Background: Histones are specifically modified in the Xenopus egg.
Results: A complex of Prmt5 and Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasmin on an arginine in a conserved motif.
Conclusion: Arginine methylation is enriched in the egg and targets chromatin-acting proteins.
Significance: Histone arginine methylation probably results in specification of the pluripotent developmental program.
Keywords: Chromatin Histone Modification, Histone Chaperone, Histone Modification, Histones, Xenopus, Mep50, Nucleoplasmin, PRMT, Prmt5, Arginine Methyltransferase
Abstract
Histone proteins carry information contained in post-translational modifications. Eukaryotic cells utilize this histone code to regulate the usage of the underlying DNA. In the maturing oocytes and eggs of the frog Xenopus laevis, histones are synthesized in bulk in preparation for deposition during the rapid early developmental cell cycles. During this key developmental time frame, embryonic pluripotent chromatin is established. In the egg, non-chromatin-bound histones are complexed with storage chaperone proteins, including nucleoplasmin. Here we describe the identification and characterization of a complex of the protein arginine methyltransferase 5 (Prmt5) and the methylosome protein 50 (Mep50) isolated from Xenopus eggs that specifically methylates predeposition histones H2A/H2A.X-F and H4 and the histone chaperone nucleoplasmin on a conserved motif (GRGXK). We demonstrate that nucleoplasmin (Npm), an exceedingly abundant maternally deposited protein, is a potent substrate for Prmt5-Mep50 and is monomethylated and symmetrically dimethylated at Arg-187. Furthermore, Npm modulates Prmt5-Mep50 activity directed toward histones, consistent with a regulatory role for Npm in vivo. We show that H2A and nucleoplasmin methylation appears late in oogenesis and is most abundant in the laid egg. We hypothesize that these very abundant arginine methylations are constrained to pre-mid blastula transition events in the embryo and therefore may be involved in the global transcriptional repression found in this developmental time frame.
Introduction
Chromatin, the compacted higher order structure of DNA packaged with proteins, is the physiological form of the genome (1). The fundamental unit of chromatin is the nucleosome, in which DNA is wrapped around an octamer of the four core histones, H2A, H2B, H3, and H4 (3). Linker histones (somatic H1 and embryonic isoforms such as B4 in the frog Xenopus) and other protein components further condense and arrange chromatin (4, 5). Post-translational modifications of histones, along with deposition of histone variants and linker histones, form a “histone code” (6) regulating the usage of the underlying DNA.
Dynamic changes in deposition of histone variants and alterations of post-translational modification patterns occur throughout the life of a multicellular organism and vary in each cell type (7). Furthermore, these dynamic alterations are specified on individual gene loci (8). The histone code is highly correlated with and predictive for active processes utilizing the DNA information in the chromatin. These include transcription, repair, and replication (9). The sum of these modifications, variants, and DNA methylation form the molecular basis of epigenetic information, the increased complexity of the genome without changes in the underlying gene sequence (10). Epigenetics has been implicated as a cause and effect in many events during cellular differentiation, animal development, and aging (11). Furthermore, reprogramming of somatic cells, both in somatic cell nuclear transfer and the generation of induced pluripotent stem cells, is ultimately driven and signaled by rewriting of the epigenetic state (12).
Arginine methylation is an underexplored histone post-translational modification. It has been implicated in regulation of many realms of biology, including development, somatic transcriptional regulation, and cancer development. The family of protein arginine methyltransferases (PRMTs)2 in metazoans includes at least 10 proteins that have diverse roles (13, 14). The majority of these enzymes are Type I enzymes that are capable of monomethylation and asymmetric dimethylation of arginine (15–17). Prmt5 is one of the few Type II enzymes capable of monomethylation and symmetric dimethylation (17). The functional difference between these methylation states has not been distinguished. Prmt5 has been identified in somatic and embryonic as well as nuclear and cytoplasmic contexts with distinct targets, including Smn proteins (survival of motor neuron) and histones H2A, H3, and H4 (17–19). It also has been identified partnered with a number of cofactors, including Blimp1 (20), MBD/NuRD (21), pICln (22), and Mep50 (the Prmt5-pICln-Mep50 complex is called the “methylosome”) (23, 24). Prmt5 was recently shown to methylate histone H2A Arg-3 in the cytoplasm of mouse embryonic stem cells (25), consistent with a global predeposition role for this modification. Finally, Prmt5 and Mep50 were both recently shown to be misregulated by aberrant phosphorylation, resulting in changed activity, in leukemia and lymphoma (26, 27).
Histone chaperones shield non-chromatin interactions of histones with other proteins or nucleic acids (28). Other roles include deposition onto and removal of histones from DNA during replication and transcription (29). The chaperone nucleoplasmin (Npm) is a homopentamer comprised of 22-kDa subunits. It forms a storage complex with multiple copies of newly synthesized H2A and H2B in Xenopus oocytes and eggs (30). Npm, one of the most abundant proteins in the frog egg, is phosphorylated during oogenesis and hyperphosphorylated upon progesterone-induced meiosis II and subsequent egg laying (31–36).
Previously, we showed that histone variant H2A.X-F is heavily enriched specifically in Xenopus oocytes, eggs, and embryos (37). H2A.X-F comprises at least 50% of the H2A protein in the frog egg. H2A.X-F is mostly identical to canonical H2A in its N terminus, including its conserved N-terminal SGRGK motif (see Fig. 1 in Ref. 37). Here we present the identification and characterization of a complex of the arginine methyltransferase Prmt5 and Mep50 isolated from Xenopus laevis eggs that specifically methylates predeposition histones H2A and H4. We also show that the Prmt5-Mep50 complex targets the histone chaperone nucleoplasmin on a conserved motif (GRGXK). We show that Npm and histones modulate their respective abilities to be methylated by Prmt5-Mep50, consistent with an in vivo role of the histone storage chaperone in regulating a predeposition global histone code. Finally, we present evidence demonstrating the in vivo presence of these modifications on Npm and histones H2A.X-F and H4.
FIGURE 1.
