Background: Human MUC1 is overexpressed and abnormally glycosylated in adenocarcinomas, correlating with inflammation and tumor growth.
Results: MUC1 activates NF-κB in tumors and, in complex with NF-κB p65, migrates to the nucleus, where it promotes transcription of proinflammatory cytokines.
Conclusion: MUC1 is an important mediator of tumor-induced inflammation.
Significance: Targeting the MUC1-p65 complex could reduce inflammation and inhibit tumor growth.
Keywords: Cancer Biology, Cytokine Induction, Inflammation, MUC1, NF-κB, IL-6, TNF-α, Variable Number of Tandem Repeats (VNTR)
Abstract
MUC1 is a transmembrane glycoprotein abnormally expressed in all stages of development of human adenocarcinomas. Overexpression and hypoglycosylation of MUC1 in cancer cells compared with normal epithelial cells are likely to alter its function and affect the behavior of cancer cells. The extracellular domain, specifically the highly O-glycosylated VNTR (variable number of tandem repeats) region, plays an important role in cell-cell communication; however, we show here that it also participates intracellularly in activation of the NF-κB pathway. Transfection of MUC1− tumor cells with cDNA encoding MUC1 with 22 tandem repeats (MUC1/22TR) or two tandem repeats (MUC1/2TR) or two isoforms that lack the VNTR region (MUC1/Z and MUC1/Y) showed that the highest expression levels of NF-κB family members correlated with the presence of VNTR and the highest number of tandem repeats. Because expression of MUC1 with VNTR on tumors was previously associated with chemotactic activity for cells of the innate immune system, we investigated the influence of MUC1 expression on the NF-κB-dependent transcriptional regulation of proinflammatory cytokines. ChIP and real-time PCR experiments revealed that MUC1/22TR up-regulated IL-6 and TNF-α expression by binding to their promoter regions in a NF-κB p65-dependent manner in both MUC1-transfected and human breast cancer cells that express endogenous MUC1. This newly detected complex of MUC1 and p65 is a novel mechanism that tumors can use to promote inflammation and cancer development.
Introduction
MUC1 (mucin 1) is a transmembrane glycoprotein normally expressed at low levels, polarized to the apical surface of epithelial cells. In most adenocarcinomas and inflammatory diseases of epithelial tissues, MUC1 is overexpressed and unpolarized. MUC1 consists of a large extracellular subunit containing a VNTR (variable number of tandem repeats) region composed of 20–200 tandem repeat (TR)2 units of 20 amino acids (rich in prolines and O-glycosylated serines and threonines), a transmembrane domain, and a cytoplasmic tail (CT). On normal epithelia, MUC1 VNTR is extensively O-glycosylated with long branched glycans, whereas on tumor cells, it is markedly hypoglycosylated with simpler and shorter glycan chains. Changes in glycosylation affect many functions of MUC1 (1–3). Of particular interest was the observation that aberrant glycosylation of MUC1 stimulates its endocytosis and intracellular accumulation (4), which we hypothesized might also alter its role in intracellular signaling in cancer cells.
The cytoplasmic tail of MUC1 (MUC1.CT) was reported to be involved in intracellular signal transduction by association with several protein partners and modulation of their function (5). Specific examples are associations with 1) ErbB growth factor receptor tyrosine kinases, thus enhancing ErbB1 signaling in breast cancer cells (6); 2) β-catenin, thus increasing its cytoplasmic and nuclear levels by inhibiting glycogen synthase kinase 3β-mediated phosphorylation and degradation (7); and 3) estrogen receptor α, thus stimulating estrogen receptor α-mediated transcription by binding directly to the estrogen receptor α DNA-binding domain (8). Moreover, MUC1.CT was shown to modulate the activity of the NF-κB pathway in breast cancer cells by interacting with and activating IκB kinase (IKK) family members and NF-κB p65 (9, 10). Much of this work was done by engineering MUC1.CT on an unrelated transmembrane protein. Although these studies confirmed the signaling properties of MUC1.CT, the fact that MUC1.CT is identical in normal and tumor cells makes it difficult to define the role, if any, of this signaling in cancer. The greatest difference in MUC1 between normal and tumor cells is in its localization pattern and the glycosylation state of the VNTR region, which increases the accessibility of the peptide backbone to other proteins and changes its three-dimensional structure (11, 12), thus potentially affecting protein-protein interactions and intracellular signaling.
Here, we report the interaction of the full-length MUC1 molecule, including the VNTR-containing extracellular domain, with the NF-κB pathway, which is activated in a large number of cancers and plays a role as a pivotal link between cancer and inflammation through its ability to up-regulate expression of tumor-promoting cytokines such as IL-6 and TNF-α. For the first time, we show that the extracellular domain of MUC1 localizes in the nucleus, promoting the expression of proinflammatory cytokines in tumors through a newly identified complex of full-length MUC1 and p65.
EXPERIMENTAL PROCEDURES
Cell Culture
The RMA mouse T cell lymphoma cell line was grown in Cellgro® DMEM (Mediatech, Inc., Herndon, VA), whereas the DM6 human melanoma cell line and the MCF-7 and MDA-MB-231 human breast cancer cell lines were grown in Cellgro® RPMI 1640 medium (Mediatech, Inc.). Both DMEM and RPMI 1640 medium were supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mmol/liter l-glutamine. MCF-10A cells were cultured in MEGM® supplemented with 52 μg/ml bovine pituitary extract, 0.5 μg/ml hydrocortisone, 10 ng/ml human EGF, and 5 μg/ml insulin (MEGM® BulletKit, Lonza, Walkersville, MD). MEGM was also supplemented with 100 ng/ml cholera toxin (Sigma). As an activating signal, RMA cells were treated with 10 μg/ml concanavalin A (ConA; Sigma), whereas human cells were treated with 20 ng/ml TNF-α (R&D Systems, Minneapolis, MN). 10 μm BAY 11-7085 (Calbiochem) was used to inhibit the NF-κB pathway.
