with time (excitation at 290 nm) was monitored through a 335-nm cutoff filter. The second-order rate constant (k2) was calculated as described elsewhere (63).
Effect of DNA on the Stoichiometry of Inhibition of PL with α2AP and on PL Activity
To determine whether or not DNA affects PL activity with a fluorogenic substrate, PL (0.5–5 nm) was preincubated with dsDNA (0–20 μg/ml) in 50 μl of 0.05 m Hepes/NaOH buffer (pH 7.4) (1 mg/ml BSA) for 15–20 min at room temperature. Then equal volumes of 1.0 mm fluorogenic substrate were added. Changes in fluorescence emission with time were detected using a Varian Eclipse spectrofluorometer. To determine the effect of DNA on the inactivation of PL by α2AP and on the stoichiometry of inhibition, the reaction was carried out in the presence of the fluorogenic PL substrate. PL (0.5–5 nm) was preincubated with dsDNA. Equal volumes of a mixture of the PL substrate (1.0 mm) and α2AP (0–40 nm) were added to start the reaction. The inactivation of PL by α2AP was monitored as a loss of PL amidolytic activity, as described previously for slow inhibition of the Factor VIIa·tissue factor complex by PAI-1 (64). Alternatively, α2AP was incubated with DNA, and a freshly made mixture of PL and the substrate was employed to start the reaction. Changes in fluorescence emission were recorded using a Varian Eclipse to monitor the inactivation of PL by α2AP. An increase in the ratio [PL]/[α2AP] for the same residual PL activity indicates an increase in the SI. To determine the effects of DNA on the rate of the second slow step of the reaction between PL and α2AP, we used an assay described by Wiman and Collen (65).
Effects of DNA on Rates of in Vitro Fibrinolysis
To model the effects of DNA on fibrinolysis, a FITC-fibrin film in a fluorescence quench and dequench assay (66) was employed. FITC-fibrin/DNA (oligonucleotide) films were formed at the bottom of 96-well plates. FITC-Fbg (Molecular Innovations) was diluted 1:1 with unlabeled Fbg at a total concentration of 0.4 mg/ml in 0.05 m Hepes/NaOH (pH 7.4) (20 mm NaCl, 5 mm CaCl2) buffer at room temperature. A solution of FITC-Fbg, with or without dsDNA (0–25 μg/ml) or oligo(dT)65 (0–9.5 μm) added, was titrated into 96-well plates (50 μl/well). Thrombin (final concentration 10 nm) was added to each well. Polymerization of Fbg was confirmed by monitoring the quenching of FITC-fluorescence emission, with time, using a Varian Eclipse. Plates were dried overnight at room temperature, in a dark enclosed space, washed three times with 0.3 ml of Hepes/NaOH buffer, and were either used immediately or stored at −20 °C. The incorporation of the dsDNA and oligonucleotide into the fibrin network was verified by monitoring their increasing levels in the solution during the cleavage of the film by PL.
Fibrinolysis was monitored via the time-dependent increase in the fluorescence emission of fluorescein due to dequenching, as described by Wu and Diamond (66). Human Glu-Plg (20–100 nm) in Hepes/NaOH buffer with 1 mg/ml BSA was added to each well, followed by the addition of tPA or uPA (0.25 nm). In control experiments, PL (0–20 nm) was added to the FITC-fibrin film. To study the effects of DNA in solution on the rate of fibrinolysis, dsDNA (0–10 μg/ml) was added to a Glu-Plg solution. Fibrinolysis was also studied in the presence of both dsDNA (0–10 μg/ml) and α2AP (15 nm). The rate of degradation of a solid FITC-fibrin film by PL depends on different parameters (66). Assuming that the rate of fibrinolysis is proportional to the concentration of active PL, which forms due to the activation of Glu-Plg by tPA and uPA, the slopes of a linear increase in FITC-fluorescence emission on square of time were used for comparison of the rates of fibrinolysis.
Data Analysis and Statistics
Stopped-flow fluorescence traces were analyzed using a Pro-Data viewer (Applied Photophysics Ltd.). Assuming pseudo first-order kinetics (the concentration of proteinases was at least 5-fold higher than that of NBD P9 PAI-1 or its complex with Vn), a single exponential equation was fit to the stopped-flow traces to calculate kobs. The quality of the fit was estimated by visual analysis of the plots of the residuals (deviation of the fitted function from the actual data). The values of kobs (average of 4–6 measurements; S.E. less than 10%) were plotted against [E]. Plots were fit by hyperbolic equation to calculate parameters (klim, Km), using nonlinear least squares fitting with the Levenberg-Marquardt algorithm (SigmaPlot 11.0 for Windows; SPSS Inc.). Correlation coefficients (r) calculated from curve fittings were used as a parameter of the goodness of fitting (r2 of the fit was greater than 0.90 for all the kinetic data).
RESULTS
dsDNA Potentiates Plg Activation by tPA and uPA
To study the effects of dsDNA on Plg activation, human recombinant tPA and uPA were preincubated for 10 min, with increasing amounts of salmon dsDNA, in 0.05 m Hepes/NaOH buffer (pH 7.4), containing 1 mg/ml BSA. Plg activation was initiated by adding an equal volume of solution of Glu- or Lys-Plg mixed either with fluorogenic or chromogenic PL substrate (0.5 mm) in the same buffer. The generation of PL from Plg was monitored by measuring increasing PL amidolytic activity, which was proportional to the rate of accumulation of the cleavage products of the fluorogenic (Fig. 1) or chromogenic (Fig. 2) substrate. The effects of dsDNA (0.3 μg/ml) on the activation of Glu- and Lys-Plg (0.2 μm) by 0.18 nm tPA and 0.09 nm uPA are shown in Fig. 1, A and B, respectively. Although dsDNA significantly increases the rate of PL generation by tPA from Glu-Plg, and to a lesser degree from Lys-Plg, its effect on uPA was at least 10-fold less pronounced (Fig. 1, A and B). Complete digestion of dsDNA with DNase I eliminates its effect on Plg activation by tPA and uPA (data not shown). The slopes of the linear dependences of the initial increase in the fluorescence emission on t2 (50–54) were used to calculate rates of PL generation at different dsDNA concentrations. Concentration dependences of the rates of PL generation (Fig. 1, insets) demonstrate that dsDNA considerably potentiates the activation of Plg by tPA. In contrast, the effect on uPA was rather moderate. A decrease in the rates of PL generation at dsDNA concentrations significantly higher than those found in pathological conditions such as sepsis (data not shown) could indicate either inhibition of the enzymatic activity or a decrease in the concentration of the ternary (plasminogen activator·DNA·Plg) complex due to competition with free template.
FIGURE 1.
Effects of salmon dsDNA on the activation of Glu-Plg (A) and Lys-Plg (B) by tctPA and tcuPA. Glu-Plg (A) or Lys-Plg (B) (0.2 μm) was incubated with 0.18 nm tPA (■) or 0.09 nm uPA (□) alone or with dsDNA (0.3 μg/ml) (●) and (○), respectively, in 0.05 m Hepes/NaOH buffer (pH 7.4) (room temperature). Plg activation was monitored by observing an increase in the fluorescence emission of 0.5 mm fluorogenic PL substrate using a Varian Eclipse spectrofluorimeter. Insets, dependences of the rate of PL generation from Glu-Plg (A) and Lys-Plg (B) by tPA (●) and uPA (○) on the concentration of dsDNA. The rates of PL generation (v) were calculated from the slopes of the linear dependences of the concentration of the product of the cleavage of fluorogenic PL substrate (Sf) on t2 that are equal to 0.5(B1)v, where B1 = kS1 [Sf]/(KS1m + [Sf]) as described under “Experimental Procedures.”
FIGURE 2.
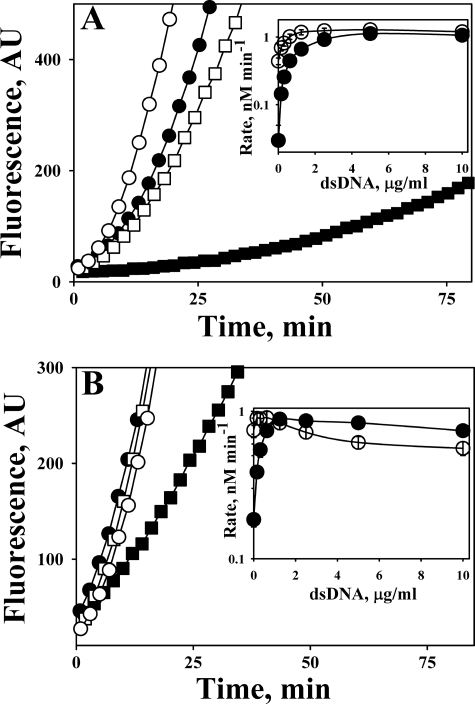
Effects of human dsDNA, ssDNA, RNA, oligo(dT)20, and total nucleic acids from PFs of patients with EMP, PN, and LC on activation of Glu-Plg (A) and Lys-Plg (B) by tPA. Plg (0.1 μm) was incubated in 0.05 m Hepes/NaOH (pH 7.4) with 0.18 nm tctPA alone or together with total nucleic acids isolated from 0.1 ml of PF of patients with EMP, PN, and LC, or with commercially available human dsDNA (1 μg/ml). A mixture of Plg with human dsDNA (1 μg/ml) was used as a negative control; n = 3 for PN; and n = 2 for all other PF samples. Inset, effect of human dsDNA (●), ssDNA (▴), RNA (♦), and oligo(dT)20 (▾) on the rate of PL generation from Glu-Plg (A) or Lys-Plg (B) by tctPA (0.18 nm). The rates of PL generation (v) were calculated from the slopes of the linear dependences of [pNA], formed due to the hydrolysis of PL chromogenic substrate (Sc), on t2 that are equal to 0.5(B2)v, where B2 = kS2 [Sc]/(KS2m + [Sc]).
Human ds- and ssDNA and Nucleic Acids from Pleural Fluids Accelerate Plg Activation
Although incubation of Glu- (Fig. 2A) and Lys- (Fig. 2B) Plg with human dsDNA (1 μg/ml) without tPA did not result in the conversion of Plg to PL, human dsDNA accelerated the activation of both Lys- and Glu-Plg by tPA (Fig. 2) in a manner similar to that observed for salmon dsDNA (Fig. 1). To determine whether or not DNA from the PFs of patients with pleural loculation affects the activation of Plg, samples of total nucleic acids were isolated from the PFs of PN or EMP patients. Control PFs were from patients with LC and CHF. Nucleic acids isolated from the PFs of patients with PN and EMP increased Glu-Plg (Fig. 2A) and Lys-Plg (Fig. 2B) activation by tPA. Notably, the amount of isolated nucleic acids (estimated by electrophoresis in agarose gel with ethidium bromide; data not shown) correlated with the observed effect on the rate of PL generation. The increase in the rate of Plg activation was considerably lower for the PFs of patients with LC (Fig. 2, A and B) and CHF (data not shown), which contained less DNA than the PFs of patients with EMP and PN.