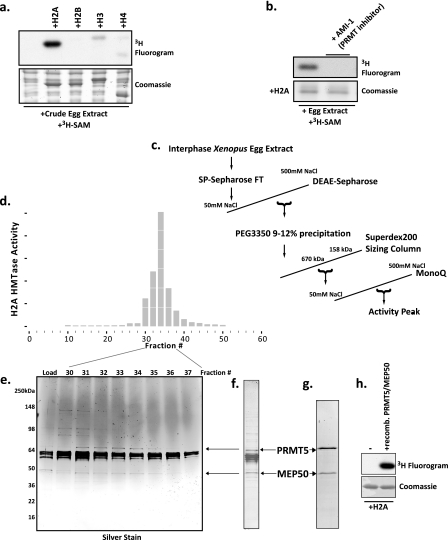
Purification and identification of an abundant histone H2A and H4 arginine 3 methyltransferase in Xenopus laevis egg extract as a complex of Prmt5 and Mep50. a, Xenopus egg extract was incubated with recombinant histones H2A, H2B, H3, and H4 and with the methyl donor [3H]AdoMet ([3H]SAM). Shown are a Coomassie-stained SDS gel (bottom) and a fluorogram (top). b, recombinant H2A was incubated with egg extract and [3H]AdoMet in the presence or absence of 50 μm AMI-1. Bottom, Coomassie-stained SDS gel; top, corresponding fluorogram. c, chromatography scheme for the purification of the H2A methyltransferase activity. d, full-length histone H2A methyltransferase activity across the fractions of the MonoQ column (final column in the purification) performed as a P81 filter binding assay; the HMTase activity histogram showed peak activity in fractions 32–34. e, silver-stained gel of the peak activity fractions. f, Coomassie-stained SDS gel of the pooled peak activity fractions 32–34. Stained bands were identified by mass spectrometry (supplemental Fig. S1), and the location of the arginine methyltransferase Prmt5 and its known cofactor Mep50 are shown with arrows on the gel and in the silver-stained gel. g, Coomassie-stained SDS gel of recombinant Prmt5 and Mep50 produced in baculovirus-infected Sf9 cells. h, methyltransferase assay demonstrating that the recombinant Prmt5-Mep50 complex methylates recombinant H2A (fluorogram, top); lane 1 did not contain enzyme, whereas lane 2 did contain enzyme, and both lanes contained H2A.
EXPERIMENTAL PROCEDURES
General Reagents and Tissue Culture
AMI-1 was purchased from EMD Chemicals, and tritiated S-adenosylmethionine ([3H]AdoMet) was purchased from GE Healthcare. Silver staining was performed with SilverSNAP stain (Pierce). XL2 and A6 cells were grown in L15 medium (Hyclone) with 10% fetal bovine serum and 10 μg/ml hypoxanthine at 27 °C.
Xenopus Extract Preparation and Histone Isolation
Xenopus interphase egg extract, nucleoplasmic extract (NPE), and sperm chromatin were prepared as described (38). Histones from S3 and A6 tissue culture cells were all acid-extracted as described (31). Egg histones were isolated as described (38).
Purification of H2A Methyltransferase Activity from Xenopus Egg Extract
15 ml of clarified interphase egg extract was applied to two connected 5-ml HiTrip SP columns. The flow-through was collected and immediately loaded onto a DEAE-FastFlow 16/10 column previously equilibrated in 50 mm Tris, pH 8.0, 10% glycerol, 1 mm EDTA, 5 mm DTT, and 50 mm NaCl. The column was washed with six column volumes of buffer and eluted with a 20-column volume linear gradient to 500 mm NaCl. Fractions were assayed immediately for H2A methyltransferase activity. Peak activity fractions were raised to 9% (w/v) polyethylene glycol 3350 and incubated on ice for 20 min, and the precipitate was pelleted at 14,000 rpm in an SS34 rotor. The supernatant was raised to 12% PEG 3350 and incubated on ice for 20 min, and the precipitate was pelleted as before. The pellets were dissolved in 50 mm NaCl buffer (above) and assayed for H2A methyltransferase activity. The majority of the activity was found in the 9–12% PEG 3350 fraction, which was then applied to two Superdex 200 10/300 columns connected in series. Eluted fractions were assayed for methyltransferase activity, and the peak fractions were then applied to a MonoQ 5/50 column. The MonoQ column was eluted with a 10-column volume linear gradient to 500 mm NaCl. The activity and protein peaks are shown in Fig. 1.
Mass Spectrometry Identification of Proteins in Peak Activity Fraction
Visible protein bands were excised from the Coomassie-stained SDS gel and destained in 55% ammonium bicarbonate (100 mm), 45% acetonitrile. Gel slices were treated with iodoacetamide (50 mm) to alkylate cysteines.
Proteins were digested in-gel with 75 ng of trypsin (Roche Applied Science) per gel band in 50 mm ammonium bicarbonate for 6 h at 37 °C. Tryptic peptides were extracted from the gel pieces with an 8-μl slurry of 1 volume of POROS R2 20 reverse-phase resin (Applied Biosystems, Foster City, CA) to 10 volumes of 5% formic acid, 0.2% trifluoroacetic acid (TFA) at 4 °C for 16 h. POROS R2 20 resin was transferred to Ziptips (Millipore, Billerica, MA), washed with 0.1% TFA, and eluted onto the matrix-assisted laser desorption ionization (MALDI) target with one-third saturated 2,5-dihydroxybenzoic acid (Lancaster Synthesis, Windham, NH) in 50% methanol, 20% acetonitrile, 0.1% TFA. Tryptic peptides were identified by MALDI mass spectrometric analysis and identified using XProteo (Chao Zhang; available on the World Wide Web).
Identification of the Site of Methylation
To identify the site of methylation, recombinant histone H2A incubated in the active MonoQ fraction was propionylated, as described (39), and digested with trypsin for 6 h. The propionylation reaction was performed again on the newly generated N termini, and the tryptic peptides were separated by online nanoflow HPLC and analyzed using an LTQ-Orbitrap mass spectrometer (Thermo Electron) operated in a data-dependent mode with a full MS scan followed by MS/MS scans. A gradient of 0–60% B (70% acetonitrile, 100 mm acetic acid in water) in 40 min was used. The mass spectra were searched against a histone database by using the SEQUEST algorithm, and the peptide assignments were manually confirmed.
Cloning of Expression Vectors and Baculovirus Production
The X. laevis prmt5 (hsl7) gene was a generous gift of S. Kornbluth (40) (GenBankTM accession number BF613396). All cloning was performed using ligation-independent cloning techniques (41). We initially subcloned it into pMCSG7 for Escherichia coli expression and then into a custom pFastBac1 vector engineered with the pMCSG7 ligation-independent cloning site for baculovirus production. We then subcloned mep50 (Open Biosystems IMAGE clone 6316264) into pMCSG7 and pFastBac1. All clones contained a His6 tag followed by a TEV protease site on the N terminus of the expressed protein. We produced recombinant baculovirus DNA in DH10Bac cells (Invitrogen) according to the manufacturer's instructions.