Plasmids and Transfection
We utilized the pcDNA3 core plasmid (Invitrogen), into which we inserted MUC1 cDNA containing either 2 or 22 tandem repeats (MUC1/2TR and MUC1/22TR) or one of two short isoforms (MUC1/Y or MUC1/Z). Transfection was performed with LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. Stable expression was achieved by selection in up to 1000 μg/ml G418 (Sigma).
Subcutaneous Tumor Growth
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Tumor growth analysis was performed on subcutaneously implanted 1 × 105 RMA-MUC1/22TR and RMA-MUC1/Y cells or untransfected RMA control cells in 0.15 ml of PBS, 3 weeks after inoculation.
Western Blotting and Immunoprecipitation
Total cell proteins were extracted using radioimmune precipitation assay buffer (150 mm NaCl, 0.5 sodium deoxycholate, 0.1% SDS, 50 mm Tris-HCl, and 1% Nonidet P-40) with commercial protease inhibitors (Complete protease inhibitor mixture, Roche Applied Science) and phosphatase inhibitors (Phosphatase Inhibitor Cocktail 2, Sigma).
Fractionated proteins were obtained using cytoplasmic lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1.5 mm MgCl2, 10 mm EGTA (pH 7.5), and 10% glycerol) and nuclear lysis buffer (20 mm HEPES (pH 8), 0.1 mm EDTA, 5 mm MgCl2, 0.5 NaCl, 20% glycerol, and 1% Nonidet P-40). Nuclear DNA-bound proteins were isolated using a subcellular protein fractionation kit (Thermo Scientific, Rockford, IL) following the manufacturer's instructions. This was accomplished by first gently collecting protein from the nucleus and then collecting the protein-chromatin complexes. These complexes were treated with micrococcal nuclease, which released chromatin-associated protein into the supernatant.
50 μg of protein from lysates was used for Western blotting, and 500 μg was used for immunoprecipitation. The following antibodies were employed: anti-IκBα, anti-IKKγ, anti-C23 (nucleolin), and anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA) for Western blotting; anti-NF-κB p65 (Santa Cruz Biotechnology) for Western blotting and immunoprecipitation; anti-phospho-IKKβ, anti-phospho-IκBα, and anti-phospho-NF-κB p65 Ser-276 (Cell Signaling Technology, Beverly, MA) for Western blotting; and anti-MUC1 3C6 (a gift from Dr. Hilgers, Free University, Brussels, Belgium) and anti-MUC1 Ab5 (Fitzgerald, Concord, MA) for Western blotting and immunoprecipitation.
For immunoprecipitation, protein lysates were first precleared with protein G-Sepharose beads (Sigma) for 3 h at 4 °C and then incubated with primary antibody at 4 °C for 16 h. The immune complexes were precipitated for 2 h at 4 °C with protein G-Sepharose 4B. In control samples, the primary Ab was substituted with control IgG (rabbit or mouse depending on the source of the primary Abs). Immunoprecipitates were washed four times with radioimmune precipitation assay buffer containing 0.5 m NaCl and 2% SDS and three times with PBS and then resuspended in Laemmli buffer.
The proteins were separated on a precast 4–20% polyacrylamide gel (Bio-Rad) and immunoblotted, and the reactivity was detected with horseradish peroxidase-conjugated secondary antibody and chemiluminescence (PerkinElmer Life Sciences). The intensity of signals was determined by densitometric scanning (Kodak Image Station 4000MM).
Quantitative Real-time PCR
Total RNA was isolated using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. A total of 2 μg of RNA was reverse-transcribed using a SuperScript first strand kit (Invitrogen). A total of 3 μl of RT products was used to amplify IL-6, IFN-γ, TNF-α, IL-2, and GAPDH as an internal control. Real-time PCR was performed using a SYBR Green PCR kit (Qiagen) and a StepOnePlus real-time PCR system (Applied Biosystems). The sequence of each primer is shown in supplemental Table S1.
Changes in the mRNA content relative to GAPDH mRNA were determined using the threshold cycle (CT) method (ABI User Bulletin 2, Applied Biosystems) to calculate changes in CT and, ultimately, -fold and percent changes. An average CT value for each RNA was obtained for replicate reactions.
Chromatin Immunoprecipitation
Cells were subjected to chromatin immunoprecipitation based on EZ-ChIP protocol 17-371 (Millipore). Briefly, treatment with 1% formaldehyde was used for cross-linking, and cell lysate was collected and sonicated to shear DNA. Soluble chromatin was rotated for 2 h at 4 °C with 60 μl of protein A-agarose-salmon sperm DNA (Millipore). Precleared lysate was incubated overnight at 4 °C with 4 μg of anti-MUC1 antibody Ab5 or 3C6. The antibody-protein-DNA complexes were precipitated with 60 μl of protein A-agarose beads at 4 °C for 2 h. Complexes were eluted in elution buffer (0.1 mm NaHCO3 and 1% SDS) before reversal of cross-links overnight at 65 °C under high salt conditions (0.5 m NaCl). After proteinase K (Sigma) digestion, DNA was extracted in 25:24:1 phenol/chloroform/isoamyl alcohol and precipitated overnight in ethanol at −20 °C, and DNA was then eluted in Tris/EDTA buffer. The presence of IL-2, IFN-γ, IL-6, and TNF-α gene promoter sequences in immunoprecipitated DNA was identified by 34 cycles of PCR using the primer sequences listed in supplemental Table S2. In control samples, primary antibody was replaced with non-immune IgG. All experiments were repeated at least three times. For re-ChIP assays, complexes from the primary chromatin immunoprecipitation were eluted with 10 mm dithiothreitol, diluted in re-ChIP buffer, and re-immunoprecipitated with anti-p65 antibody.