The magnitude of the effect of ds- and ssDNA on the activation of Plg by tPA was substantially higher for Glu-Plg (Fig. 2A, inset) than that for Lys-Plg (Fig. 2B, inset). In contrast to DNA, neither RNA (Fig. 2, insets) nor short ss-oligonucleotides (oligo(dT)20 (Fig. 2, inset), and oligo(dAT)10 (data not shown)) affected the activation of Glu-Plg (Fig. 2A, inset) or Lys-Plg (Fig. 2B, inset) by tPA. Low molecular weight dsDNA (0.2–1.0 kb), obtained by partial DNase I digestion of dsDNA, and longer oligonucleotides (oligo(dT)65 and oligo(dAT)33) potentiated the activation of Plg in a manner similar to that observed for high molecular weight dsDNA (data not shown).
dsDNA Increases Specificity of Plg Activation
The effects of dsDNA on the kinetic parameters (kcat and Km) for the activation of Glu- and Lys-Plg by tPA and uPA were determined next as described under “Experimental Procedures.” The values of kcat and Km, which were calculated from the dependences of the rates of generation of PL from Plg by tPA or uPA, with or without dsDNA, are shown in Table 1. dsDNA induced a significant increase in the specificity (kcat/Km) of Plg activation by tPA by ∼480 and 71 times for Glu- and Lys-Plg, respectively. However, the effect of DNA on the activation of Plg by uPA was smaller (10.7- and 17.0-fold increase, respectively, Table 1). Therefore, dsDNA likely serves as a macromolecular template for fibrin-independent Plg activation, bringing plasminogen activator and Plg into close physical proximity. However, the binding of the ligand (oligonucleotide greater than 60 bases in length) could result in the activation of the enzyme toward its substrate Plg. To further elucidate DNA-mediated Plg activation, the interaction of DNA with fibrinolytic enzymes was studied.
TABLE 1.
Effect of dsDNA on the kinetic parameters of activation of Glu- and Lys-Plg by tPA and uPA
kcat and Km were calculated from double-reciprocal plots of the rate of PL formation by tPA or uPA and Plg concentration (0.025–1.0 μm) as described under “Experimental Procedures.” A linear equation was fit to the data; x and y axis intercepts were −1/Km and 1/Vm, respectively. The values of kcat were calculated as Vm/[E].
| Enzyme | Glu-Plg |
Lys-Plg |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| dsDNA | Km | kcat | kcat/Km | Increase | Km | kcat | kcat/Km | Increase | |
| μg/ml | μm | s−1 | μm−1s−1 | -fold | μm | s−1 | μm−1s−1 | -fold | |
| tctPA | 0 | 9.0 ± 2.5 | 0.009 ± 0.002 | 0.001 | 1 | 6.2 ± 4 | 0.040 ± 0.004 | 0.007 | 1 |
| tcuPA | 0 | 0.90 ± 0.22 | 0.60 ± 0.07 | 0.66 | 1 | 4.5 ± 0.9 | 1.2 ± 0.2 | 0.27 | 1 |
| tctPA | 5.0 | 0.025 ± 0.007 | 0.012 ± 0.002 | 0.48 | 480 | 0.03 ± 0.01 | 0.015 ± 0.002 | 0.50 | 71 |
| tcuPA | 5.0 | 0.12 ± 0.04 | 0.85 ± 0.12 | 7.08 | 10.7 | 0.20 ± 0.10 | 0.92 ± 0.13 | 4.60 | 17.0 |
Gel Shift Analysis of Interactions between Fibrinolytic Enzymes and DNA
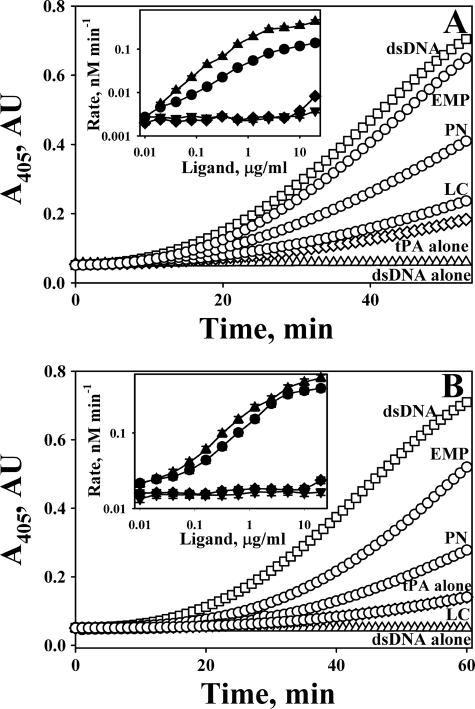
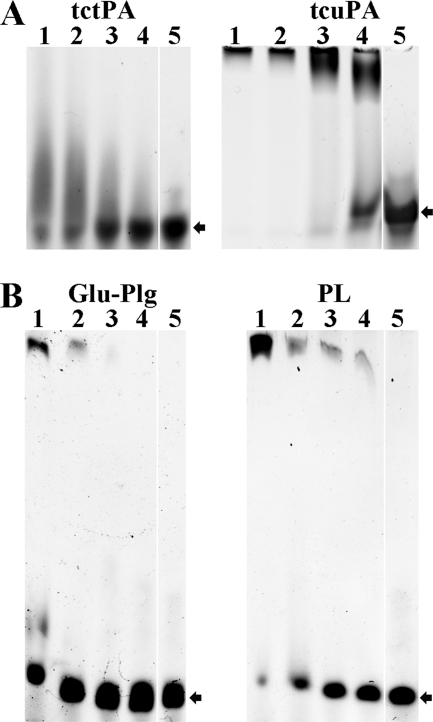
Direct binding of tcuPA, tctPA, PL, and Plg to oligonucleotides was demonstrated by EMSA. Mixtures of oligonucleotide (0.08–0.4 μm) with enzyme (0.25–10 μm) were subjected to electrophoresis in 6% DNA-retardation PAGE (Invitrogen). Unlabeled oligonucleotides were visualized with ethidium bromide (Fig. 3). The effect of tctPA and tcuPA (1.25–10.0 μm) on the electrophoretic mobility of 400 nm oligo(dAT)33 is shown in Fig. 3A. Although the majority of the oligonucleotide was localized to the top of the tcuPA gel (Fig. 3A, right gel), where tcuPA was identified by protein staining (data not shown), an oligo(dAT)33 smear was detected on the tctPA gel (Fig. 3A, left gel), indicating relative instability of the tctPA·oligonucleotide complex under the EMSA conditions. tPA and uPA used in the same concentrations affected the mobility of the longer oligonucleotides to a greater extent (Fig. 3A) than that of the oligo(dAT)10 (data not shown). However, although both tctPA and tcuPA bind long oligo(dAT)33 (Fig. 3A) and short oligo(dAT)10 or oligo(dT)20 (data not shown) oligonucleotides, only longer ones (oligo(dAT)33 and oligo(dT)65) potentiated Plg activation. Effects of Glu-Plg and PL on the electrophoretic mobility of oligo(dAT)33 are shown in Fig. 3B. Although both the proenzyme and enzyme bind to the oligonucleotide, the complex of PL with oligo(dAT)33 apparently is more stable under the EMSA conditions.
FIGURE 3.
Effects of tcuPA, tctPA, Glu-Plg, and PL on the electrophoretic mobility of oligo(dAT)33. A, effects of tctPA (left panel) and tcuPA (right panel) on the electrophoretic mobility of 400 nm oligo(dAT)33. The concentrations of plasminogen activators in the reaction mixture (10 μl) loaded on the gel were 10.0 (lane 1), 5.0 (lane 2), 2.5 (lane 3), and 1.25 (lane 4) μm, respectively. Lane 5 represents the electrophoretic mobility of the free probe (oligonucleotide without protein) in the same gel. Arrows indicate the position of the free oligonucleotide. The positions of the proteins were verified by staining with SimplyBlue (Invitrogen). B, effects of Glu-Plg (left panel) and PL (right panel) on the electrophoretic mobility of oligo(dAT)33. The concentrations of proteins in the reaction mixture (10 μl) were 10.0 (lane 1), 5.0 (lane 2), 2.5 (lane 3), and 1.25 (lane 4) μm, respectively. Lane 5 represents the electrophoretic mobility of the free probe in the same gel. Arrows indicate the position of the free oligonucleotide. The positions of the proteins were verified by staining with SimplyBlue (Invitrogen). Oligo(dAT)33 mixed with increasing amounts of the enzyme (0–10 μm) in 10 μl of 10 mm Hepes (pH 7.4) were resolved by gel electrophoresis using a 6% DNA retardation gel from Invitrogen. Gels were developed in ethidium bromide, and the position of the oligonucleotide was visualized using a Molecular Imager supplemented with Quantity One (version 4.2.3) software (Bio-Rad) as described under “Experimental Procedures.”