Nucleoplasmin was subcloned from pET16 (42) into pMCSG7 using ligation-independent cloning. The long N-terminal tag present on the original construct was eliminated and replaced with the His6TEV tag in pMCSG7. Npm C-terminal mutations and truncations were created using 3′ PCR primers containing the mutations. This clone produced the 200-amino acid protein isoform (GenBankTM accession number 128910) (43). The numbering scheme we use for the C terminus, however, refers to the 196-amino acid protein found in vivo.3
Purification of Recombinant Prmt5-Mep50
X. laevis Prmt5 and Mep50 protein complex was expressed in Hi5 cells by infecting 600 ml of 2 × 106 cells/ml culture in MaxXP medium (BD Biosciences) with both Prmt5 and Mep50 viruses for 48 h, shaking at 27 °C. Cells were pelleted and suspended in a cold hypotonic lysis buffer (40 mm Hepes, pH 7.8, and 5 mm KCl with 5 mm β-mercaptoethanol and protease inhibitors) and lysed by Dounce homogenizing with 15 strokes. The suspension was raised to 1 m NaCl to extract the nuclei and then sonicated and centrifuged. The supernatant was immediately applied to Ni2+-NTA resin in the presence of 10 mm imidazole and 250 mm NaCl, and the complex was eluted with 300 mm imidazole. The eluate was spin-concentrated and applied to a Superose 6 column, and the peak complex fractions were pooled and used for all experiments.
Other Protein Production and Purification
Recombinant nucleoplasmin was expressed in E. coli and purified in batch via Ni2+-NTA followed by Superdex 200 16/60 chromatography on an ATKA Purifier 10 (GE Healthcare). Peak fractions were spin-concentrated to 1 mg/ml final concentration (Costar). GST-Prmt1 (rat) (44) was expressed in E. coli and purified in batch on glutathione-Sepharose followed by MonoQ chromatography. Core histones and histone dimers and octamers were expressed and purified as described (45). Oocyte and egg nucleoplasmin were purified as described (46).
Antibody Production and Other Antibodies
Recombinant His6-tagged Xenopus Mep50 purified via urea solubilization from E. coli inclusion bodies was injected into rabbits (Proteintech), and the resulting serum was used for all immunoblots. Commercial Mep50 antibodies against the mammalian protein do not recognize the Xenopus Mep50 protein. Xenopus Prmt5 is recognized by a commercial Prmt5 antibody (Upstate/Millipore 07-405). Two different sets of antibodies recognizing arginine methylation were used for this work; their characterization is shown in supplemental Fig. S3. The first set was produced by Upstate Biotechnology. The antibody recognizing monomethylarginine is referred to as anti-R3me1, labeled 8039, whereas anti-R3me2s, labeled 8040, specifically recognizes symmetrically dimethylated arginine. Both antibodies also recognize either mono- or dimethylated Npm. The second set of antibodies was produced by Proteintech; these are referred to as R3me1 and R3me2. These sera recognize H2A and H4 Arg-3 and Npm Arg-187 mono- or dimethylation. R3me1 is specific for the monomethylation, and R3me2 is specific for dimethylation, although it cannot distinguish between symmetric and asymmetric dimethylation.
Endogenous Immunoprecipitation
Protein A-Sepharose beads (GE Healthcare) were coupled to antibodies in egg lysis buffer (10 mm HEPES, pH 7.7, 250 mm sucrose, 2.5 mm MgCl2, 50 mm KCl, 1 mm DTT, 1 mm PMSF) for 2 h. After washing in egg lysis buffer, the antibody-coupled beads were incubated with egg extract for 3 h at 4 °C. The washed beads were resuspended in 1× Laemmli buffer, and the samples were resolved by SDS-PAGE and analyzed via Western blot.
Histone Methyltransferase Assay
All assays were conducted at 30 °C.
p81 Filter Binding Assay
The reaction mix was spotted on P81 filter paper and air-dried. After washing five times with 100 ml of sodium carbonate buffer (0.1 m, pH 8.5) and a final wash with acetone, the filter papers were air-dried and put into scintillation vials containing 2.5 ml of scintillation liquid. Counts per minute (cpm) were measured via a scintillation counter (Beckman).
SDS-PAGE Fluorography
Tritiated labeled proteins were run on SDS-PAGE, stained with Coomassie R-250 dye, soaked in Amplify solution (GE Healthcare) for 30 min, vacuum-dried onto filter paper, and exposed to preflashed x-ray film (Eastman Kodak Co.); exposures ranged from overnight to 1 week at −80 °C.
RNA Isolation and RT-PCR
Embryos were staged according to Nieuwkoop and Faber (47), whereas early stage embryos were collected according to the number of observed cell divisions. RNA was isolated from five embryos at each stage in TRIzol. Reverse transcription was performed using Superscript III (Invitrogen) and random hexamer primers. The cDNA was amplified with 21 cycles of PCR with [32P]dATP and exposed to a PhosphorImager plate.
RESULTS
We initially tested the histone methyltransferase activity of Xenopus laevis cell-free egg extract by adding exogenous recombinant core histone proteins and [3H]AdoMet, followed by electrophoresis and fluorography. As shown (Fig. 1a), we observed pronounced activity directed toward histone H2A, with minor activity toward H3 and H4. To identify the enzyme responsible for this activity, we treated the extract with AMI-1, an arginine methyltransferase inhibitor, which completely abrogated the activity (Fig. 1b), suggesting that the enzyme was an arginine methyltransferase. We then used conventional chromatography to purify this protein. We followed full-length H2A methyltransferase activity by spotting reactions on P81 filter paper, washing untransferred tritium, and scintillation counting. The purification scheme is outlined in Fig. 1c. The peak activity from fractions of the final MonoQ column is shown in Fig. 1d. We then silver-stained the peak fractions (Fig. 1e) and pooled fractions 31–34. We ran a larger mass of the pooled fractions on an SDS-polyacrylamide gel, and the Coomassie-stained bands (Fig. 1f) were subjected to mass spectrometry analysis (supplemental Fig. S1). The arrows in Fig. 1, e and f, indicate the location of the most likely candidate for the methyltransferase activity, a potential complex of the arginine methyltransferase Prmt5 (also known as Hsl7), and the WD-repeat protein Mep50. We cloned these genes into baculovirus and co-infected Sf9 cells. We purified the recombinant protein complex via Ni2+-NTA chromatography and size exclusion chromatography (supplemental Fig. S2). The purified complex is shown in Fig. 1g.
The recombinant Prmt5-Mep50 complex recapitulated the H2A methyltransferase activity in the purified peak from the egg extract (Fig. 1h). It also eluted from a sizing column at the same molecular weight as the endogenous protein complex. Therefore, we concluded that Prmt5-Mep50 was the enzyme complex responsible for our observed methyltransferase activity in the egg extract. We cannot rule out the possibility that other polypeptides in the peak fraction contribute to or modulate the activity. However, the Prmt5 and Mep50 bands track tightly with the activity profile (Fig. 1, d and e), and none of the other proteins in the peak fraction are known to bind to or regulate enzyme activity (supplemental Fig. S1).