Interleukin Detection by ELISA
Subconfluent mouse cells were left untreated or were treated for 24 h with 10 μm BAY 11-7085 and/or 10 μg/ml ConA for 24 h. Human cells were treated for 24 h with 10 μm BAY 11-7085 and/or 20 ng/ml TNF-α for 2 h. After that, the cells were counted, and the conditioned medium was collected. Mouse IL-6 and TNF-α concentrations in the conditioned medium were measured using a mouse or human ELISA kit (BD Biosciences). A phospho-IκBα ELISA kit was purchased from Cell Signaling Technology. The standard curve was done using 0, 15.6, 31.2, 62.5, 125, 250, 500, 1000, and 2000 pg/ml interleukin. Interleukin concentrations detected in the conditioned medium were within the range of the standard curve. The concentrations are expressed as picograms/ml per 1 × 105 cells. All points were done in triplicate, and the experiments were repeated three times.
Flow Cytometry Analysis
To detect intracellular MUC1 and p65, cells were fixed and permeabilized with BD Cytofix/CytopermTM buffer (BD Bioscience) for 20 min on ice, washed, and resuspended in staining buffer (2% fetal bovine serum in PBS). Cells were stained with Armenian hamster Ab5 antibody or rabbit anti-phospho-p65 monoclonal antibody (Cell Signaling Technology) for 1 h at 4 °C, followed by phycoerythrin-conjugated anti-hamster or Alexa Fluor 488-conjugated anti-rabbit secondary antibody (Invitrogen). Cells were analyzed on a BD Biosciences LSR II flow cytometer running FACSDiva software.
Immunofluorescence Confocal Microscopy
Cells were grown on a 4-well Millcell EZ slide (Millipore) and treated with TNF-α or left untreated. Cells were fixed with 4% paraformaldehyde for 20 min and permeabilized in 0.1% Triton X-100 for 20 min. The fixed cells were incubated with FITC-labeled mouse anti-human MUC1 antibody CD227 or anti-MUC1 antibody VU4H5 (Santa Cruz Biotechnology) for 2 h at room temperature and the latter followed by Cy3-labeled secondary anti-mouse antibody (Invitrogen). Nuclei were stained with mounting medium for fluorescence with DAPI (Vector Labs).
Statistical Analysis
The correlations were evaluated by Student's t test. p values <0.05 were considered statistically significant.
RESULTS
Extracellular Domain of MUC1 Interacts with p65 and Enhances the NF-κB Pathway
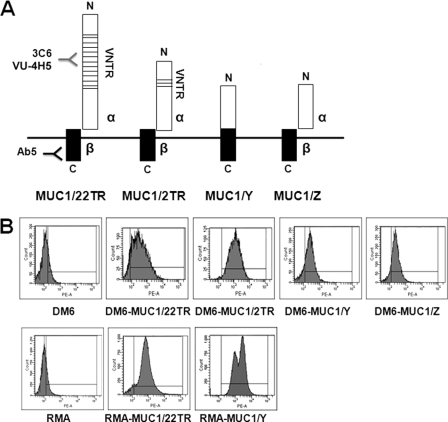
To assess whether the extracellular domain of MUC1 affects NF-κB signaling, we used four different isoforms of MUC1 schematically represented in Fig. 1A: full-length MUC1 containing the VNTR region composed of either 22 (MUC1/22TR) or 2 (MUC1/2TR) tandem repeats and two short forms (MUC1/Y and MUC1/Z) that completely lack the VNTR segment. MUC1/22TR, MUC1/2TR, and MUC1/Z were proteolytically cleaved soon after their synthesis, generating two subunits, α (containing the VNTR region) and β (composed primarily of the cytoplasmic tail), whereas MUC1/Y lacks the cleavage site, resulting in a single protein. These four MUC1 isoforms were stably transfected into the mouse tumor cell line RMA (T cell lymphoma) and into the human melanoma cell line DM6, which do not express MUC1. Flow cytometry analysis showed that MUC1 was expressed in 80–90% of the transfected cells (Fig. 1B).
FIGURE 1.
MUC1 isoforms and MUC1 expression in transfected cells. A, schematic of MUC1 isoforms used for transfection: MUC1/22TR, containing 22 tandem repeats in the VNTR region; MUC1/2TR, containing two tandem repeats; MUC1/Y and MUC1/Z, which lack the VNTR region. MUC1/22TR, MUC1/2TR, and MUC1/Z are composed of two noncovalently linked subunits, α and β. MUC1/Y is expressed as a single protein. All forms have the same C-terminal region (black boxes) composed of the transmembrane domain and the cytoplasmic tail recognized by anti-MUC1 antibody Ab5, but they differ in the N-terminal region (white boxes). The VNTR region is recognized by antibodies 3C6 and VU4H5 (hypoglycosylated VNTR). B, flow cytometry analysis showing MUC1 expression in transfected cells as detected by antibody Ab5.
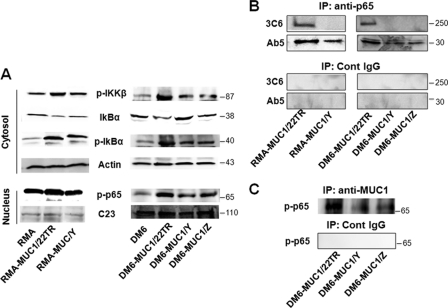
After stimulation with ConA for 24 h to activate RMA cells, total proteins were extracted from RMA-MUC1/22TR and RMA-MUC1/Y cells and from the untransfected parental control. We analyzed the expression levels of IκBα, phospho-IκBα (Ser-32/Ser-36), and phospho-IKKβ (Ser-181) proteins by Western blotting (Fig. 2A). Phospho-IKKβ and phospho-IκBα were significantly increased in both RMA-MUC1/22TR and RMA-MUC1/Y cells compared with untransfected cells. As a consequence, unphosphorylated IκBα levels were drastically reduced. The presence of VNTR appeared to contribute to stronger activation in RMA-MUC1/22TR cells compared with RMA-MUC1/Y cells. The levels of phospho-IκBα protein were also tested by ELISA so that the results could be better quantified. They confirmed that the levels of phospho-IκBα increased by 2-fold in RMA-MUC1/22TR cells compared with untransfected cells and by 1.5-fold compared with RMA-MUC1/Y cells (supplemental Fig. S1A). Similarly, phospho-NF-κB p65 levels in the nucleus were higher in RMA-MUC1/22TR cells compared with RMA-MUC1/Y and untransfected cells (Fig. 2A).