Binding of dsDNA and Oligonucleotides to Fibrinolytic Enzymes
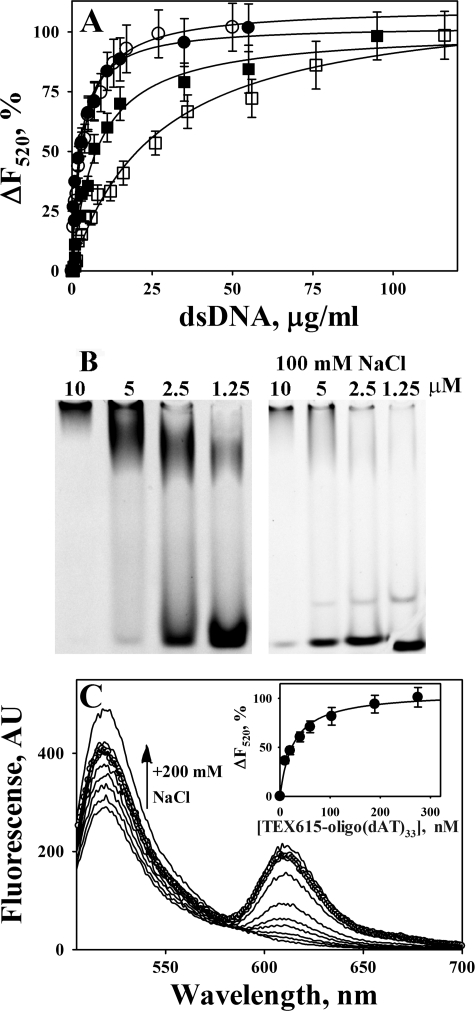
The affinities of fibrinolytic enzymes to dsDNA and oligonucleotides were measured under equilibrium conditions, using either the changes in the intrinsic tryptophan fluorescence as described previously (60, 61, 67) or by monitoring the quenching of the fluorescein fluorescence emission that resulted from the interaction of DNA and FITC-labeled proteins. Equilibrium fluorescence titration of tPA and uPA (data not shown), as well as their FITC-derivatives (Fig. 4A), by dsDNA was employed to estimate the affinity of enzyme/DNA interactions. dsDNA induces concentration-dependent quenching of both the intrinsic tryptophan fluorescence emission of WT enzymes at 340 nm and FITC-fluorescence emission at 520 nm. The dependences of changes in the fluorescence at 520 nm (excitation at 493 nm) of FITC-tcuPA and FITC-tctPA with and without 200 mm NaCl on concentration of dsDNA are shown in Fig. 4A. As expected from the results of EMSA analysis, the affinity of tPA and uPA to dsDNA decreases with an increase in the ionic strength of the solution and the effect of NaCl on uPA is stronger than that for tPA (Fig. 4A; Table 2). The values of Kd for tPA and uPA to dsDNA were 2.7 ± 0.5 and 4.2 ± 0.6 μg/ml, which approximately correspond to 70–80 and 100–140 nm of ds-60-mer, respectively. The values of Kd for oligo(dT)65 (intrinsic tryptophan quenching) were 70 ± 20 and 170 ± 20 nm for tPA and uPA, respectively (Table 2).
FIGURE 4.
Binding of WT and FITC-tctPA and FITC-tcuPA to oligonucleotides. A, dependence of the changes in the fluorescence emission at 520 nm (ΔF520) (excitation at 493 nm) on the dsDNA concentration for the equilibrium fluorescence titration of 20 nm FITC-tctPA and FITC-tcuPA in 0.05 m Hepes/NaOH buffer (pH 7.4). FITC-tctPA (●) and FITC-tcuPA (○) with 20 mm NaCl, FITC-tctPA (■), FITC-tcuPA (□) with 200 mm NaCl. ΔF520 were calculated as 100·(F − Fmin)/(F0 − Fmin), where F0, F, and Fmin are fluorescence emission at 520 nm without dsDNA, after the addition of dsDNA, and at saturation, respectively. Solid lines represent the best fit (r2 >0.90) of the hyperbolic equation ΔF520 = 100·(dsDNA)/(Kd + (dsDNA)), where (dsDNA) is the concentration of dsDNA in μg/ml, to the data using SigmaPlot 11.0. The values of the apparent dissociation constant Kd, which correspond to the concentration of dsDNA that induced half of the maximal quenching of the fluorescence emission, were 2.7 ± 0.5 and 4.2 ± 0.6 μg/ml for tctPA and tcuPA with 20 mm NaCl, and 14.3 ± 1.6; 32.0 ± 4.0 μg/ml for tctPA and tcuPA with 200 mm NaCl, respectively (Table 2). B, EMSA analysis of the interaction of TEX615-oligo(dAT)33 with tcuPA. Mixtures of 400 nm of TEX615-oligo(dAT)33 with increasing amounts of the enzyme (0–10 μm) in 10 μl of 10 mm Hepes (pH 7.4) (left panel) or 0.5× TBE (pH 8.0) buffer with 100 mm NaCl (right panel) were resolved by gel electrophoresis using a 6% DNA retardation gel, Invitrogen. The position of the oligonucleotides was visualized using a Molecular Imager supplemented with Quantity One (version 4.2.3) software (Bio-Rad) as described under “Experimental Procedures.” The upper band represents TEX615-oligo(dAT)33 co-migrating with tcuPA. The position of tcuPA was verified by staining with SimplyBlue, Invitrogen. EMSA analysis of mixtures of TEX615-oligo(dAT)33 with FITC-tctPA, FITC-tcuPA, and FITC-Plg directly demonstrated the co-migration of labeled protein and oligonucleotide in the top band (data not shown). The lower band represents free oligo(dAT)33. The concentration of the enzyme is indicated above the lanes. C, changes in the fluorescence emission spectrum of 20 nm FITC-tcuPA in the presence of increasing concentrations (0–270 nm) of TEX615-oligo(dAT)33. An arrow indicates the effect of an increase in the ionic strength on the fluorescence emission at 520 nm (200 mm NaCl was added to the mixture of 20 nm FITC-tcuPA and 270 nm of TEX615-oligo(dAT)33 (○)). Inset, dependence of the changes in the fluorescence emission at 520 nm (ΔF520) (●) (excitation at 493 nm) on [TEX615-oligo(dAT)33] for the equilibrium fluorescence titration of FITC-tcuPA. The values of ΔF520 were calculated as 100·(F − Fmin)/(F0 − Fmin), where F0, F, and Fmin are fluorescence emission at 520 nm without TEX615-oligo(dAT)33, after the addition of TEX615-oligo(dAT)33, and at saturation, respectively. The solid lines represent the best fit (r2 = 0.96) of the hyperbolic equation ΔF520 = 100·[TEX615-oligo(dAT)33]/(Kd + [TEX615-oligo(dAT)33]) to the data, using SigmaPlot 11.0. The value of the apparent dissociation constant Kd, which corresponds to the concentration of TEX615-oligo(dAT)33 that induced half of the maximal quenching, was 31 ± 5 nm (Table 2).
TABLE 2.
Affinities of dsDNA and oligonucleotides to WT and FITC-modified tctPA and tcuPA in 0.05 m Hepes/NaOH buffer (pH 7.4) at 25 °C
| Enzyme | dsDNA |
oligo(dT)65 20 mm NaCl | TEX615-oligo(dAT)33 20 mm NaCl | |
|---|---|---|---|---|
| 20 mm NaCl | 200 mm NaCl | |||
| μg/ml | nm | nm | ||
| tctPAa | 2.7 ± 0.5 | 14.3 ± 1.6 | 70 ± 20 | NDb |
| tcuPAa | 4.2 ± 0.6 | 32.0 ± 4.0 | 170 ± 20 | ND |
| FITC-tctPAc | 2.3 ± 0.3 | 7.6 ± 1.1 | ND | 25 ± 9 |
| FITC-tcuPAc | 3.4 ± 0.4 | 26.5 ± 3.6 | ND | 31 ± 5d |
a The values of apparent Kd were determined from measurements of the enzyme's intrinsic tryptophan quenching by dsDNA and oligo(dT)65.
b ND means not determined.
c The values of apparent Kd were determined from measurements of the changes in FITC-enzyme fluorescence emission at 520 nm (excitation at 493 nm) induced by dsDNA (Fig. 4A) or TEX615-oligo(dAT)33 (Fig. 4C).
d The value of the apparent Kd for the interaction of FITC-uPA with unlabeled oligo(dAT)33 was 27 ± 4 nm.
TEX615-oligo(dAT)33, an oligonucleotide with a single TEX615 dye molecule attached to the 5′ end, was employed to further elucidate the mechanism of interaction between DNA and plasminogen activators. uPA (Fig. 4B), tPA, Plg, and PL (data not shown) affect the electrophoretic mobility of TEX615-oligo(dAT)33 in a manner similar to that found for oligo(dAT)33 (Fig. 3). An increase in NaCl concentration affects stability of the tcuPA·oligonucleotide complex (Fig. 4B, right panel) and tctPA (data not shown). Similarly, an increase in the ionic strength destabilized oligonucleotide·plasminogen activator complexes with unlabeled oligo(dAT)33 (data not shown). Therefore, electrostatic interactions at the enzyme/DNA interface and possibly a hydrophobic effect contribute to the binding of oligonucleotides to plasminogen activators. TEX615-oligo(dAT)33 quenches the fluorescence emission of FITC-tcuPA (Fig. 4C) and FITC-tctPA (data not shown) in a manner similar to that observed for dsDNA (Fig. 4A). The values of Kd calculated for the interaction of TEX615-oligo(dAT)33 with FITC-tcuPA and FITC-tctPA are shown in the Table 2. Thus, an increase in the ionic strength decreased the affinity of tcuPA and tctPA (as well as FITC-tcuPA and FITC-tctPA) to dsDNA and TEX615-oligo(dAT)33 (Fig. 4; Table 2).
Mechanism of Interaction of DNA with Plasminogen Activators
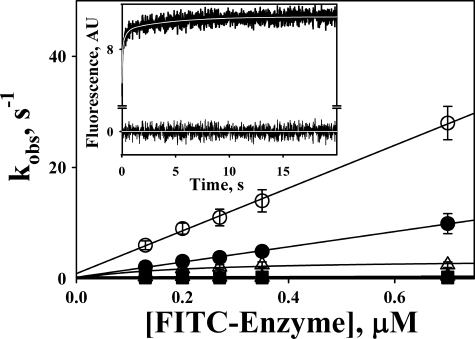
Interaction between FITC-enzymes and TEX615-oligo(dAT)33 results in a decrease in the fluorescence emission of fluorescein at 520 nm (excitation at 493 nm) and an increase in the fluorescence of TEX615, probably due to FRET between FITC on the enzyme and TEX615. A time course of the increase in the fluorescence emission monitored through a 650-nm cutoff filter (excitation at 493 nm) after fast mixing of FITC-tcuPA and TEX615-oligo(dAT)33 using a stopped-flow spectrofluorimeter is shown in Fig. 5, inset. A three-exponential equation was fit to the data as described under “Experimental Procedures.” The dependences of kobs on [FITC-enzyme] are shown in Fig. 5. The mechanism of interaction of both tcuPA and tctPA with TEX615-oligo(dAT)33 most likely includes a fast bimolecular step (kobs,1 increased linearly with increasing [FITC-enzyme]) with second-order rate constants as k1 = 39 ± 4 and 14 ± 2 μm−1 s−1, respectively. The corresponding values of the dissociation rate constant (k-1) for the first step of the interaction were 0.9 ± 0.2 and 0.3 ± 0.1 s−1, for FITC-tcuPA and FITC-tctPA, respectively. The tPA mechanism also includes two monomolecular transformations with k2 = 0.35 ± 0.10 and k3 = 0.10 ± 0.02 s−1, which were independent of [FITC-tctPA] (Fig. 5). In contrast, the hyperbolic dependence of kobs,2 on [FITC-tcuPA] (Fig. 5) could reflect the interaction of the second molecule of the enzyme with TEX615-oligo(dAT)33 with K0.5 = 0.21 ± 0.12 μm. Nevertheless, similar to tctPA, formation of the FITC-tcuPA·DNA complex was followed by two transformations, with k2 = 3.5 ± 0.7 and k3 = 0.30 ± 0.05 s−1, respectively (Fig. 5). Therefore, the simplest mechanism of interaction of the plasminogen activators with TEX615-oligo(dAT)33 likely consists of three reversible steps: a fast formation of the initial enzyme·oligonucleotide complex followed by two relatively slow transformations, most likely due to an induced fit between molecules of the enzyme and DNA.