We did attempt to produce recombinant Prmt5 and Mep50 individually. Prmt5 was completely insoluble expressed by itself in baculovirus-infected cells and in E. coli. Mep50 was soluble when expressed alone in baculovirus-infected cells (data not shown) (supplemental Fig. S2, second elution peak).
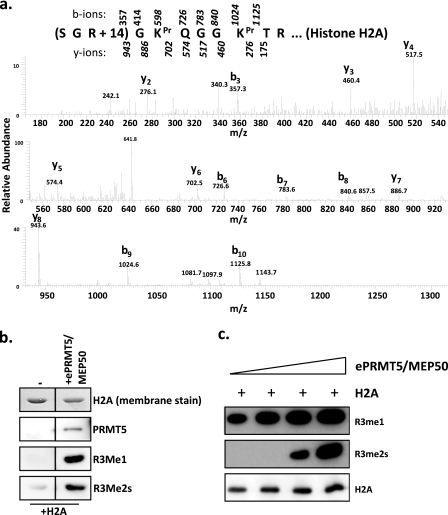
Next, we investigated the targeted residue for methylation on the substrate. We incubated purified egg Prmt5-Mep50 with recombinant histone H2A and subjected the protein to mass spectrometry analysis (Fig. 2a) and immunoblotting (Fig. 2b). Mass spectrometry showed the presence of an ion with a +14 mass on arginine 3, consistent with methylation of this residue. There was no +28 species detectable. Immunoblotting of the H2A sample treated with endogenous Prmt5-Mep50 showed that Prmt5 promoted monomethylation and symmetric dimethylation of Arg-3 on H2A. The specificity of these antibodies for R3me1 and R3me2s is shown in supplemental Fig. S3. Finally, we demonstrated that this activity is distributive, because only high relative concentrations of Prmt5-Mep50 enzyme complex led to dimethylation (Fig. 2c).
FIGURE 2.
Prmt5-Mep50 monomethylates and symmetrically dimethylates H2A at Arg-3. a, the peak activity fraction from the MonoQ was incubated with H2A and [3H]AdoMet and then subjected to mass spectrometry analysis. The MS/MS spectrum shown confirms that the activity resulted in H2A Arg-3 methylation; masses from y ions are underneath sequence, and b ions are above. Pr, propionyl. b, the peak activity fraction from the MonoQ (ePrmt5-Mep50) was incubated with H2A and AdoMet and then immunoblotted with antibodies directed against R3me1 and R3me2s (Upstate), demonstrating that Prmt5 methylates H2A on arginine. The figure is cropped for clarity from the same gel and exposure. c, egg-purified (e) Prmt5-Mep50 (in the peak MonoQ fraction from Fig. 1) was titrated into a methyltransferase reaction with a constant amount of H2A (1 μg), and the reaction products were immunoblotted with the Upstate antibodies against R3me1 (top), R3me2s (middle), and H2A (bottom).
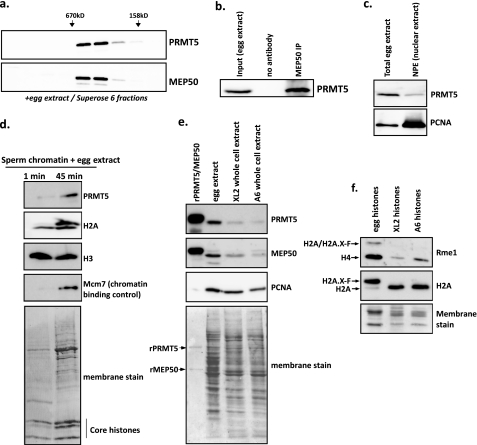
To determine if Prmt5 and Mep50 are only in complex in vivo, we immunoblotted fractions from total egg extract separated on a Superose 6 gel filtration column for Prmt5 and Mep50 (Fig. 3a). We only observed Prmt5 and Mep50 in the identical column fractions with no other peaks of elution. Specificity of these antibodies is shown in supplemental Fig. S3. We then performed an immunoprecipitation with Mep50 antiserum from egg extract and specifically enriched for Prmt5 (Fig. 3b). To determine the subcellular localization of Prmt5, we immunoblotted total egg extract and NPE. We found Prmt5 modestly enriched in the NPE, although not to the extent that PCNA was enriched, suggesting that Prmt5 may mainly act in the cytoplasm. However, we did isolate pronuclear chromatin after remodeling by egg extract and found that Prmt5 was enriched on the chromatin, along with H2A.X-F (top band in the H2A panel) and Mcm7, a known chromatin-binding protein control (48) (Fig. 3d).
FIGURE 3.
Prmt5 exists only in complex with Mep50 in vivo and is found in the nucleus and on chromatin. a, total clarified egg extract was applied to a Superose 6 sizing column, and every other fraction was immunoblotted for Prmt5 and Mep50. Elution positions of 670 and 158 kDa mass markers are shown above. b, endogenous co-immunoprecipitation of Prmt5 with α-Mep50 in egg extract. c, 0.5 μl of clarified egg extract and 0.5 μl of NPE were immunoblotted for Prmt5 and PCNA (a nuclear marker). A small proportion of Prmt5 was found in the nuclear extract. d, sperm pronuclei were assembled in egg extract and collected immediately after incubation (1 min; first lane) or after 45 min (second lane). Chromatin was isolated through a sucrose cushion and immunoblotted for Prmt5, H2A, H3, and Mcm7. e, rPrmt5-Mep50, total egg extract, XL2 whole cell extract, and A6 whole cell extract were immunoblotted for Prmt5, Mep50, and PCNA. The bottom panel shows the total protein content in each lane by Direct Blue 71 staining of the membrane. The protein contents for the egg and whole cell extracts were normalized by a Bradford assay, with an identical total mass of protein loaded in each lane. r, recombinant. f, purified histones from Xenopus eggs, XL2 cells, and A6 cells were immunoblotted for H2A (as control) and R3me1 (Proteintech). The antibodies recognize both H2A and H2A.X-F in the egg extract; for the R3me1 antibody, we refer to both as H2A/H2A.X-F because we cannot distinguish between H2A and its variant in this case. Equal masses of histones were loaded in each lane as shown by Coomassie staining (bottom). IP, immunoprecipitation.
Next, we wanted to determine if Prmt5 and Mep50 were enriched in Xenopus eggs as compared with somatic and cultured cells. We ran recombinant Prmt5-Mep50 and identical masses of total egg extract, Xenopus XL2 whole cell extract, and A6 whole cell extract on a gel and immunoblotted for Prmt5, Mep50, and PCNA. XL2 cells are derived from tadpole epithelium, and A6 cells are derived from adult kidney (49). As shown in Fig. 3e, Prmt5 and Mep50 were substantially more abundant in the egg extract as compared with the cultured cell extracts, consistent with Prmt5-Mep50 playing a bigger role in early embryogenesis. We immunoblotted histones isolated from the egg, XL2, and A6 cells for R3me1 and demonstrated that H2AR3me1 was only present in the egg, whereas H4R3me1, in contrast, was also present in the cultured cells but substantially more abundant in the egg (Fig. 3f).