FIGURE 2.
MUC1 transfection into RMA and DM6 cell lines induces NF-κB activation and interaction with p65. A, RMA cells (left) were transfected with MUC1/22TR (RMA-MUC1/22TR) or MUC1/Y (RMA-MUC1/Y) or left untransfected (RMA) and stimulated with ConA. DM6 cells (right) were transfected with MUC1/22TR (DM6-MUC1/22TR), MUC1/Y (DM6-MUC1/Y), or MUC1/Z (DM6-MUC1/Z) or left untransfected (DM6) and stimulated with TNF-α. Expression of indicated proteins was assessed by Western blotting. Protein loading was controlled by reprobing membranes for the expression of actin and nucleolin (C23) as indicated. B, lysates from RMA-MUC1/22TR, RMA-MUC1/Y, DM6-MUC1/22TR, DM6-MUC1/Y, and DM6-MUC1/Z cells were immunoprecipitated (IP) with anti-p65 Ab or a control (Cont) IgG as described under “Experimental Procedures” and immunoblotted with anti-MUC1.CT antibody Ab5 and anti-MUC1 VNTR antibody 3C6. C, whole cell lysates from DM6-MUC1/22TR, DM6-MUC1/Y, and DM6-MUC1/Z cells were immunoprecipitated with antibody Ab5 or control IgG as described under “Experimental Procedures.” The precipitates were immunoblotted with anti-phospho-p65 antibody.
We performed the same experiments with the DM6 human melanoma cell line stably transfected with MUC1/22TR, MUC1/Y, or MUC1/Z (Fig. 1A). We activated DM6 cells with TNF-α for 2 h and tested the abundance of cytosolic IκBα, phospho-IκBα (Ser-32/Ser-36), phospho-IKKβ (Ser-181), and nuclear phospho-p65 (Ser-276) (Fig. 2A). Our results show that transfection with VNTR-containing MUC1/22TR increased significantly cytosolic IKKβ, phospho-IκBα, and nuclear phospho-p65 proteins levels compared with DM6 cells transfected with the MUC1/Z and MUC1/Y isoforms lacking VNTR.
The effect of MUC1 on enhancing phospho-p65 expression was confirmed also by flow cytometry. Full-length MUC1/22TR induced phospho-p65 expression by 3.7- and 2.7-fold compared with untransfected RMA and RMA-MUC1/Y, respectively (supplemental Fig. S1B). In DM6 cells, MUC1/22TR increased phospho-p65 expression by 4.4-fold compared with untransfected cells and by 1.7- and 2.2-fold compared with DM6-MUC1/Y and DM6-MUC1/Z cells, respectively (supplemental Fig. S1C).
We next investigated whether there might be a physical association between NF-κB p65 and MUC1 and whether there was a difference in binding affinity between NF-κB p65 and different MUC1 isoforms. Upon stimulation with ConA, proteins from RMA-MUC1/22TR, RMA-MUC1/Y, and untransfected RMA cells were immunoprecipitated with anti-p65 antibody or a control IgG antibody. The immunoprecipitated proteins were resolved on the gel and immunoblotted with anti-MUC1 antibody Ab5, which recognizes the cytoplasmic tail. We found that MUC1 coprecipitated with NF-κB p65 (Fig. 2B) and that MUC1/22TR interacted more strongly with NF-κB p65 compared with MUC1 isoforms lacking the extracellular domain. The same experiment was performed using proteins extracted from DM6-MUC1/22TR, DM6-MUC1/Y, DM6-MUC1/Z, and DM6 cells (Fig. 2B), and the same results were obtained. These data indicate that full-length MUC1/22TR not only induced a higher expression of p65 but also showed stronger binding to p65 compared with the shorter forms. These results were confirmed in experiments in which anti-MUC1 precipitates were immunoblotted with phospho-p65 (Fig. 2C). To determine whether the extracellular domain (in particular, the VNTR region) is still part of the MUC1 molecule that interacts with NF-κB p65, the phospho-p65 proteins were immunoblotted with anti-MUC1 antibody 3C6, which is specific for the VNTR region (13). We found that the full-length MUC1 containing the VNTR region bound NF-κB p65 in both RMA and DM6 cells (Fig. 2B).