FIGURE 5.
Stopped-flow kinetics of interaction of TEX615-oligo(dAT)33 with FITC-tctPA (filled symbols) and FITC-tcuPA (open symbols). The dependences of kobs,1 (circles); kobs,2 (triangles), and kobs,3 (squares) obtained from the best fit of the three-exponential equation to stopped-flow traces (inset) on [FITC-enzyme] are shown. Solid lines represent the best fit of linear equations kobs,1 = k−1 + k+1·[FITC-enzyme] (○ and ●); kobs = const (▴, □, and ■); and the best fit of a hyperbolic equation kobs = klim·[FITC-enzyme]/(K0.5 + [FITC-enzyme] (Δ), where klim is kobs at infinite [FITC-Enzyme] and K0.5 is the [FITC-enzyme] at kobs = klim/2. The values of the rate constants for the first, second, and third step of the mechanism of interaction between TEX615-oligo(dAT)33 with FITC-tctPA were 14 ± 2 μm−1s−1 and 0.35 ± 0.10 and 0.10 ± 0.02 s−1 and with FITC-tcuPA were 39 ± 4 μm−1s−1 and 3.5 ± 0.7 and 0.30 ± 0.05 s−1, respectively. Inset, time dependence of the changes in fluorescence emission, monitored through a 650-nm cutoff filter (excitation at 493 nm), due to the interaction of FITC-uPA (0.35 μm) and TEX615-oligo(dAT)33 (30 nm). Pro-Data Viewer software (Applied Photophysics Ltd.) was employed to fit single, two-, and three-exponential equations to the data. The white line inside the trace represents the best fit of a three-exponential equation, Ft = F∞ + A1·e−(kobs,1)t + A2·e−(kobs,2)t + A3·e−(kobs,3)t (where Ft is the fluorescence emission at time t; F∞ is the final fluorescence (AU); kobs and A are observed rate constants and amplitudes, respectively), to the data. Residuals are shown below the trace.
Effects of DNA on Inhibition of tPA and uPA by PAI-1 and PAI-1·Vn Complexes
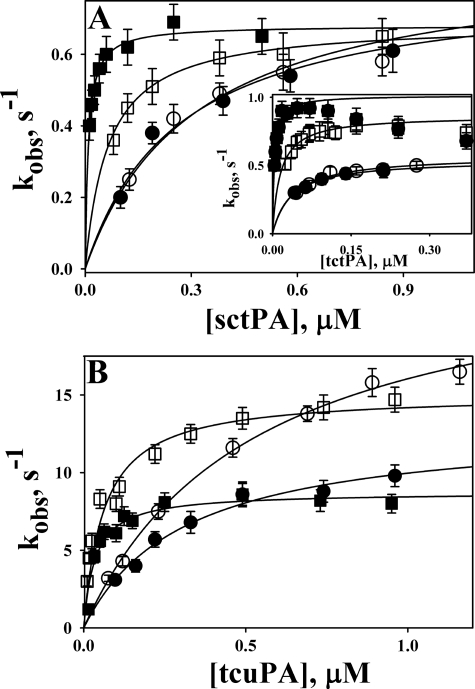
To determine the effects of DNA on the kinetics of the reaction between PAI-1 and its target proteinases, NBD P9 PAI-1 was employed. NBD P9 PAI-1 reports the insertion of RCL by a significant increase in the fluorescence emission of the NBD group (57). The values of kobs for the RCL insertion were calculated from time traces of the increase in the fluorescence emission of NBD P9 PAI-1 reacting with tPA or uPA in the presence of dsDNA (Fig. 6A). The dependences of kobs on the proteinase concentration demonstrated saturation for tctPA and sctPA (Fig. 6A) and tcuPA (Fig. 6B). The values of klim and Km were calculated by fitting a hyperbolic equation to the data (Table 3). dsDNA alone induced an 1.5-fold decrease in the klim for the reaction of NBD P9 PAI-1 with uPA but did not significantly affect the klim for the reaction with sc- and tctPA (Fig. 6). Conversely, dsDNA induced a significant decrease in the Km value for each enzyme (Table 3), which resulted in a corresponding increase in the efficiency of RCL insertion (klim/Km). Therefore, the effects of DNA on the kinetics of the reaction between plasminogen activators and PAI-1 were similar to those observed for the reaction of tPA and uPA with Plg.
FIGURE 6.
Effects of dsDNA on the inactivation of tctPA, sctPA (A), and tcuPA (B) by NBD P9 PAI-1 and its complexes with Vn. The dependences of kobs on the [proteinase] for the reactions of NBD P9 PAI-1 (open symbols) or its complex with Vn (filled symbols) with (squares) or without (circles) dsDNA (10 μg/ml) are shown. The values of kobs were calculated by fitting a single exponential equation to the time traces of an increase in NBD-fluorescence emission. The lines represent the best fit (r2 >0.98) of a hyperbolic equation kobs = klim·[E]/(Km + [E]) to the data shown. The values of klim (the limiting rate of RCL insertion) and Km (concentration of the enzyme at kobs = klim/2) for the reactions of tPA and uPA with NBD P9 PAI-1 and its complex with Vn are shown in Table 3.
TABLE 3.
The kinetic parameters and stoichiometry of inhibition for the reaction of sctPA, tctPA, and tcuPA with NBD P9 PAI-1 and its Vn complexes in the presence of dsDNA
klim and Km were calculated by fitting an equation kobs = klim [E]/(Km + [E]) to plots of kobs versus [E] using SigmaPlot 11.0 (SPSS, Inc.) as described under “Experimental Procedures.” The stoichiometry of inhibition (SI), the number of moles of PAI-1 required to inhibit 1 mol of the enzyme, was estimated from the results of a gradient SDS-PAGE of the products of the reaction of recombinant WT PAI-1 and its complexes with Vn with the proteinase with or without dsDNA (10 μg/ml).
| Enzyme | dsDNA |
Vn |
dsDNA + Vn |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| klim | Km | klim/Km | SI | klim | Km | klim/Km | SI | klim | Km | klim/Km | SI | |
| s−1 | nm | μm−1s−1 | s−1 | nm | μm−1s−1 | s−1 | nm | μm−1s−1 | ||||
| tctPA | 0.87 ± 0.03 | 16 ± 4 | 54.4 | 1.1 ± 0.2 | 0.54 ± 0.04 | 34 ± 6 | 15.9 | 1.0 ± 0.1 | 1.04 ± 0.04 | 4 ± 3 | 260 | 1.1 ± 0.1 |
| sctPA | 0.70 ± 0.04 | 60 ± 10 | 11.7 | 1.0 ± 0.1 | 0.86 ± 0.05 | 300 ± 20 | 2.9 | 1.0 ± 0.1 | 0.68 ± 0.03 | 10 ± 4 | 68 | 1.1 ± 0.2 |
| tcuPA | 15.2 ± 0.6 | 70 ± 10 | 218 | 1.2 ± 0.1 | 13.0 ± 0.2 | 300 ± 30 | 43.3 | 1.1 ± 0.2 | 8.6 ± 0.2 | 30 ± 5 | 280 | 1.2 ± 0.2 |
Vn, an endogenous PAI-1 cofactor that, like Plg, is present in the bloodstream in micromolar concentrations (68, 69) stabilizes the active PAI-1 conformation (70, 71) and affects the kinetics of the reaction between PAI-1 (PAI-1·mAb complexes) and tcuPA or tctPA (72, 73). The effects of Vn on the kinetics of RCL insertion for the reactions of NBD P9 PAI-1 with sc- and tctPA and tcuPA and values of klim, Km, and klim/Km are shown in Fig. 6 and in Table 3, respectively. Vn induced a 1.8-fold decrease in the klim for uPA but did not affect the klim for tPA (Table 3) (72). Vn significantly potentiated the effect of DNA on the specificity of PAI-1 inhibition of both sc- and tctPA, considerably decreasing the Km for the reaction and having a lesser effect on the reaction with tcuPA, affecting both klim and Km (Table 3).
SDS-PAGE analysis of the products of the reaction (59) was employed to determine the effects of ds- and ssDNA, RNA, oligo(dT)20, and oligo(dT)65 on the SI for WT PAI-1. Fluorescence titration (58, 72) was employed to determine the SI for the reactions of tPA and uPA with NBD P9 PAI-1. Neither of the ligands significantly affected the partitioning between the inhibitory and the substrate branches of the PAI-1 mechanism for the reactions of PAI-1 with the target proteinases. Vn, which by itself does not affect the SI but potentiates PAI-1 neutralization by monoclonal antibodies (72), caused no effect on the reactions of PAI-1 with tPA and uPA (Table 3) in the presence of DNA.
Effect of DNA on Inhibition of PL by α2AP
The mechanism of inhibition of PL by α2AP includes the fast formation of the Michaelis-like complex (PL* α2AP; Scheme 2) with low nanomolar affinity, followed by slow inactivation of the enzyme (74, 75).
SCHEME 2.