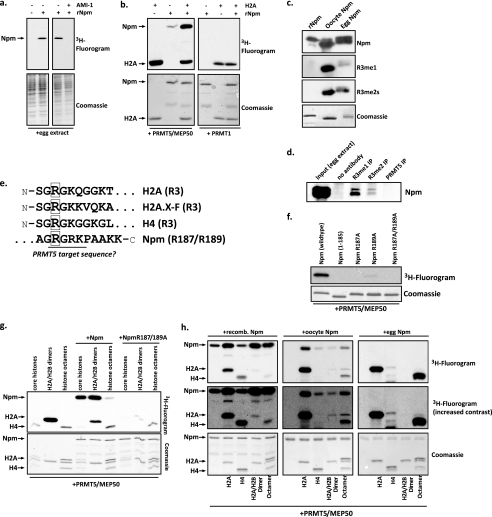
Because we hypothesized that Prmt5 targeted undeposited histones in the egg, we probed a potential role for Npm, the H2A-H2B storage chaperone, in modulation of the methyltransferase activity. Surprisingly, when we first added excess recombinant Npm (rNpm) to the egg extract with tritiated AdoMet, we observed methyltransferase activity on it. Furthermore, the addition of 50 μm AMI-1 abrogated this activity, suggesting that Npm may itself be the target of a PRMT (Fig. 4a). To determine if Prmt5-Mep50 may be the enzyme complex responsible for this activity, we incubated H2A and Npm with recombinant Prmt5-Mep50 (rPrmt5-Mep50) and with recombinant rat Prmt1, a Type I enzyme and the most abundant arginine methyltransferase in most cells (14). As shown in Fig. 4b, Prmt5-Mep50 specifically methylated Npm, and the activity was enhanced in the presence of H2A, whereas Prmt1 was only able to methylate H2A.
FIGURE 4.
Prmt5 methylates the histone chaperone nucleoplasmin on a conserved motif in its C-terminal tail. a, egg extract incubated with or without 1 μg of recombinant Npm and 1 mm AMI-1 in the presence of [3H]AdoMet. Reactions were run on an SDS gel, stained with Coomassie Blue, and exposed for fluorography. rNpm, recombinant nucleoplasmin. b, recombinant Prmt5-Mep50 complex (left) or recombinant Prmt1 was incubated in with H2A, nucleoplasmin, or both and with [3H]AdoMet; the reactions were Coomassie-stained (bottom) and exposed for fluorography (top). c, purified recombinant Npm, oocyte Npm, and egg Npm were immunoblotted for Npm, monomethylarginine, and symmetric dimethylarginine; the Upstate antibodies that recognize Arg-3 methylation were used for Npm. The Coomassie-stained SDS gel of the purified proteins is shown at the bottom. d, egg extract (immunoprecipitation (IP) input), protein A resin without antibody, resin coupled to R3me1 or R3me2 (both from Proteintech), or Prmt5 antibody incubated in egg extract were blotted for Npm. e, the first 10 amino acids of histones H2A, H2A.X-F, and H4 and the C-terminal amino acids of nucleoplasmin are shown. The known (histones) and putative (nucleoplasmin) Prmt5-targeted arginines are boxed. f, recombinant wild type and mutant (1–185 C-terminal truncation, R187A, R189A, and R187A/R195A) nucleoplasmin were incubated with 0.2 μg of recombinant Prmt5-Mep50 and [3H]AdoMet; the reactions were Coomassie-stained (bottom) and exposed for fluorography (top). g, recombinant wild type or mutant R187A/R189A nucleoplasmin were incubated with all four core histones, H2A-H2B dimers, or histone octamers and with 0.2 μg of recombinant Prmt5-Mep50 and [3H]AdoMet; the reactions were Coomassie-stained (bottom) and exposed for fluorography (top). h, recombinant Prmt5-Mep50 complex and [3H]AdoMet were incubated alone or with H2A, H4, or H2A-H2B dimers or H2A-H2B-H3-H4 histone octamers and also incubated with recombinant nucleoplasmin (first column of panels), oocyte nucleoplasmin (second column of panels), or egg nucleoplasmin (third column of panels). The reactions were Coomassie-stained (bottom) and exposed for fluorography (top).
Next, we used the R3me1 and R3me2s antibodies (Upstate) to probe purified recombinant, oocyte, and egg Npm. As shown in Fig. 4c, endogenous oocyte and egg Npm were recognized by the methylarginine antibodies, whereas the recombinant Npm was not recognized. Note that the gel shift in the purified oocyte and egg Npm proteins is due to phosphorylation. In Fig. 4d, we used the methylarginine antibodies (Proteintech) to immunoprecipitate Npm, whereas a control Prmt5 antibody did not precipitate Npm. These data show that endogenous Npm is arginine-methylated.
To determine the site of methylation, we inspected the primary amino acid sequence of Npm. The extreme C terminus of Npm contains a “GRGRK” motif, reminiscent of the N terminus of H2A and H4 (Fig. 4e). To test if one of these arginines was the target of Prmt5 methyltransferase activity, we produced recombinant Npm either truncated on the last 10 amino acids (Npm 1–185) or site-specifically mutated in the Arg-187 or Arg-189 sites. We then incubated these recombinant proteins with recombinant Prmt5-Mep50 and [3H]AdoMet. As shown in Fig. 4f, deletion or mutation of Arg-187 completely abrogated the activity, whereas residual activity was apparent in the NpmR189A mutant. These data confirmed that Arg-187 was the target of Prmt5. We further corroborated this result with mass spectrometry analysis of Npm.4
To further analyze the activity of Prmt5-Mep50 directed toward Npm and histones, we incubated rPrmt5-Mep50 with core histones, H2A-H2B dimers, or histone octamers in the presence or absence of Npm or NpmR187A/R189A and [3H]AdoMet. Intriguingly, the presence of Npm, particularly the unmethylatable Npm mutant, dramatically reduced the methyltransferase activity directed toward histones H2A and H4 (Fig. 4g). We also note that Prmt5-Mep50 only methylates H4 in octamers (Fig. 4g) (data not shown). Finally, to test the potential cross-talk between modifications on Npm and Prmt5-Mep50 activity toward histones, we incubated rPrmt5-Mep50 with H2A, H4, H2A-H2B dimers, or histone octamers in the presence of rNpm, oocyte Npm, or egg Npm (Fig. 4h). The presence of unmodified rNpm reduced the Prmt5-Mep50 activity directed toward H2A-H2B dimers or octamers (left-hand panels). However, endogenous oocyte Npm, although not a potent substrate for Prmt5 because it is already partly methylated (Fig. 4c) (mass spectrometry data not shown), did substantially modulate Prmt5 activity toward histone H2A alone and in octamers, as well as H4 in octamers (middle panels). Egg nucleoplasmin was completely unmethylatable by Prmt5 but dramatically stimulated the H2A and octamer H4 activity by Prmt5 (right-hand panels). Egg Npm is almost completely methylated on Arg-187 as shown by mass spectrometry (data not shown).