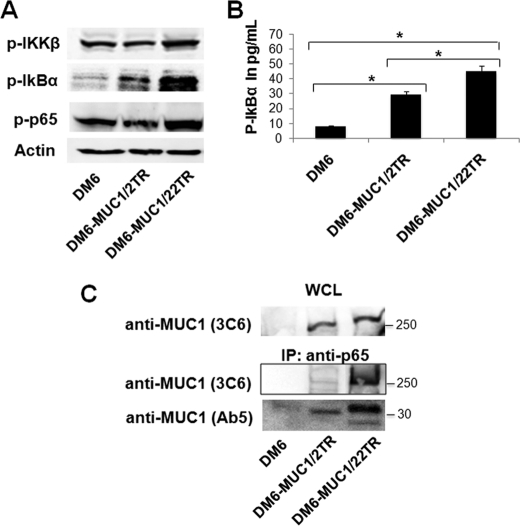
To assess whether the length of the VNTR region in particular influences NF-κB activation, DM6 cells were stably transfected with MUC1 containing two tandem repeats, generating the DM6-MUC1/2TR cell line, and these cells were compared with the DM6-MUC1/22TR cell line. Western blotting (Fig. 3A) and ELISA (Fig. 3B) revealed higher phospho-IκBα and phospho-p65 levels in DM6-MUC1/22TR cells compared DM6-MUC1/2TR and untransfected DM6 cells. We also tested whether the number of tandem repeats influences the interaction between p65 and MUC1. Total protein extracts from DM6-MUC1/22TR, DM6-MUC1/2TR, and DM6 cells were immunoprecipitated with anti-p65 antibody and then immunoblotted with anti-MUC1.CT antibody Ab5 and anti-MUC1 VNTR antibody 3C6. MUC1/22TR showed a stronger association with p65 compared with MUC1/2TR (Fig. 3C). Although NF-κB activation had been previously ascribed to MUC1.CT, our results above show that a higher number of tandem repeats increases activation efficiency and point to the VNTR region as the important region in the extracellular domain with a similar NF-κB-stimulating activity.
FIGURE 3.
VNTR region enhances NF-κB pathway activation. DM6, DM6-MUC1/2TR, and DM6-MUC1/22TR cells were activated with TNF-α, and the levels of the indicated proteins were evaluated by Western blotting (A) and ELISA (B). *, statistically significant difference (p < 0.05). C, whole cell lysates (WCL) from DM6-MUC1/22TR and DM6-MUC1/2TR cells were immunoblotted with anti-MUC1 antibody 3C6 as a positive control (upper) or immunoprecipitated (IP) with anti-p65 antibody and immunoblotted for MUC1 with anti-MUC1 VNTR antibody 3C6 (middle) or anti-MUC1.CT antibody Ab5 (lower).
MUC1-p65 Complex Is Found on the IL-2, IL-6, IFN-γ, and TNF-α Promoters in Vitro and in Vivo
The association of MUC1 with p65 suggested that this could be a transcriptionally active complex. Because we and others had shown previously that MUC1 has proinflammatory activity (14–16) and because NF-κB is one of the most important transcriptional activators of proinflammatory cytokine genes in the cancer microenvironment, we investigated whether the MUC1-p65 complex might be responsible for transcriptional regulation of inflammatory cytokines.
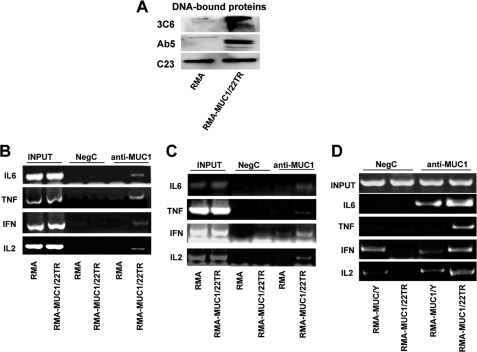
After ConA stimulation, RMA-MUC1/22TR and RMA cells were subjected to subcellular fractionation, and nuclear proteins bound to chromatin were isolated as a separate fraction from total soluble nuclear proteins as described under “Experimental Procedures.” Chromatin-bound nuclear proteins were immunoblotted with anti-MUC1.CT antibody Ab5 and anti-MUC1 VNTR antibody 3C6. Surprisingly, considering the size of full-length MUC1 with 22 tandem repeats, we found the entire molecule included in the VNTR region interacting with chromatin (Fig. 4A).
FIGURE 4.
MUC1 and p65 colocalize on cytokine gene promoters. A, nuclear DNA-bound proteins were isolated as described under “Experimental Procedures” and immunoblotted with anti-MUC1 antibodies Ab5 and 3C6. Nucleolin (C23) was included as a loading control. B, ChIP assay. Soluble chromatin was immunoprecipitated with anti-MUC1.CT antibody Ab5 or control IgG (NegC) and analyzed for κB consensus sites of promoters for the indicated cytokines. C, re-ChIP assay. Soluble chromatin was first immunoprecipitated with anti-MUC1.CT antibody Ab5 and then re-immunoprecipitated with anti-p65 antibody. D, in vivo ChIP assay. Soluble chromatin from tumor tissue was immunoprecipitated with anti-MUC1.CT Ab5 or control IgG and analyzed for κB consensus sites on promoters of the indicated cytokines.
To further confirm this association, we performed a ChIP assay. Chromatin was precipitated from ConA-activated RMA-MUC1/22TR and RMA cells with anti-MUC1.CT antibody Ab5. MUC1-DNA complexes were isolated, and the promoter regions containing the consensus NF-κB-binding sites (GGGRNNYYCC, where R is purine, Y is pyrimidine, and N is any base) of IL-2 (−206 to −195, GGGATTTCACC) (17), the non-canonical κB site of IFN-γ (−278 to −268, AAAAATTTCC) (18), IL-6 (−75 to −63, GGGATTTTCC) (19), and two NF-κB consensus sites (κB2, −657 to −638, GTGAATTCCC; and κB3, −510 to −488, GGGGCTTTCC) of TNF-α (20) were amplified by PCR (Fig. 4B) using the primers listed in supplemental Table S2. Additionally, we performed a re-ChIP assay to verify that MUC1 associated with the cytokine promoters was simultaneously associated with NF-κB p65. As shown in Fig. 4C, all four promoters displayed MUC1-p65 co-occupancy in RMA-MUC1/22TR cells.