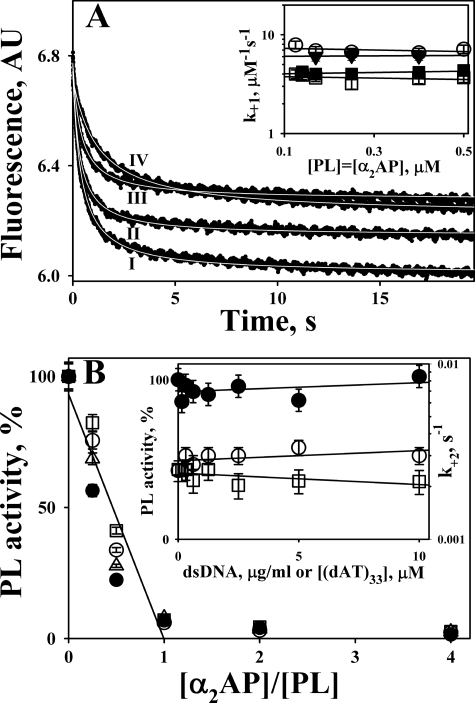
To test whether or not DNA affects the rate of the first step of the reaction between PL and α2AP, changes in the intrinsic tryptophan fluorescence emission that accompany the formation of the Michaelis complex between the two proteins (63) were monitored using stopped-flow fluorimetry (Fig. 7A). Although the amplitude of the fluorescence emission quenching decreased in the presence of dsDNA or oligo(dAT)33 (Fig. 7A), ligands did not significantly affect the values of the second-order rate constants (Fig. 7A, inset). Even at 100 μg/ml, dsDNA only slightly affected the rate of formation of the Michaelis complex (Fig. 7A, traces I and IV). The values of the second-order rate constant (k+1) (Scheme 2) without DNA but with 1.7 μm oligo(dAT)33, 3.4 μm oligo(dAT)33, 30 μg/ml dsDNA, and 100 μg/ml dsDNA were 7.2 ± 0.6, 6.1 ± 0.4, 5.1 ± 0.4, 4.1 ± 0.4, and 3.6 ± 0.2 μm−1 s−1 (Fig. 7A). Therefore, it is unlikely that DNA interacts with the active site or the kringle 1 domain of PL, which plays a major role in the interaction with α2AP (76).
FIGURE 7.
DNA and oligo(dAT)33 do not affect significantly interaction between PL and α2AP. A, changes in the intrinsic tryptophan fluorescence emission (excitation at 290 nm) after mixing equal volumes of 0.25 μm PL and 0.25 μm α2AP in the presence of 100 μg/ml dsDNA (IV), 3.4 μm oligo(dAT)33 (III), 1.7 μm oligo(dAT)33 (II), and without DNA (I), using stopped-flow fluorimeter. The second-order rate constants (k+1 (Scheme 2)) were calculated as described elsewhere (63). The white lines inside the traces represent the best fit. Inset, values of k+1 measured at different [PL] = [α2AP] without DNA (○), with 30 μg/ml dsDNA (■), with 100 μg/ml dsDNA (□), and with 1.7 μm oligo(dAT)33 (▾). B, DNA does not affect the dependence of the residual PL amidolytic activity on [α2AP]/[PL]. PL (2.0 nm) was preincubated with 0 (●), 0.625 (○), 2.5 (Δ), and 10 (□) μg/ml dsDNA. An equal volume of mixture of the PL substrate (1.0 mm) and α2AP (0–40 nm) was added to start the reaction. Residual amidolytic PL activity was plotted against the ratio of [α2AP]/[PL] to determine the SI. A solid line represents the best fit of the linear equation to all data observed for 0 ≤ [α2AP]/[PL] ≤ 1. Inset, left axis, dsDNA does not affect the amidolytic PL activity (●); PL (2 nm) was preincubated with dsDNA (0–10 μg/ml). Then an equal volume of 1.0 mm fluorogenic substrate was added, and changes in the fluorescence emission with time were detected using a Varian Eclipse spectrofluorimeter. The relative amidolytic activity of PL (% of PL activity without dsDNA) was calculated from the slopes of linear increase in the fluorescence emission with time. Inset, right axis, dependence of the first-order rate constant (k+2) for the second slow PL inactivation (Scheme 2) on the concentration of dsDNA (○) or oligo(dAT)33 (□). To determine the effects of DNA and oligo(dAT)33 on the rate of the second slow step of the reaction between PL and α2AP, an assay described by Wiman and Collen (65) was employed.
To test whether or not DNA (0–10 μg/ml) affects the stoichiometry of inhibition of PL by α2AP, the reaction between the enzyme (2 nm) and the serpin (0–80 nm) was carried out in the presence of the fluorogenic PL substrate as described under “Experimental Procedures.” Changes in PL activity were monitored for 0–300 min. The residual amidolytic activity of PL (after a 60-min incubation) was plotted against [PL]/[α2AP] (Fig. 7B). Neither dsDNA nor oligo(dAT)33 significantly affected the stoichiometry of inhibition of PL by α2AP (Fig. 7B) or the amidolytic activity of PL (Fig. 7B, inset, filled symbols, left axis).
Finally, the effect of dsDNA (0–10 μg/ml) and oligo(dAT)33 (0–10 μm) on the rate constant for the second slow step of the reaction (k+2; Scheme 2) was studied by monitoring the residual PL amidolytic activity after mixing high concentrations of PL (0.1–0.5 μm) with equimolar amounts of α2AP in the presence of the fluorogenic substrate, as described previously (63, 77). Although the values of k+2 did not change with an increase in the dsDNA concentration and slightly decreased with an increase in the [oligo(dAT)33] (Fig. 7B, inset, open symbols, right axis), the effect was rather moderate (the values of k+2 without ligands and with 10 μm oligo(dAT)33 and with 10 μg/ml dsDNA were 2.5 ± 0.4, 2.1 ± 0.3, and 3.0 ± 0.4 × 10−3 s−1, respectively; Fig. 7B, inset).
These results demonstrate that despite accelerating Plg activation by tPA and uPA, DNA and oligonucleotides, unlike fibrin, do not protect tPA, uPA, or PL from inactivation by serpins. Therefore, similar to protein aggregates (45, 46), EC DNA is able to form a macromolecular template, which could compete for fibrinolytic enzymes with fibrin and therefore affect fibrinolysis.
Effects of DNA in Solution on Fibrinolysis in Vitro
The quenching-dequenching of fluorescence emission during the polymerization of FITC-Fbg and its cleavage by PL, respectively (66), was used to monitor the effect of DNA on the rates of fibrinolysis. FITC-fibrin films were formed in 96-well plates as described under “Experimental Procedures.” Pre-formed FITC-fibrin films (66) were incubated with either 20 nm PL or with a mixture of 100 nm Glu-Plg with 0.25 nm plasminogen activator (tPA or uPA) in 0.05 m Hepes buffer (pH 7.4) (20 mm NaCl, 1 mg/ml BSA) with or without dsDNA (0–25 μg/ml). Fibrinolysis was monitored as an increase in the fluorescence emission at 520 nm (excitation 493 nm) using a Varian Eclipse spectrofluorimeter (Fig. 8A). Although dsDNA at low concentrations (0.05–0.5 μg/ml) did not significantly affect fibrinolysis by PL, higher levels of DNA caused inhibition (Fig. 8A, inset). In contrast to PL, 0.1–1.0 μg/ml dsDNA increased the rates of fibrinolysis by mixtures of 100 nm Glu-Plg with tPA and uPA by almost 5-fold (Fig. 8A, inset). The observed increase in the rate of fibrinolysis in the presence of dsDNA most likely reflects the contribution of the template mechanism to Plg activation. However, dsDNA at higher concentrations (1.0–25 μg/ml) inhibited fibrinolysis by Glu-Plg with tPA or uPA, similar to its effect on fibrinolysis by PL (Fig. 8A, inset). Thus, despite the fact that DNA in solution did not affect PL amidolytic activity (Fig. 7B, inset, left axis), it is able to compete with fibrin for fibrinolytic enzymes, slow down fibrinolysis, and make enzymes susceptible to mechanism-based inhibition by serpins (Figs. 6 and 7). To test this hypothesis, the rate of fibrinolysis of FITC-fibrin film in the presence of dsDNA by a mixture of Glu-Plg with tPA and uPA was studied in the presence of 15 nm α2AP (Fig. 8A, inset). Because fibrin protects bound PL from inactivation by α2AP, the fibrinolysis of FITC-fibrin was detected. However, there was no activation of fibrinolysis by 0.1–1.0 μg/ml dsDNA in the presence of α2AP (Fig. 8A, inset). Therefore, α2AP likely inactivated the PL bound to DNA in solution.
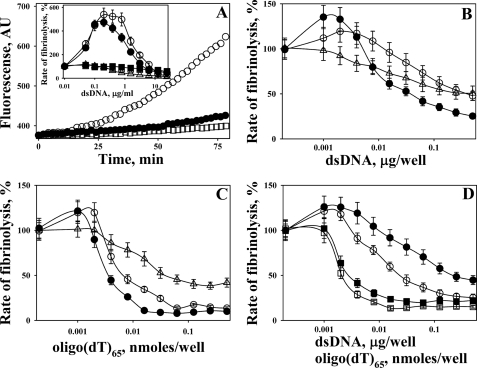
FIGURE 8.
Effects of DNA in solution and DNA incorporated into a FITC-fibrin film on fibrinolysis. A, changes in the fluorescence emission at 520 nm (excitation 493 nm) during the incubation of FITC-fibrin film with 100 μl of a mixture of 100 nm Glu-Plg with 0.25 nm tctPA without DNA (●) and with 1.0 (○) and 25 (□) μg/ml dsDNA in 0.05 m Hepes/NaOH buffer (pH 7.4) (with 20 mm NaCl and BSA 1 mg/ml). Inset, dependences of rates of fibrinolysis of FITC-fibrin film by 20 nm PL (Δ), mixtures of 100 nm Glu-Plg with 0.25 nm tctPA (●), or 0.25 nm tcuPA (○), 100 nm Plg/tctPA with 15 nm α2AP (■) and 100 nm Plg/tcuPA with 15 nm α2AP (□) on the concentration of dsDNA in the solution. The rates of fibrinolysis were calculated as slopes of a linear (r2 >0.9) increase in the fluorescein fluorescence emission with time (PL) or the square of time (tctPA/Plg and tcuPA/Plg) and expressed as the % of the control reaction (no DNA added). B, effects of dsDNA incorporated into a FITC-fibrin film on the rates of fibrinolysis by 20 nm PL (Δ) and a mixture of 100 nm Glu-Plg with 0.25 nm tctPA (●) or 0.25 nm tcuPA (○). The indicated amounts of dsDNA (μg/well) were added to 20 μg of FITC-Fbg before its polymerization. The rates of fibrinolysis were calculated as slopes of a linear (r2 >0.9) increase in the fluorescein fluorescence emission with time (PL) or the square of time (tctPA/Plg) and expressed as % of the control reaction (no DNA added). C, effects of oligo(dT)65 incorporated into FITC-fibrin film on the rates of fibrinolysis by 20 nm PL (Δ) and a mixture of 100 nm Glu-Plg with 0.25 nm tctPA (●) or 0.25 nm tcuPA (○). The indicated amounts of nanomoles of oligo(dT)65 per well were added to 20 μg of FITC-Fbg before its polymerization. Rates of fibrinolysis were calculated as slopes of a linear (r2 >0.9) increase in the fluorescein fluorescence emission with time (PL) or the square of time (tctPA/Plg) and expressed as % of the control reaction (no oligo(dT)65 added). D, effects of α2AP (15 nm) on the rates of fibrinolysis of FITC-fibrin films with incorporated dsDNA (circles) or oligo(dT)65 (squares) by 100 nm Plg/tctPA (filled symbols) and 100 nm Plg/tcuPA (open symbols). The indicated amounts of dsDNA (μg/well) or oligo(dT)65 (nanomoles/well) were added to 20 μg of FITC-Fbg before its polymerization. The rates of fibrinolysis were calculated as slopes of a linear (r2 >0.9) increase in the fluorescein fluorescence emission with the square of time and expressed as % of the control reaction (no dsDNA or oligo(dT)65 added).