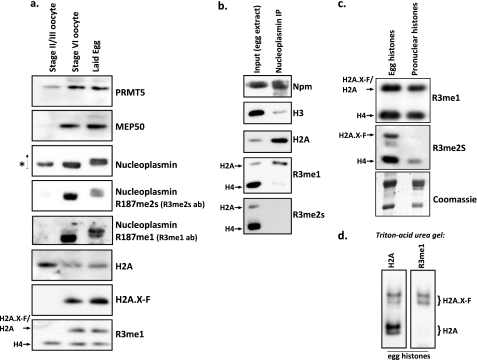
Histones are synthesized during oogenesis in the frog and stored in complex with chaperones, in particular Npm. To determine the developmental timing of Npm and histone arginine methylation, we prepared blots with total lysate from early stage II/III pools of oocytes, late stage VI oocytes, and laid eggs. We blotted with antibodies, as shown in Fig. 5a. Prmt5 and Mep50 appeared in late oogenesis. Npm was present in stage II/III oocytes but was only methylated in stage VI oocytes and laid eggs. Note the gel shift of Npm, due to phosphorylation (33), as indicated by the up arrow. We observed H4 methylation in all three stages of oogenesis, whereas H2A methylation appeared only in stage VI and laid eggs.
FIGURE 5.
H2A.X-F is preferentially methylated, whereas symmetrically dimethylated H2A does not bind to nucleoplasmin. a, total stage II/III and stage VI oocyte extract and laid egg extract were immunoblotted as shown. b, nucleoplasmin was immunoprecipitated from crude egg extract (lane 2; lane 1 contains 0.5% of the input egg extract) and immunoblotted for nucleoplasmin, histones H3 and H2A, and R3me1 and R3me2s (Upstate). c, isolated histones from chaperone-bound complexes in the egg (lane 1) and from acid-extracted pronuclear chromatin (lane 2) were immunoblotted against R3me1 and R3me2s (Upstate); the bottom panel was Coomassie-stained. d, isolated egg histones were run on a Triton-acid urea gel and immunoblotted for H2A (left) and R3me1 (right; Upstate). The migration positions of H2A.X-F and H2A are noted, as shown previously (37).
There are at least eight PRMT genes in Xenopus. We extracted prmt5 transcription abundance data from a recent RNA-Seq analysis of Xenopus tropicalis oocytes (50) and microarray analysis of developing X. laevis embryos (51). As shown in supplemental Fig. S4, prmt1 was by far the most abundant transcript, whereas prmt5 was the second most abundant transcript in the egg, and the expression level dropped off after the mid-blastula transition. prmt7, the only other Type II enzyme found in Xenopus capable of symmetric dimethylation, was substantially less abundant in X. laevis eggs and early embryos. We also performed RT-PCR on RNA extracted from staged embryos with primers for prmt5, mep50, and maternal and zygotic mcm6 (52) to mark zygotic gene activation. These experiments, consistent with data extracted from the microarray study (51), showed that prmt5 and mep50 RNA abundance drops after the mid-blastula transition (supplemental Fig. S5).
Next, we asked if H2A and H4 containing Arg-3 methylation were deposited into pronuclear chromatin assembled in egg extract. First, we performed Npm immunoprecipitation with polyclonal Npm antisera. As expected and shown in Fig. 5b, the Npm IP enriched for H2A but not H3. Intriguingly, H2AR3me1 was enriched, but no H2AR3me2s was precipitated at all. Npm-bound histones are used for pronuclear chromatin assembly. Consistent with this pathway, we only observed H2AR3me1, but no H2AR3me2s, in deposited histones on pronuclei (Fig. 5c). H4R3me1 and H4R3me2s were both found in pronuclear histones.
DISCUSSION
Here, we described the identification of a potent H2A methyltransferase activity in X. laevis eggs. Following conventional chromatography, we isolated a peak fraction containing Prmt5 and Mep50, a known enzyme complex. We showed that recombinant Prmt5-Mep50 methylated histone H2A and histone H4. Surprisingly, we also showed that Prmt5-Mep50 methylated the histone storage chaperone nucleoplasmin on a conserved motif on its unstructured C terminus. We showed that nucleoplasmin modulated the Prmt5 activity directed toward histones, suggesting an in vivo mechanism by which global histone modification may be regulated. Finally, we showed that symmetrically dimethylated histone H2A is not bound by Npm and is not deposited into pronuclear chromatin in contrast to monomethylated Npm.
The stoichiometry of the Prmt5-Mep50 complex is currently unknown, although previous reports show oligomer formation for Prmt5 (53). Endogenous and recombinant Prmt5-Mep50 complexes elute from sizing columns at high apparent molecular weight (Fig. 3a and supplemental Fig. S2). Therefore, we assume that Prmt5-Mep50 forms a larger structure than a simple 1:1 ratio of Prmt5 and Mep50.
Intriguingly, not every egg extract preparation exhibited this methyltransferase activity toward full-length histone H2A; nor did oocyte extract, whereas all preparations did target an H2A N-terminal 1–21 peptide. However, the activity was still present in the extract, because chromatography through a gel filtration column always recovered the activity coincident with Prmt5-Mep50. There is likely to be a stoichiometric inhibitor of Prmt5 in the oocyte and egg extract, because the addition of a fraction from the gel filtration chromatography to egg extract or recombinant Prmt5-Mep50 inhibited the H2A-directed methyltransferase activity (data not shown). The nature of this inhibitor is still unclear because we have been unable to further purify it.
The function of Npm as a substrate and a modulator of Prmt5-Mep50 activity directed toward histones is intriguing, both biochemically and biologically. Because endogenous H2A is only found bound to Npm in the egg, the likely endogenous histone substrate for Prmt5-Mep50 is bound to Npm. Npm binding to histones possibly alters the conformation so that the unstructured tails of both Npm and histones may be no longer accessible for binding and methyltransferase activity (54). Furthermore, endogenous modifications on Npm, including phosphorylation and arginine methylation, substantially alter the Prmt5 activity toward histones as well. This suggests that Npm phosphorylation, in particular, alters its interactions with histones; we are currently testing this hypothesis. Our assays shown in Fig. 4 did demonstrate altered activity of Prmt5-Mep50 toward histone octamers and only modest changes upon binding to histone H2A-H2B dimers. The actual endogenous histone binding partner for Npm is still unclear, and neither dimers nor octamers may be the true partner. Regardless, our observations are consistent with a significant role for Npm in modulation of Prmt5 activity toward H2A. Furthermore, as shown in Fig. 4g, the presence of histones substantially stimulates the activity of Prmt5 toward Npm itself, suggesting that this substrate modulation activity is mutual.