We also confirmed MUC1 interaction with the cytokine promoters during tumor growth in vivo. 1 × 105 RMA-MUC1/22TR and RMA-MUC1/Y (no VNTR) cells were subcutaneously injected into mice (as described under “Experimental Procedures), and after 3 weeks of growth, tumors were harvested, and ChIP assay was performed using anti-MUC1.CT antibody Ab5. Greater MUC1 occupancy of the IL-2, IFN-γ, IL-6, and TNF-α promoters was observed in the RMA-MUC1/22TR tumors compared with RMA-MUC1/Y tumors (Fig. 4D). These data are in agreement with our Western blot results (Fig. 2), indicating that the extracellular VNTR region of MUC1 is important in either facilitating MUC1-p65 complex formation or its binding to the promoter DNA.
MUC1 Stimulates Pro-inflammatory Cytokine Expression in a p65-dependent Manner
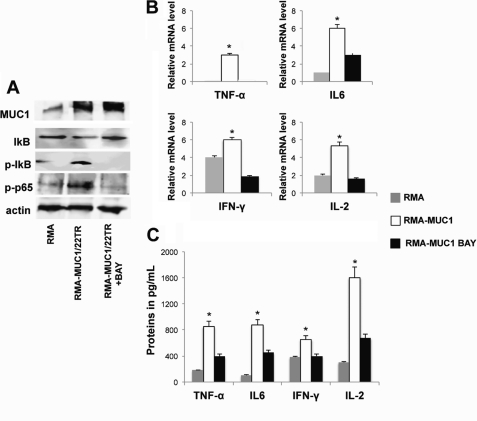
To confirm the ability of MUC1 to activate the transcriptional activity of interleukins via NF-κB activation, we activated RMA-MUC1/22TR and RMA cells with ConA in the presence or absence of BAY 11-7085, an inhibitor of IκB phosphorylation and consequently of p65 nuclear translocation. In the presence of BAY 11-7085, IκBα and p65 phosphorylation was inhibited, whereas MUC1 protein levels remained unchanged (Fig. 5A). Interleukin RNA levels were measured by quantitative real-time PCR (Fig. 5B). Relative to untransfected RMA, there was a significant increase in the mRNAs of IFN-γ (1.5-fold), IL-6 (5-fold), and IL-2 and TNF-α (3-fold each), whereas in the presence of BAY 11-7085, MUC1-stimulated expression of IL-2, IFN-γ, and TNF-α was completely abolished, and IL-6 expression was reduced by 50% (Fig. 5B). By ELISA, we measured significantly increased levels of secreted IL-2 (8-fold), IFN-γ (1.5-fold), IL-6 (4-fold), and TNF-α (5-fold), which could be partially reduced by BAY 11-7085 treatment (Fig. 5C).
FIGURE 5.
Negative effect of p65 inhibitor on MUC1-mediated activation of cytokine expression. A, RMA and RMA-MUC1/22TR cells were activated with ConA, and RMA-MUC1/22TR cells were either treated (+BAY) or not with BAY 11-7085 as described under “Experimental Procedures.” Total cell proteins were blotted with the indicated antibodies. Actin was used as a loading control. B, real-time PCR. The graphs represent the increase in cytokine mRNAs relative to the increase in GAPDH mRNA in the same sample ± S.D. of three independent experiments. *, p < 0.05. C, concentrations of secreted interleukins were measured by ELISA. *, statistically significant difference (p < 0.05).
DM6 cells do not make all of the abovementioned cytokines, so we tested the effect of MUC1 on IL-6 expression in DM6-MUC1/22TR cells compared with untransfected cells and activated or not with TNF-α. We found that DM6-MUC1/22TR cells expressed significantly higher levels of IL-6 compared with MUC1− DM6 cells (supplemental Fig. S2).
Endogenous MUC1 in Breast Cancer Cells Interacts with IL-6 and TNF-α Promoters
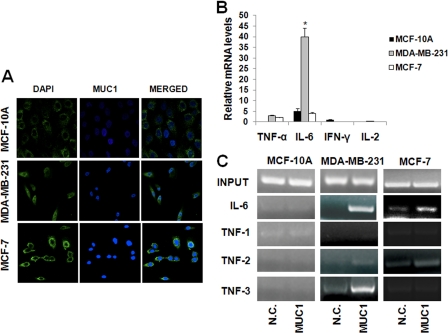
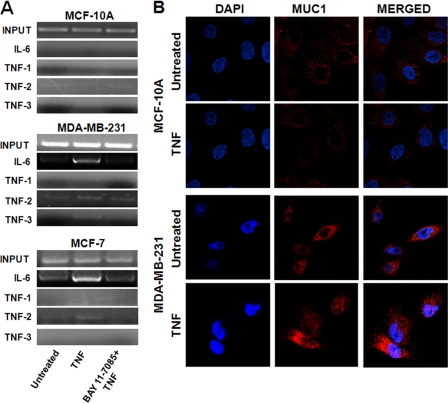
The results described above were obtained from MUC1-transfected cells, and thus, we wanted to confirm that endogenous MUC1 would have a similar effect on inflammatory cytokine expression in human cancer. We examined its interaction with interleukin promoters in two human breast cancer cell lines (MCF-7 and MDA-MB-231) and in the untransformed epithelial cell line MCF-10A, all of which express MUC1 on the membrane and in the cytosol (Fig. 6A). None of these cells make IL-2 and IFN-γ, but we found that MCF-7 and MDA-MB-231 cells produced TNF-α and IL-6 mRNAs (Fig. 6B). Thus, we investigated MUC1 binding to human IL-6 and TNF-α promoters. ChIP with anti-MUC1.CT antibody Ab5 revealed that, upon TNF-α stimulation, elevated binding of MUC1 occurred at the NF-κB consensus site (−75 to −63) of the IL-6 promoter in MDA-MB-231 cells, whereas low abundance was found in MCF-7 cell lines (Fig. 6C). The human TNF-α promoter contains multiple NF-κB-like consensus sites, and we analyzed three of them: TNF-1, containing κB1 (−873 to −864); TNF-2, containing κB2 (−627 to −618) and κB2a (−598 to −589); and TNF-3, containing κB3 (−98 to −89) (21). Our results reveal that MUC1 did not bind the TNF-1 region, but strong binding was noted at TNF-3 in MDA-MB-231 cells and a lower abundance at TNF-2 in MDA-MB-231 and MCF-7 cells. MUC1 did not interact with the TNF-α and IL-6 promoters in the untransformed MCF-10A cells (Fig. 6C). We then investigated whether the native MUC1 that interacts with cytokine promoters contains the VNTR region. Precipitation of chromatin from MDA-MB-231, MCF-7, and MCF-10A cells was performed with anti-MUC1 antibody 3C6, and we found that, as was the case in transfected cells (Fig. 4A), both the cytoplasmic tail and the extracellular VNTR region were found associated with the promoters (Fig. 7A). However, this association was drastically reduced when the NF-κB pathway was inhibited by BAY 11-7085 treatment (Fig. 7A).