Effects of DNA in the Fibrin Matrix on Fibrinolysis in Vitro
To test the effects of DNA on fibrin degradation in vitro, FITC-fibrin films, containing different amounts of dsDNA or oligo(dT)65, were obtained as described under the “Experimental Procedures.” The effects of dsDNA (and oligo(dT)65) incorporated into the FITC-fibrin film on the rates of fibrinolysis by 20 nm PL and 100 nm Glu-Plg with 0.25 nm tPA or uPA are shown in Fig. 8, B and C, respectively. Although at the lowest amounts per well both ligands demonstrated a slight (less than 1.5-fold) increase in the rate of fibrinolysis by mixtures of Glu-Plg with tPA or uPA, an increase in the DNA in FITC-fibrin film resulted in inhibition of fibrinolysis (Fig. 8, B and C).
Finally, the fibrinolytic activity of 100 nm Glu-Plg (with 0.25 nm tPA or uPA) toward FITC-fibrin/DNA films was studied in the presence of 15 nm α2AP (Fig. 8D). In the presence of α2AP, low concentrations of oligo(dT)65 did not increase the rate of fibrinolysis, although dsDNA did (by ∼1.2-fold; Fig. 8D). The latter observation suggests that Plg activation on the dsDNA template, which is incorporated into a fibrin film, could be partially protected from α2AP in solution.
DISCUSSION
This study reports newly recognized interactions between DNA and tPA, uPA, Plg, PL, as well as their endogenous inhibitors PAI-1 and α2AP. Fibrinolytic enzymes bind DNA with submicromolar affinity (Table 2). Increasing the ionic strength of the solution induces a decrease in the affinity of tPA and uPA to DNA, indicating the contribution of electrostatic interactions to the formation of the enzyme·DNA complex. The mechanism of interaction of tPA and uPA with DNA includes a fast binding step, followed by two monomolecular transformations most likely reflecting conformational changes resulting in a mature enzyme·DNA complex. DNA and oligonucleotides greater than 60 bases in length act as a template for Plg activation by tPA and uPA. The DNA template promotes the assembly of a productive enzyme·Plg complex, resulting in an increase in the specificity of the reaction (kcat/Km; Table 1). DNA induces a significant increase in kcat/Km for tPA, mostly due to a dramatic decrease in Km for both Glu- and Lys-Plg (360 and 200 times, respectively). Thus, the increased affinity of Plg to DNA/enzyme templates promotes formation of productive ternary (DNA·enzyme·Plg) complexes. However, the higher apparent affinity of PL to DNA (Fig. 3B) could result in competition between DNA and fibrin for PL and decrease the rate of fibrinolysis (Fig. 8). This decrease in Km value is smaller for uPA (7.5- and 22-fold, respectively). Notably, the effects of dsDNA on Plg activation by tPA were within the same order of magnitude as that described for the fibrin template-mediated mechanism (50, 55). Such an increase in the efficiency of Plg activation could reflect the stabilization of the ternary complex due to conformational changes in the enzyme bound to DNA that bring the substrate closer to the enzyme. As expected for the template mechanism, the length of DNA is an essential factor when forming the ternary complex. Thus, EC DNA could act as a macromolecular template for plasminogen activators and Plg and therefore promote fibrin-independent Plg activation.
Moreover, the DNA template competing with fibrin for fibrinolytic enzymes increases their susceptibility to serpins. Although fibrin protects tPA from inactivation by PAI-1 (79), DNA markedly potentiates the inactivation of sc- and tctPA and tcuPA by PAI-1 (Table 3). Vn, an endogenous ligand of active PAI-1, further increased the efficiency of mechanism-based inhibition of tctPA, sctPA, and tcuPA by PAI-1 (by 4.8, 5.8, and 1.3 times, respectively). As a result, in the presence of both DNA and Vn, the specificity constants for inactivation of tctPA and tcuPA by PAI-1 became almost identical (260 and 280 μm−1 s−1, respectively, see Table 3). Therefore, the interactions of the somatomedin B domain of Vn with the α-helices F and E and strand 1 of β-sheet A of PAI-1 (80) induce conformational changes that could further stabilize DNA·enzyme·PAI-1 complexes. Neither DNA nor DNA with Vn affected the SI for the interaction of PAI-1 with any enzyme (Table 3). Notably, DNA and Vn did not significantly affect the difference in specificity constants between sc- and tctPA (Table 3). In contrast, the rates of inhibition of sc- and tctPA bound to fibrin by PAI-1 were equal (79). DNA and Vn did not affect the reaction between PAI-1 and pro-urokinase (data not shown), which is limited by a slow transition from the inactive to active form of scuPA (59). Thus, it is unlikely that interaction with DNA affects the equilibrium between inactive and active species of the proenzyme.
In contrast to fibrin, which protects PL from inactivation by α2AP (76), DNA only slightly decreases the rate of tight binding of the enzyme to the serpin (Fig. 7A). DNA causes no effect on the rate of the second step of the PL/α2AP reaction (Fig. 7B) or on the stoichiometry of interaction (Fig. 7B). Because dsDNA did not affect PL amidolytic activity, and did not protect PL from inactivation by α2AP (Fig. 7), it is unlikely that interaction between PL and DNA template involves the kringle 1 domain or the active site of the enzyme. Thus, unlike fibrin, DNA does not protect plasminogen activators or PL from endogenous serpins. The latter mechanism is similar to that described for the reaction between cathepsin V and SERP (81).
Thus EC DNA in vivo could both compete with fibrin for fibrinolytic enzymes and induce fibrin-independent Plg activation and promote the inhibition of fibrinolytic enzymes by endogenous serpins. Indeed, a decreased level of Plg in plasma (up to 60% in septic shock (82)) together with increased levels of PAI-1 (41–44) and EC DNA (18, 19) are predictors of the severity of sepsis (83, 84). Moreover, the median Plg level in the PFs of patients with PN is 2.2–4.0 times less than that in the PFs of patients with CHF and LC (85), which correlates with the relative concentrations of EC DNA observed in PFs this study.
The observed effects of DNA on fibrinolysis of FITC-fibrin film support the putative mechanism of DNA interaction with fibrinolytic enzymes. Indeed, although dsDNA (0.1–1.0 μg/ml) in solution caused no effect on the degradation of FITC-fibrin by PL, it markedly potentiated fibrinolysis by Plg/tPA or Plg/uPA, most likely due to the accelerated fibrin-independent activation of Plg. α2AP, which rapidly inactivates PL both in solution and bound to DNA (Fig. 7A) but does not affect the activity of PL bound to fibrin (76), inhibited fibrinolysis that originated from DNA template-mediated Plg activation. Further increasing the DNA concentration (up to 25 μg/ml) resulted in the inhibition of fibrinolysis by both PL or Plg/tPA or Plg/uPA, most likely due to the competition of DNA with fibrin for PL.
The role of the structure of fibrin in fibrinolysis is incompletely understood, and the degradation of a pure fibrin network depends on its structure and thickness (86, 87). The entrapment of DNA could further stabilize the fibrin meshwork and affect both Plg activation and subsequent fibrinolysis. Polyanions such as unfractionated (but not low molecular weight) heparin (88) and polyphosphates (89) affect fibrin structure inducing formation of thicker fibrin fibrils. It is known that myosin (90), other proteins (86, 91, 92), as well as polyphosphates (89) affect fibrinolysis when incorporated into fibrin. Conversely, heparin, which potentiates Plg activation by tPA (47, 88) through a so-called semi-template mechanism (48), does not affect fibrinolysis (48, 93). Recently, the DNA of neutrophil extracellular traps was shown to promote thrombosis and be incorporated into a fibrin structure (1). Indeed, an increase in the dsDNA (Fig. 8B) or oligo(dT)65 (Fig. 8C) entrapped in FITC-fibrin slows fibrinolysis with both PL and Plg/tPA or uPA. Only a small (1.2–1.5-fold) stimulation of fibrinolysis was detected at low concentrations of ligands (Fig. 8, B and C). The increased fibrinolysis with the addition of dsDNA that was observed in the presence of α2AP could reflect the resistance of DNA-mediated Plg activation by DNA/fibrin surfaces to serpins. However, the lack of a similar effect for oligo(dT)65 (Fig. 8D) could indicate that there is a fast dissociation of short oligonucleotides into solution during fibrinolysis.
A two-faceted mechanism of contributions of soluble EC DNA to fibrinolysis suggests the following: (i) the profibrinolytic role of EC DNA-mediated Plg activation under conditions of neutralization by endogenous serpins; (ii) the profibrotic effects of EC DNA under conditions of overexpression of endogenous serpins. The former mechanism could potentially contribute to the success of fibrinolytic therapy with a combination of tPA and DNase in an animal model (94) and in humans with complicated organizing pleural effusions (95). However, the salutary effects of the DNase/tPA treatment have been attributed to decreasing the viscosity of pleural effusions (94, 95). In a related vein, the use of DNase alone should result in an increase in the concentration of soluble EC DNA and products of its degradation, which would promote inhibition of endogenous fibrinolytic enzymes by serpins. Such a mechanism could contribute to the increased mortality and need for surgery that was observed in the DNase group in MIST2 (95). Moreover, high levels of EC DNA intrapleurally, in combination with high serpin activity, could also contribute to the lack of efficiency of the plasminogen activator alone, as observed in both the MIST1 and MIST2 clinical trials (78, 95).