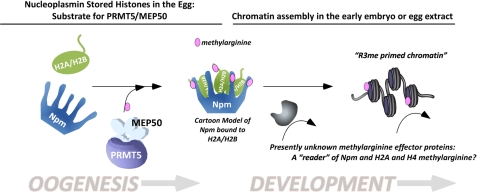
We propose the model shown in Fig. 6. During the months-long process of oogenesis in the ovary of the female frog, massive pools of histones are synthesized and captured by the storage chaperones Npm and N1 (only Npm is illustrated in Fig. 6 for clarity). Prmt5 and Mep50 are synthesized late in oogenesis. This enzyme probably functions in the cytoplasm outside of the oocyte germinal vesicle to methylate the Npm-H2A complex as well as histone H4 bound to N1. We have not tested the function of N1 in modulating Prmt5 activity toward H4, although it is possible that it may play a similar role to Npm. However, N1 itself is not a substrate for Prmt5 (data not shown). Upon fertilization, Npm releases its histone load, which is then deposited onto the growing chromatin by downstream chaperones. Intriguingly, only monomethylated H2A is deposited into chromatin, whereas dimethylated H2A appears to be specifically excluded from deposition. We hypothesize that distinct effectors, such as Tudor domain-containing proteins, recognize the methylarginine on Npm and on the histones deposited into chromatin. These effector proteins then act or recruit other proteins to perform particular biological outcomes, such as modulation of nucleosome spacing. We are currently investigating potential effector proteins.
FIGURE 6.
A schematic model for the function of Prmt5-dependent methylation of histones H2A and H4 and the histone chaperone nucleoplasmin. During oogenesis, core histone proteins H2A, H2B, H3, and H4 are synthesized and stored by the storage chaperones Npm and N1. For clarity, in this figure we only show Npm and its known interacting partners H2A and H2B. The Prmt5-Mep50 complex is produced later in oogenesis, at which time we hypothesize that it methylates Npm and H2A/H2A.X-F and H4. Methylated Npm then acts upon fertilization to deposit histones, specifically monomethylated H2A.X-F and H4, in the growing chromatin. Specific effector proteins recognize the methylarginine in Npm and histones, during oogenesis and/or during early development. We hypothesize that these putative “readers” of methylarginine play a significant role in developmental regulation of gene expression and plasticity. Mep50 has six WD40 domains, illustrated as a single polypeptide (24).
Our discoveries are consistent with a wide literature demonstrating a significant role for arginine methylation in early embryogenesis of metazoans (25, 55). First of all, we showed that Prmt5 and Mep50 were substantially enriched in the egg compared with somatic cultured cells. We also demonstrated that prmt5 and mep50 gene expression drops off substantially after zygotic gene activation. Furthermore, our discovery of nucleoplasmin as a novel substrate for Prmt5 is consistent with a particular early embryogenesis function for Prmt5-catalyzed arginine methylation. Nucleoplasmin protein expression is limited to oocytes, eggs, and early embryos (data not shown). Our unpublished observations4 show that the vast majority of endogenous nucleoplasmin is methylated on Arg-187. Furthermore, we previously showed that egg and pronuclear H4 is methylated on Arg-3 (7, 56). Additional unpublished observations4 show that egg and pronuclear H2A, primarily the abundant variant H2A.X-F (37), are highly methylated on Arg-3. As we had previously demonstrated, H2A.X-F expression and protein levels decline substantially after the mid-blastula transition, and the protein is not present in somatic cells (37). Histone H2A and H2B are more labile in nucleosomes in general than H3 and H4, and H2A-H2B post-translational modifications are likely to be more transient (57). These observations in general are strongly consistent with a particularly specific biological function for Npm and H2A arginine methylation in early embryogenesis. We hypothesize that these very abundant arginine methylations are constrained to pre-midblastula transition events in the embryo and therefore may be involved in the global transcriptional repression found in this developmental time frame (58).
Supplementary Material
Acknowledgments
The hsl7 (prmt5) gene was a gift of S. Kornbluth. The pET16 nucleoplasmin expression vector was a gift of J. Kalinich. Purified Npm antiserum was a gift of S. Dilworth. GST-rat Prmt1 expression vector was a gift of S. Coonrod. XL2 cells were a gift of N. Kikyo. L. Peshkin provided the normalized PRMT expression data from Xenopus developmental profiles. T. Swigut provided initial assistance with embryology. S. Stadler contributed tests of the methylarginine antibodies. C. Goldberg assisted with Xenopus tissue culture and histone isolation. M. C. Ho provided assistance and consultation with protein purification. We are grateful to C. David Allis, in whose laboratory this work was initiated.
This work was supported, in whole or in part, by National Institutes of Health, Grants GM075486 (to D. S.), RR00862 (to B. T. C.), and GM037537 (to D. F. H.). This work was also supported by startup funds from the Albert Einstein College of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Unpublished mass spectrometry evidence; Joshua J. Nicklay and Donald F. Hunt, GenBankTM accession number 46249828.
Joshua J. Nicklay, Takashi Onikubo, Jeffrey Shabanowitz, Donald F. Hunt, and David Shechter, manuscript in preparation.
- PRMT
- protein arginine methyltransferase
- AdoMet
- S-adenosylmethionine
- NPE
- nucleoplasmic extract
- rPrmt5-Mep50
- recombinant Prmt5-Mep50
- rNpm
- recombinant Npm
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Shechter D., Allis C. D. (2007) Nat. Rev. Genet. 8, S23 [Google Scholar]
- 2. Deleted in proof.