FIGURE 6.
MUC1 modulates the transcriptional activity of IL-6 and TNF-α in breast cancer cell lines. A, cells were fixed and stained with FITC-labeled mouse anti-human MUC1 antibody CD227 (green). Nuclei were stained with DAPI (blue) B, real-time PCR measurement of mRNA levels of the indicated interleukins in breast cancer cell lines. Interleukin mRNA levels are relative to the increase in GAPDH mRNA in the same samples ± S.D. of three independent experiments. *, p < 0.05. C, ChIP assay. Soluble chromatin from the indicated cell lines was immunoprecipitated with anti-MUC1.CT antibody Ab5 or control IgG (N.C.) and analyzed for indicated the κB consensus sites.
FIGURE 7.
MUC1 containing the VNTR region interacts with cytokine promoters in a p65-dependent manner in breast cancer cells. A, cells were activated with TNF-α with or without BAY 11-7085 or left untreated. Soluble chromatin was immunoprecipitated with anti-MUC1 antibody 3C6 and analyzed for the indicated κB consensus sites. B, MCF-10A and MDA-MB-231 cells were activated with TNF-α or left untreated, fixed, and stained with anti-MUC1 antibody 4H5 (red). Nuclei were stained with DAPI (blue).
MCF-10A, MDA-MB-231 and MCF-7 cells were also stained with antibody VU4H5, which binds the tandem repeat units of the hypoglycosylated tumor form of MUC1. Fluorescence imaging showed a low abundance of MUC1 in the cytosol and its absence in the nucleus in MCF-10A cells (Fig. 7B). Interestingly, the hypoglycosylated form of MUC1, which is highly expressed in MDA-MB-231 cells, showed nuclear localization following TNF-α stimulation (Fig. 7B). Similar results were found in MCF-7 cells (data not show).
DISCUSSION
In the past decade, the importance of chronic inflammation within the tumor microenvironment has been correlated with increased invasiveness and poor prognosis in many types of cancer. We propose that abnormal expression of MUC1 (overexpression and hypoglycosylation of the VNTR region), known to occur early in cancer development, activates production of proinflammatory cytokines by tumor cells, leading to their accumulation in the tumor microenvironment and chronic inflammation, favoring tumor progression.
Using a mouse lymphoma cell line and a human melanoma cell line activated with ConA or TNF-α, respectively, both naturally negative for MUC1 expression, we observed that transfection with full-length MUC1 and two short MUC1 isoforms lacking the VNTR region in the extracellular domain resulted in activation of the NF-κB pathway. Our results reveal that full-length MUC1 induced the highest expression of phospho-IκBα and NF-κB p65 compared with the short isoforms. This implies that the VNTR-containing extracellular domain participates actively in intracellular signaling. In MUC1-expressing RMA cells, we detected formation of the novel MUC1-p65 complex, which bound to interleukin promoters containing κB consensus sites and, in particular, the IL-2, IFN-γ, TNF-α, and IL-6 promoters. Subsequent analysis of the RNA and protein levels of these interleukins revealed that full-length MUC1 induced their transcriptional activity. Importantly, interaction with the IL-6 and TNF-α promoters was also observed for endogenous MUC1 in the human breast cancer cell lines MDA-MB-231 and MCF-7 but not in untransformed MCF-10A cells. IL-6 and TNF-α are considered to be protumor cytokines involved in inflammatory pathways that promote tumorigenesis. As critical mediators of the inflammatory response, IL-6 and TNF-α provide a molecular link between chronic inflammation and tumor progression (22). In a previous report, our laboratory showed that the proinflammatory cytokines IL-6 and TNF-α characterize the microenvironment of MUC1 tumors and that unglycosylated epitopes in the tandem repeat region of MUC1 are potent chemoattractants for circulating immature myeloid dendritic cells and are able to subvert their immunostimulatory function (23).
To understand whether association with NF-κB p65 is required for MUC1-induced interleukin expression, we treated cells with BAY 11-7085, an inhibitor of IκBα phosphorylation. MUC1-induced IL-6 expression was reduced by 50%, and expression of IL-2, IFN-γ, and TNF-α was completely abolished (Fig. 5). BAY 11-7085 also reduced MUC1 occupancy on the promoters (Fig. 7A). MUC1 contains no known DNA-binding motif, and thus, its action depends on other DNA-binding proteins. We still do not know the specific MUC1 sequence that interacts with p65, but our results point to the VNTR region in the extracellular domain as important for cytokine regulation. In cells expressing MUC1 containing the VNTR region, cytokine transcription was significantly increased compared with cells expressing a short MUC1 isoform lacking the VNTR region.