In summary, our results support the concept that fibrinolytic enzymes are prone to distinct molecular interactions in an environment enriched with EC DNA and serpins, as occurs in severe sepsis, complicated parapneumonic effusions, EMP, and acute respiratory distress syndrome. Competition between the fibrin and non-fibrin Plg-activating template formed by protein aggregates for tPA have been recently shown in injured neurons (46). The results of this study demonstrate another macromolecular template, which not only competes with fibrin for tPA, uPA, and PL but also promotes the inhibition of the fibrinolytic enzymes by endogenous serpins. The novel interactions we describe here have potentially important clinical ramifications. This study provides a rationale to determine how DNA/fibrinolysin interactions contribute to the outcome of fibrinolytic therapy in future clinical studies.
Acknowledgments
We thank Genentech for providing sctPA and Abbott for the tcuPA that was employed in this work. We thank Sophia Karandashova for help in the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants PO-1 076406 and P50 HL107186-02 from NHLBI. This work was also supported by the Texas Lung Injury Institute.
- EC
- extracellular
- α2AP
- α2-antiplasmin
- AU
- arbitrary units
- CHF
- congestive heart failure
- EMP
- empyema
- Fbg
- fibrinogen
- Glu-Plg
- native human Plg (residues 1–790) with N-terminal glutamic acid
- LC
- lung cancer
- Lys-Plg
- plasmin cleaved human Plg (residues 77–790)
- MI
- myocardial infarction
- NBD
- N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-3-diazole
- oligo(dAT)n
- ss-2n-mer oligonucleotide containing n AT repeats
- PAI-1
- plasminogen activator inhibitor-1
- PF
- pleural fluid
- PL
- plasmin
- Plg
- plasminogen
- PN
- pneumonia
- pNA
- p-nitroanilide
- RCL
- reactive center loop
- sc
- single chain
- Sc
- chromogenic substrate
- Sf
- fluorogenic substrate
- SI
- stoichiometry of inhibition
- ss
- single stranded
- tc
- two chain
- tPA
- tissue type plasminogen activator
- TEX615
- a red fluorophore, an analog of Texas Red
- uPA
- urokinase type plasminogen activator
- Vn
- vitronectin.
REFERENCES
- 1. Fuchs T. A., Brill A., Duerschmied D., Schatzberg D., Monestier M., Myers D. D., Jr., Wrobleski S. K., Wakefield T. W., Hartwig J. H., Wagner D. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15880–15885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rogers W. J., Bowlby L. J., Chandra N. C., French W. J., Gore J. M., Lambrew C. T., Rubison R. M., Tiefenbrunn A. J., Weaver W. D. (1994) Circulation 90, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 3. Hedstrom L. (2002) Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 4. Albers G. W., Amarenco P., Easton J. D., Sacco R. L., Teal P. (2008) Chest 133, 630S-669S [DOI] [PubMed] [Google Scholar]
- 5. Allport L. E., Butcher K. S. (2008) J. Clin. Neurosci. 15, 917–920 [DOI] [PubMed] [Google Scholar]
- 6. Kim H. S., Patra A., Paxton B. E., Khan J., Streiff M. B. (2006) Cardiovasc. Intervent. Radiol. 29, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 7. Parikh S., Motarjeme A., McNamara T., Raabe R., Hagspiel K., Benenati J. F., Sterling K., Comerota A. (2008) J. Vasc. Interv. Radiol. 19, 521–528 [DOI] [PubMed] [Google Scholar]
- 8. Bouros D., Tzouvelekis A., Antoniou K. M., Heffner J. E. (2007) Pulm. Pharmacol. Ther. 20, 616–626 [DOI] [PubMed] [Google Scholar]
- 9. Idell S. (2005) Clin. Pulm. Med. 12, 184–190 [Google Scholar]
- 10. Hardaway R. M., Harke H., Williams C. H. (1994) Adv. Ther. 11, 43–51 [PubMed] [Google Scholar]
- 11. Hardaway R. M., Williams C. H., Marvasti M., Farias M., Tseng A., Pinon I., Yanez D., Martinez M., Navar J. (1990) Crit. Care Med. 18, 1413–1418 [DOI] [PubMed] [Google Scholar]
- 12. Lo Y. M. (2001) Ann. N.Y. Acad. Sci. 945, 1–7 [DOI] [PubMed] [Google Scholar]
- 13. Wu T. L., Zhang D., Chia J. H., Tsao K. H., Sun C. F., Wu J. T. (2002) Clin. Chim. Acta 321, 77–87 [DOI] [PubMed] [Google Scholar]
- 14. Chang C. P., Chia R. H., Wu T. L., Tsao K. C., Sun C. F., Wu J. T. (2003) Clin. Chim. Acta 327, 95–101 [DOI] [PubMed] [Google Scholar]
- 15. Rainer T. H., Wong L. K., Lam W., Yuen E., Lam N. Y., Metreweli C., Lo Y. M. (2003) Clin. Chem. 49, 562–569 [DOI] [PubMed] [Google Scholar]
- 16. Lam N. Y., Rainer T. H., Wong L. K., Lam W., Lo Y. M. (2006) Resuscitation 68, 71–78 [DOI] [PubMed] [Google Scholar]
- 17. Lo Y. M., Rainer T. H., Chan L. Y., Hjelm N. M., Cocks R. A. (2000) Clin. Chem. 46, 319–323 [PubMed] [Google Scholar]
- 18. Rhodes A., Wort S. J., Thomas H., Collinson P., Bennett E. D. (2006) Crit. Care 10, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saukkonen K., Lakkisto P., Pettilä V., Varpula M., Karlsson S., Ruokonen E., Pulkki K. (2008) Clin. Chem. 54, 1000–1007 [DOI] [PubMed] [Google Scholar]
- 20. Vargo J. S., Becker D. M., Philbrick J. T., Schoonover F. W., Davis J. S. (1990) Chest 97, 63–68 [DOI] [PubMed] [Google Scholar]
- 21. Uzuelli J. A., Dias-Junior C. A., Izidoro-Toledo T. C., Gerlach R. F., Tanus-Santos J. E. (2009) Clin. Chim. Acta 409, 112–116 [DOI] [PubMed] [Google Scholar]
- 22. Benlloch S., Martí-Ciriquián J. L., Galbis-Caravajal J. M., Martín C., Sánchez-Payá J., Rodríguez-Paniagua J. M., Romero S., Massutí B. (2006) Clin. Lung Cancer 8, 140–145 [DOI] [PubMed] [Google Scholar]
- 23. Chan M. H., Chow K. M., Chan A. T., Leung C. B., Chan L. Y., Chow K. C., Lam C. W., Lo Y. M. (2003) Clin. Chem. 49, 740–745 [DOI] [PubMed] [Google Scholar]
- 24. Wijeratne S., Butt A., Burns S., Sherwood K., Boyd O., Swaminathan R. (2004) Ann. N.Y. Acad. Sci. 1022, 232–238 [DOI] [PubMed] [Google Scholar]
- 25. Saukkonen K., Lakkisto P., Varpula M., Varpula T., Voipio-Pulkki L. M., Pettilä V., Pulkki K. (2007) Intensive Care Med. 33, 1624–1627 [DOI] [PubMed] [Google Scholar]
- 26. Idell S., Girard W., Koenig K. B., McLarty J., Fair D. S. (1991) Am. Rev. Respir. Dis. 144, 187–194 [DOI] [PubMed] [Google Scholar]
- 27. Philip-Joët F., Alessi M. C., Philip-Joët C., Aillaud M., Barriere J. R., Arnaud A., Juhan-Vague I. (1995) Eur. Respir. J. 8, 1352–1356 [DOI] [PubMed] [Google Scholar]
- 28. Iglesias D., Alegre J., Alemán C., Ruíz E., Soriano T., Armadans L. I., Segura R. M., Anglés A., Monasterio J., de Sevilla T. F. (2005) Eur. Respir. J. 25, 104–109 [DOI] [PubMed] [Google Scholar]
- 29. Pedersen O. D., Gram J., Bagger H., Keller N., Jespersen J. (1994) Coron. Artery Dis. 5, 617–623 [DOI] [PubMed] [Google Scholar]
- 30. Held C., Hjemdahl P., Rehnqvist N., Wallén N. H., Björkander I., Eriksson S. V., Forslund L., Wiman B. (1997) Circulation 95, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 31. Thögersen A. M., Jansson J. H., Boman K., Nilsson T. K., Weinehall L., Huhtasaari F., Hallmans G. (1998) Circulation 98, 2241–2247 [DOI] [PubMed] [Google Scholar]
- 32. Wiman B. (1999) Scand. J. Clin. Lab. Invest. Suppl. 230, 23–31 [PubMed] [Google Scholar]
- 33. Johansson L., Jansson J. H., Boman K., Nilsson T. K., Stegmayr B., Hallmans G. (2000) Stroke 31, 26–32 [DOI] [PubMed] [Google Scholar]
- 34. Nordenhem A., Leander K., Hallqvist J., de Faire U., Sten-Linder M., Wiman B. (2005) Thromb. Res. 116, 223–232 [DOI] [PubMed] [Google Scholar]
- 35. Zeerleder S., Hack C. E., Wuillemin W. A. (2005) Chest 128, 2864–2875 [DOI] [PubMed] [Google Scholar]
- 36. Haj M. A., Robbie L. A., Croll A., Adey G. D., Bennett B. (1998) Intensive Care Med. 24, 258–261 [DOI] [PubMed] [Google Scholar]
- 37. Watanabe R., Wada H., Miura Y., Murata Y., Watanabe Y., Sakakura M., Okugawa Y., Nakasaki T., Mori Y., Nishikawa M., Gabazza E. C., Shiku H., Nobori T. (2001) Clin. Appl. Thromb. Hemost. 7, 229–233 [DOI] [PubMed] [Google Scholar]
- 38. Dawson S., Henney A. (1992) Atherosclerosis 95, 105–117 [DOI] [PubMed] [Google Scholar]
- 39. Grimaudo V., Bachmann F., Hauert J., Christe M. A., Kruithof E. K. (1992) Thromb. Haemost. 67, 397–401 [PubMed] [Google Scholar]