- 3. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 4. Ausió J. (2006) Brief Funct. Genomic. Proteomic. 5, 228–243 [DOI] [PubMed] [Google Scholar]
- 5. Woodcock C. L., Skoultchi A. I., Fan Y. (2006) Chromosome Res. 14, 17–25 [DOI] [PubMed] [Google Scholar]
- 6. Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 7. Shechter D., Nicklay J. J., Chitta R. K., Shabanowitz J., Hunt D. F., Allis C. D. (2009) J. Biol. Chem. 284, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 9. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 10. Turner B. M. (2007) Nat. Cell Biol. 9, 2–6 [DOI] [PubMed] [Google Scholar]
- 11. Bernstein B. E., Meissner A., Lander E. S. (2007) Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 12. Shafa M., Krawetz R., Rancourt D. E. (2010) BioEssays 32, 791–799 [DOI] [PubMed] [Google Scholar]
- 13. Cheng D., Yadav N., King R. W., Swanson M. S., Weinstein E. J., Bedford M. T. (2004) J. Biol. Chem. 279, 23892–23899 [DOI] [PubMed] [Google Scholar]
- 14. Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bedford M. T., Richard S. (2005) Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 16. Bedford M. T. (2007) J. Cell Sci. 120, 4243–4246 [DOI] [PubMed] [Google Scholar]
- 17. Wolf S. S. (2009) Cell Mol. Life Sci. 66, 2109–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wysocka J., Allis C. D., Coonrod S. (2006) Front. Biosci. 11, 344–355 [DOI] [PubMed] [Google Scholar]
- 19. Lee Y. H., Stallcup M. R. (2009) Mol. Endocrinol. 23, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. (2006) Nat. Cell Biol. 8, 623–630 [DOI] [PubMed] [Google Scholar]
- 21. Le Guezennec X., Vermeulen M., Brinkman A. B., Hoeijmakers W. A., Cohen A., Lasonder E., Stunnenberg H. G. (2006) Mol. Cell Biol. 26, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001) Mol. Cell Biol. 21, 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 24. Furuno K., Masatsugu T., Sonoda M., Sasazuki T., Yamamoto K. (2006) Biochem. Biophys. Res. Commun. 345, 1051–1058 [DOI] [PubMed] [Google Scholar]
- 25. Tee W. W., Pardo M., Theunissen T. W., Yu L., Choudhary J. S., Hajkova P., Surani M. A. (2010) Genes Dev. 24, 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu F., Zhao X., Perna F., Wang L., Koppikar P., Abdel-Wahab O., Harr M. W., Levine R. L., Xu H., Tefferi A., Deblasio A., Hatlen M., Menendez S., Nimer S. D. (2011) Cancer Cell 19, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aggarwal P., Vaites L. P., Kim J. K., Mellert H., Gurung B., Nakagawa H., Herlyn M., Hua X., Rustgi A. K., McMahon S. B., Diehl J. A. (2010) Cancer Cell 18, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eitoku M., Sato L., Senda T., Horikoshi M. (2008) Cell Mol. Life Sci. 65, 414–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akey C. W., Luger K. (2003) Curr. Opin. Struct. Biol. 13, 6–14 [DOI] [PubMed] [Google Scholar]
- 30. Arnan C., Saperas N., Prieto C., Chiva M., Ausió J. (2003) J. Biol. Chem. 278, 31319–31324 [DOI] [PubMed] [Google Scholar]
- 31. Cotten M., Sealy L., Chalkley R. (1986) Biochemistry 25, 5063–5069 [DOI] [PubMed] [Google Scholar]
- 32. Sealy L., Cotten M., Chalkley R. (1986) Biochemistry 25, 3064–3072 [DOI] [PubMed] [Google Scholar]
- 33. Leno G. H., Mills A. D., Philpott A., Laskey R. A. (1996) J. Biol. Chem. 271, 7253–7256 [DOI] [PubMed] [Google Scholar]
- 34. Tamada H., Van Thuan N., Reed P., Nelson D., Katoku-Kikyo N., Wudel J., Wakayama T., Kikyo N. (2006) Mol. Cell Biol. 26, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bañuelos S., Omaetxebarria M. J., Ramos I., Larsen M. R., Arregi I., Jensen O. N., Arizmendi J. M., Prado A., Muga A. (2007) J. Biol. Chem. 282, 21213–21221 [DOI] [PubMed] [Google Scholar]
- 36. Taneva S. G., Muñoz I. G., Franco G., Falces J., Arregi I., Muga A., Montoya G., Urbaneja M. A., Bañuelos S. (2008) Biochemistry 47, 13897–13906 [DOI] [PubMed] [Google Scholar]
- 37. Shechter D., Chitta R. K., Xiao A., Shabanowitz J., Hunt D. F., Allis C. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Banaszynski L. A., Allis C. D., Shechter D. (2010) Methods 51, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., Hunt D. F. (2007) Nat. Protoc. 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamada A., Duffy B., Perry J. A., Kornbluth S. (2004) J. Cell Biol. 167, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 42. Kalinich J. F., McClain D. E. (1994) Protein Expr. Purif. 5, 324–330 [DOI] [PubMed] [Google Scholar]
- 43. Bürglin T. R., Mattaj I. W., Newmeyer D. D., Zeller R., De Robertis E. M. (1987) Genes Dev. 1, 97–107 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., Wysocka J., Sayegh J., Lee Y. H., Perlin J. R., Leonelli L., Sonbuchner L. S., McDonald C. H., Cook R. G., Dou Y., Roeder R. G., Clarke S., Stallcup M. R., Allis C. D., Coonrod S. A. (2004) Science 306, 279–283 [DOI] [PubMed] [Google Scholar]
- 45. Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 46. Sealy L., Burgess R. R., Cotten M., Chalkley R. (1989) Methods Enzymol. 170, 612–630 [DOI] [PubMed] [Google Scholar]
- 47. Faber J., Niuwkoop P. D. (1994) Normal Table of Xenopus Laevis (Daudin), Garland Publishing, Inc., New York [Google Scholar]
- 48. Shechter D., Gautier J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10845–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anizet M. P., Huwe B., Pays A., Picard J. J. (1981) In Vitro 17, 267–274 [DOI] [PubMed] [Google Scholar]
- 50. Simeoni I., Gilchrist M. J., Garrett N., Armisen J., Gurdon J. B. (2011) Stem Cells Dev., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yanai I., Peshkin L., Jorgensen P., Kirschner M. W. (2011) Dev. Cell 20, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sible J. C., Erikson E., Hendrickson M., Maller J. L., Gautier J. (1998) Curr. Biol. 8, 347–350 [DOI] [PubMed] [Google Scholar]
- 53. Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., Im D. S. (2001) J. Biol. Chem. 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 54. Ramos I., Martín-Benito J., Finn R., Bretaña L., Aloria K., Arizmendi J. M., Ausió J., Muga A., Valpuesta J. M., Prado A. (2010) J. Biol. Chem. 285, 33771–33778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nagamatsu G., Kosaka T., Kawasumi M., Kinoshita T., Takubo K., Akiyama H., Sudo T., Kobayashi T., Oya M., Suda T. (2011) J. Biol. Chem. 286, 10641–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nicklay J. J., Shechter D., Chitta R. K., Garcia B. A., Shabanowitz J., Allis C. D., Hunt D. F. (2009) J. Biol. Chem. 284, 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wyrick J. J., Parra M. A. (2009) Biochim. Biophys. Acta 1789, 37–44 [DOI] [PubMed] [Google Scholar]
- 58. Newport J., Kirschner M. (1982) Cell 30, 687–696 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.