In normal cells, the VNTR region is extensively glycosylated with long, branched, O-linked carbohydrate chains, whereas in malignant cells, glycosylation is consistently altered, yielding a MUC1 molecule that is hypoglycosylated and that carries primarily short glycans. We propose that tumor-specific short O-glycans or no glycans on the tandem repeats of the VNTR region reduce the size of the molecule and its three-dimensional configuration, allowing its binding to transcriptional coactivators such as p65 and trafficking to the nucleus. This is not expected to occur in normal epithelial cells, where MUC1 is strictly polarized to the luminal surface, and long, branched oligosaccharides conceal the VNTR protein and increase tremendously the size of the molecule. Our data also show that degradation and/or cleavage of MUC1 is not required for NF-κB activation and interleukin induction (Fig. 2, 3, and 7). Endocytosis and intracellular accumulation of full-length MUC1 without its degradation or cleavage have been reported in tumor cells (4, 24), and here, we associated this process with a specific function. The ability of MUC1 to induce proinflammatory cytokines and the specificity that this activity might have for cancer cells versus normal cells are important findings that could explain the mechanism by which MUC1 promotes tumor progression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA56103 (to O. J. F.) from NCI. This work was also supported by the Fondazione Ri.Med (to S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- TR
- tandem repeat
- CT
- cytoplasmic tail
- IKK
- IκB kinase
- ConA
- concanavalin A.
REFERENCES
- 1. Burchell J. M., Mungul A., Taylor-Papadimitriou J. (2001) J. Mammary Gland Biol. Neoplasia 6, 355–364 [DOI] [PubMed] [Google Scholar]
- 2. Kufe D. W. (2009) Nat. Rev. Cancer 9, 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hattrup C. L., Gendler S. J. (2008) Annu. Rev. Physiol. 70, 431–457 [DOI] [PubMed] [Google Scholar]
- 4. Altschuler Y., Kinlough C. L., Poland P. A., Bruns J. B., Apodaca G., Weisz O. A., Hughey R. P. (2000) Mol. Biol. Cell 11, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh A. P., Chauhan S. C., Bafna S., Johansson S. L., Smith L. M., Moniaux N., Lin M. F., Batra S. K. (2006) Prostate 66, 421–429 [DOI] [PubMed] [Google Scholar]
- 6. Pochampalli M. R., el Bejjani R. M., Schroeder J. A. (2007) Oncogene 26, 1693–1701 [DOI] [PubMed] [Google Scholar]
- 7. Huang L., Chen D., Liu D., Yin L., Kharbanda S., Kufe D. (2005) Cancer Res. 65, 10413–10422 [DOI] [PubMed] [Google Scholar]
- 8. Wei X., Xu H., Kufe D. (2006) Mol. Cell 21, 295–305 [DOI] [PubMed] [Google Scholar]
- 9. Ahmad R., Raina D., Joshi M. D., Kawano T., Ren J., Kharbanda S., Kufe D. (2009) Cancer Res. 69, 7013–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmad R., Raina D., Trivedi V., Ren J., Rajabi H., Kharbanda S., Kufe D. (2007) Nat. Cell Biol. 9, 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fontenot J. D., Mariappan S. V., Catasti P., Domenech N., Finn O. J., Gupta G. (1995) J. Biomol. Struct. Dyn. 13, 245–260 [DOI] [PubMed] [Google Scholar]
- 12. Taylor-Papadimitriou J., Burchell J. M., Plunkett T., Graham R., Correa I., Miles D., Smith M. (2002) J. Mammary Gland Biol. Neoplasia 7, 209–221 [DOI] [PubMed] [Google Scholar]
- 13. Price M. R., Rye P. D., Petrakou E., Murray A., Brady K., Imai S., Haga S., Kiyozuka Y., Schol D., Meulenbroek M. F., Snijdewint F. G., von Mensdorff-Pouilly S., Verstraeten R. A., Kenemans P., Blockzjil A., Nilsson K., Nilsson O., Reddish M., Suresh M. R., Koganty R. R., Fortier S., Baronic L., Berg A., Longenecker M. B., Hilgers J., et al. (1998) Tumour Biol. 19, Suppl. 1, 1–20 [DOI] [PubMed] [Google Scholar]
- 14. Beatty P. L., Narayanan S., Gariepy J., Ranganathan S., Finn O. J. (2010) Cancer Prev. Res. 3, 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beatty P. L., Plevy S. E., Sepulveda A. R., Finn O. J. (2007) J. Immunol. 179, 735–739 [DOI] [PubMed] [Google Scholar]
- 16. Tinder T. L., Subramani D. B., Basu G. D., Bradley J. M., Schettini J., Million A., Skaar T., Mukherjee P. (2008) J. Immunol. 181, 3116–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoyos B., Ballard D. W., Böhnlein E., Siekevitz M., Greene W. C. (1989) Science 244, 457–460 [DOI] [PubMed] [Google Scholar]
- 18. Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., Young H. A. (1997) J. Biol. Chem. 272, 30412–30420 [DOI] [PubMed] [Google Scholar]
- 19. Libermann T. A., Baltimore D. (1990) Mol. Cell. Biol. 10, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collart M. A., Baeuerle P., Vassalli P. (1990) Mol. Cell. Biol. 10, 1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Udalova I. A., Knight J. C., Vidal V., Nedospasov S. A., Kwiatkowski D. (1998) J. Biol. Chem. 273, 21178–21186 [DOI] [PubMed] [Google Scholar]
- 22. Szlosarek P. W., Balkwill F. R. (2003) Lancet Oncol. 4, 565–573 [DOI] [PubMed] [Google Scholar]
- 23. Carlos C. A., Dong H. F., Howard O. M., Oppenheim J. J., Hanisch F. G., Finn O. J. (2005) J. Immunol. 175, 1628–1635 [DOI] [PubMed] [Google Scholar]
- 24. Merlin J., Stechly L., de Beauce S., Monte D., Leteurtre E., van Seuningen I., Huet G., Pigny P. (2011) Oncogene, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.