- 40. Ioannidou-Papayannaki E., Lefkos N., Boudonas G., Efthimiadis A., Psirropoulos D., Vogas V., Papadopoulos I., Klonizakis I. (2000) Acta Cardiol. 55, 247–253 [DOI] [PubMed] [Google Scholar]
- 41. Gando S., Nakanishi Y., Tedo I. (1995) Crit. Care Med. 23, 1835–1842 [DOI] [PubMed] [Google Scholar]
- 42. Iba T., Kidokoro A., Fukunaga M., Sugiyama K., Sawada T., Kato H. (2005) Shock 23, 25–29 [DOI] [PubMed] [Google Scholar]
- 43. Raaphorst J., Johan Groeneveld A. B., Bossink A. W., Erik Hack C. (2001) Thromb. Haemost. 86, 543–549 [PubMed] [Google Scholar]
- 44. Hack C. E. (2001) Semin. Thromb. Hemost. 27, 633–638 [DOI] [PubMed] [Google Scholar]
- 45. Gebbink M. F., Bouma B., Maas C., Bouma B. N. (2009) FEBS Lett. 583, 2691–2699 [DOI] [PubMed] [Google Scholar]
- 46. Samson A. L., Borg R. J., Niego B., Wong C. H., Crack P. J., Yongqing T., Medcalf R. L. (2009) Blood 114, 1937–1946 [DOI] [PubMed] [Google Scholar]
- 47. Andrade-Gordon P., Strickland S. (1986) Biochemistry 25, 4033–4040 [DOI] [PubMed] [Google Scholar]
- 48. Liang J. F., Li Y., Yang V. C. (2000) Thromb. Res. 97, 349–358 [DOI] [PubMed] [Google Scholar]
- 49. Pâques E. P., Stöhr H. A., Heimburger N. (1986) Thromb. Res. 42, 797–807 [DOI] [PubMed] [Google Scholar]
- 50. Hoylaerts M., Rijken D. C., Lijnen H. R., Collen D. (1982) J. Biol. Chem. 257, 2912–2919 [PubMed] [Google Scholar]
- 51. Zamarron C., Lijnen H. R., Collen D. (1984) J. Biol. Chem. 259, 2080–2083 [PubMed] [Google Scholar]
- 52. Fears R., Hibbs M. J., Smith R. A. (1985) Biochem. J. 229, 555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nieuwenhuizen W., Voskuilen M., Vermond A., Hoegee-de Nobel B., Traas D. W. (1988) Eur. J. Biochem. 174, 163–169 [DOI] [PubMed] [Google Scholar]
- 54. Horrevoets A. J., Pannekoek H., Nesheim M. E. (1996) J. Biol. Chem. 272, 2183–2191 [DOI] [PubMed] [Google Scholar]
- 55. Johnsen L. B., Ravn P., Berglund L., Petersen T. E., Rasmussen L. K., Heegaard C. W., Rasmussen J. T., Benfeldt C., Fedosov S. N. (1998) Biochemistry 37, 12631–12639 [DOI] [PubMed] [Google Scholar]
- 56. Kvassman J. O., Shore J. D. (1995) Fibrinolysis 9, 215–221 [Google Scholar]
- 57. Shore J. D., Day D. E., Francis-Chmura A. M., Verhamme I., Kvassman J., Lawrence D. A., Ginsburg D. (1995) J. Biol. Chem. 270, 5395–5398 [DOI] [PubMed] [Google Scholar]
- 58. Komissarov A. A., Declerck P. J., Shore J. D. (2002) J. Biol. Chem. 277, 43858–43865 [DOI] [PubMed] [Google Scholar]
- 59. Komissarov A. A., Mazar A. P., Koenig K., Kurdowska A. K., Idell S. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L568–L577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Komissarov A. A., Deutscher S. L. (1999) Biochemistry 38, 14631–14637 [DOI] [PubMed] [Google Scholar]
- 61. Komissarov A. A., Marchbank M. T., Calcutt M. J., Quinn T. P., Deutscher S. L. (1997) J. Biol. Chem. 272, 26864–26870 [DOI] [PubMed] [Google Scholar]
- 62. Kvassman J. O., Verhamme I., Shore J. D. (1998) Biochemistry 37, 15491–15502 [DOI] [PubMed] [Google Scholar]
- 63. Christensen U., Bangert K., Thorsen S. (1996) FEBS Lett. 387, 58–62 [DOI] [PubMed] [Google Scholar]
- 64. Sen P., Komissarov A. A., Florova G., Idell S., Pendurthi U. R., Vijaya Mohan Rao L. (2011) J. Thromb. Haemost. 9, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wiman B., Collen D. (1978) Eur. J. Biochem. 84, 573–578 [DOI] [PubMed] [Google Scholar]
- 66. Wu J. H., Diamond S. L. (1995) Thromb. Haemost. 74, 711–717 [PubMed] [Google Scholar]
- 67. Komissarov A. A., Calcutt M. J., Marchbank M. T., Peletskaya E. N., Deutsher S. L. (1996) J. Biol. Chem. 271, 12241–12246 [DOI] [PubMed] [Google Scholar]
- 68. Tomasini B. R., Mosher D. F. (1991) Prog. Hemost. Thromb. 10, 269–305 [PubMed] [Google Scholar]
- 69. Preissner K. T., Seiffert D. (1998) Thromb. Res. 89, 1–21 [DOI] [PubMed] [Google Scholar]
- 70. Declerck P. J., De Mol M., Alessi M. C., Baudner S., Pâques E. P., Preissner K. T., Müller-Berghaus G., Collen D. (1988) J. Biol. Chem. 263, 15454–15461 [PubMed] [Google Scholar]
- 71. Preissner K. T., Grulich-Henn J., Ehrlich H. J., Declerck P., Justus C., Collen D., Pannekoek H., Müller-Berghaus G. (1990) J. Biol. Chem. 265, 18490–18498 [PubMed] [Google Scholar]
- 72. Komissarov A. A., Andreasen P. A., Bødker J. S., Declerck P. J., Anagli J. Y., Shore J. D. (2005) J. Biol. Chem. 280, 1482–1489 [DOI] [PubMed] [Google Scholar]
- 73. Komissarov A. A., Zhou A., Declerck P. J. (2007) J. Biol. Chem. 282, 26306–26315 [DOI] [PubMed] [Google Scholar]
- 74. Christensen U., Clemmensen I. (1977) Biochem. J. 163, 389–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Christensen U., Clemmensen I. (1978) Biochem. J. 175, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Anonick P. K., Gonias S. L. (1991) Biochem. J. 275, 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Christensen S., Sottrup-Jensen L., Christensen U. (1995) Biochem. J. 305, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maskell N. A., Davies C. W., Nunn A. J., Hedley E. L., Gleeson F. V., Miller R., Gabe R., Rees G. L., Peto T. E., Woodhead M. A., Lane D. J., Darbyshire J. H., Davies R. J. (2005) N. Engl. J. Med. 352, 865–874 [DOI] [PubMed] [Google Scholar]
- 79. Thelwell C., Longstaff C. (2007) J. Thromb. Haemost. 5, 804–811 [DOI] [PubMed] [Google Scholar]
- 80. Zhou A., Huntington J. A., Pannu N. S., Carrell R. W., Read R. J. (2003) Nat. Struct. Biol. 10, 541–544 [DOI] [PubMed] [Google Scholar]
- 81. Ong P. C., McGowan S., Pearce M. C., Irving J. A., Kan W. T., Grigoryev S. A., Turk B., Silverman G. A., Brix K., Bottomley S. P., Whisstock J. C., Pike R. N. (2007) J. Biol. Chem. 282, 36980–36986 [DOI] [PubMed] [Google Scholar]
- 82. Mavrommatis A. C., Theodoridis T., Economou M., Kotanidou A., El Ali M., Christopoulou-Kokkinou V., Zakynthinos S. G. (2001) Intensive Care Med. 27, 1853–1859 [DOI] [PubMed] [Google Scholar]
- 83. Gallimore M. J., Aasen A. O., Erichsen N. S., Larsbraaten M., Lyngaas K., Amundsen E. (1980) Thromb. Res. 18, 601–608 [DOI] [PubMed] [Google Scholar]
- 84. Duboscq C., Quintana I., Bassilotta E., Bergonzelli G. E., Porterie P., Sassetti B., Haedo A. S., Wainsztein N., Kruithof E. K., Kordich L. (1997) Thromb. Haemost. 77, 1090–1095 [PubMed] [Google Scholar]
- 85. Alemán C., Alegre J., Monasterio J., Segura R. M., Armadans L., Anglés A., Varela E., Ruiz E., Fernández de Sevilla T. (2003) Clin. Sci. 105, 601–607 [DOI] [PubMed] [Google Scholar]
- 86. Kolev K., Longstaff C., Machovich R. (2005) Curr. Med. Chem. Cardiovasc. Hematol. Agents 3, 341–355 [DOI] [PubMed] [Google Scholar]
- 87. Longstaff C., Thelwell C., Williams S. C., Silva M. M., Szabo L., Kolev K. (2010) Blood 117, 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parise P., Morini M., Agnelli G., Ascani A., Nenci G. G. (1993) Blood Coagul. Fibrinolysis 4, 721–727 [PubMed] [Google Scholar]
- 89. Smith S. A., Morrissey J. H. (2008) Blood 112, 2810–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kolev K., Tenekedjiev K., Ajtai K., Kovalszky I., Gombas J., Váradi B., Machovich R. (2003) Blood 101, 4380–4386 [DOI] [PubMed] [Google Scholar]
- 91. Torbet J. (1986) Biochemistry 25, 5309–5314 [DOI] [PubMed] [Google Scholar]
- 92. Merkle D. L., Cheng C. H., Castellino F. J., Chibber B. A. (1996) Blood Coagul. Fibrinolysis 7, 650–658 [DOI] [PubMed] [Google Scholar]
- 93. Weitz J. I., Kuint J., Leslie B., Hirsh J. (1991) Thromb. Haemost. 65, 541–544 [PubMed] [Google Scholar]
- 94. Zhu Z., Hawthorne M. L., Guo Y., Drake W., Bilaceroglu S., Misra H. L., Light R. W. (2006) Chest 129, 1577–1583 [DOI] [PubMed] [Google Scholar]
- 95. Rahman N. M., Maskell N. A., West A., Teoh R., Arnold A., Mackinlay C., Peckham D., Davies C. W., Ali N., Kinnear W., Bentley A., Kahan B. C., Wrightson J. M., Davies H. E., Hooper C. E., Lee Y. C., Hedley E. L., Crosthwaite N., Choo L., Helm E. J., Gleeson F. V., Nunn A. J., Davies R. J. (2011) N. Engl. J. Med. 365, 518–526 [DOI] [PubMed] [Google Scholar]