Background: CRABP-II delivers retinoic acid (RA) to the nucleus to activate the transcription factor RAR.
Results: Apo-CRABP-II is associated with endoplasmic reticulum and it dissociates from this location upon RA-induced SUMOylation at residue K102.
Conclusion: SUMOylation of CRABP-II enables its mobilization to the nucleus in response to RA binding.
Significance: This is a novel mechanism for regulating the transcriptional activity of RA.
Keywords: Nuclear Receptors, Retinoid, Retinoid-binding Protein, Sumoylation, Transcription Factors
Abstract
Cellular retinoic acid-binding protein II (CRABP-II) undergoes nuclear translocation upon binding of retinoic acid (RA). In the nucleus, CRABP-II directly binds to the nuclear receptor RAR to form a complex through which RA is “channeled” from the binding protein to the receptor. CRABP-II thus facilitates the ligation of RAR and markedly enhances its transcriptional activity. The primary sequence of CRABP-II contains three putative SUMOylation sites, centered at K45, K87, and K102. We show here that RA induces interactions of CRABP-II with the E2 SUMO ligase Ubc9 and triggers SUMOylation of the protein both in vitro and in cultured cells. Mutagenesis analyses demonstrate that K102 is the sole CRABP-II residue to be SUMOylated in response to RA. Mutation of this residue abolishes the ability of CRABP-II to undergo nuclear translocation in response RA and thus impairs CRABP-II-mediated activation of RAR. Additional observations demonstrate that apo-CRABP-II is associated with endoplasmic reticulum (ER), and that RA triggers the dissociation of CRABP-II from this location. Furthermore, we show that RA-induced dissociation of CRABP-II from the ER requires SUMOylation of K102. Hence, SUMOylation of K102 in response to RA binding is critical for dissociation of CRABP-II from ER and, consequently, for mobilization of the protein to nucleus and for its cooperation with RAR.
Introduction
The vitamin A metabolite retinoic acid (RA)5 regulates gene transcription by activating several members of the nuclear receptor family of ligand-activated transcription factors: the classical RA receptors RARα, RARβ, and RARγ (1) and the peroxisome proliferator-activated receptor β/δ (PPARβ/δ) (2–5). The partitioning of the hormone between its receptors is regulated by two intracellular lipid-binding proteins, cellular retinoic acid-binding protein type II (CRABP-II), which delivers RA to RAR, and fatty acid-binding protein type 5 (FABP5), which shuttles it to PPARβ/δ (3, 4, 6–10). These proteins reside in the cytosol in the absence of an activating ligand and they undergo nuclear localization upon binding of RA. In the nucleus, RA-bound CRABP-II and FABP5 associate with RAR and PPARβ/δ, respectively, to form a complex through which RA is “channeled” from the binding protein to the receptor. These binding proteins thus markedly enhance the transcriptional activity of their cognate receptors.
We previously showed that the nuclear localization signal (NLS) of CRABP-II is comprised of residues K20, R29, and K30. These residues do not cluster together in the protein primary sequence but they assemble in the three-dimensional structure of the protein to form a functional NLS. We showed further that the side-chain of the NLS residues are directed toward the protein in apo- (non-liganded) CRABP-II, and that RA binding results in changes in their configuration rendering them accessible to the solution. The NLS formed upon ligation of CRABP-II is then recognized by the nuclear import protein importin α, which mobilizes it to the nucleus (9). Additional structure/function analyses revealed that the region of CRABP-II, which mediates its interactions with RAR consists of residues Q75, P81, and K102. These spatially aligned residues, located on a protrusion above the entrance to the ligand-binding pocket of the protein, form a surface patch that was found to be necessary and sufficient for mediating the transfer of RA from CRABP-II to RAR and for allowing CRABP-II to enhance receptor transcriptional activity (11). Interestingly, examination of the primary sequence of CRABP-II revealed that the protein contains three consensus SUMOylation sequences, centered at K45, K87, and K102, the latter of which also participates in the makeup of the RAR interaction domain of the protein. We thus set out to investigate whether these residues are SUMOylated in cells and to explore the significance of the modification for CRABP-II function.
The human genome contains four SUMO proteins: SUMO1–SUMO4. Among these, SUMO1–SUMO3 are ubiquitously expressed whereas SUMO4 is expressed mainly in the kidney, lymph node and spleen (12, 13). SUMOylation, the reaction by which a SUMO protein is conjugated to a lysine residue within the consensus sequence ψKXE, (ψ, hydrophobic residue; X, any residue) in a target protein, is a multistep process. In the first step, the immature SUMO protein is cleaved and activated by sentrin-specific protease (SENP). The mature SUMO is then attached to a heterodimer, comprised of UBA2 and AOS1, which functions as an E1-activating enzyme to transfer SUMO to the E2-conjugating enzyme Ubc9. In the final step, facilitated by E3 ligases, Ubc9 transfers SUMO to the substrate by linking the C-terminal Gly residue of SUMO to the side chain of the target lysine (13). A large number of proteins were reported to be modified by SUMOylation and the functional consequences of the modification are extremely diverse. These include effects on the localization and activity, and, in some cases, effects on the stability of the target proteins (12, 13). The observations described in this report reveal that binding of RA to CRABP-II induces SUMOylation at residue K102. The data further show that the modification triggers dissociation of CRABP-II from the endoplasmic reticulum (ER) and thus enables the RA-induced mobilization of the protein to the nucleus. Hence, SUMOylation is critical for the ability of CRABP-II to cooperate with the transcription factor RAR in mediating the transcriptional activities of RA.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies against SUMO2/3 was purchased from Cell Signaling (cat 4971S). Antibodies against HA (cat. H9658) and Myc (cat. 9106) was obtained from Sigma and Abcam, respectively. Antibodies against CRABP-II were provided by Cecile Rochette-Egly (IGBMC, Strasbourg). Retinoic acid was purchased from Calbiochem. Anti-Flag M2-agarose beads (Cat. A2220), 3XFlag elution peptide (Cat. F4799) and N-ethylmaleimide (Cat. E3876) were obtained from Sigma Co. Protein A/G-agarose beads (Cat. sc-2003) were purchased from Santa Cruz Biotechnology. In vitro SUMOylation kit (SUMOlink, Cat. 40220) was purchased from Active Motif. Trizol was purchased from Molecular Research Centre (MRC).
Vectors
CRABP-II was subcloned into a pCMVFlag Tag 2B and EGFPC2 expression vector respectively. Plasmids encoding HA-Ubc9 and Myc-SUMO2 were provided by Hung-Ying Kao (Case Western Reserve University, Cleveland) and Jun-Lin Guan (University of Michigan, Ann Arbor), respectively. The bacterial expression vector for GST-importinαΔIBB construct was provided by Karsten Weis (University of California, Berkeley).
Cells
MCF-7, HEK 293T and COS-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Immunoblotting
Cells were lysed in buffer containing 150 mm NaCl, 10 mm Tris, pH 8.0, 0.1% SDS, 0.1% Triton X-100, 5 mm EDTA, 20 mm N-ethyl-maleimide, protease, and phosphatase inhibitor mixture. Protein concentrations were determined by the Bradford assay. 30–50 μg protein/lane were resolved by SDS-PAGE and probed by immunoblots using indicated antibodies. Site-directed mutagenesis was carried out using a site-directed mutagenesis kit (Stratagene cat. 200521) following the manufacturer's protocol. Transient transfections of MCF7 and HEK293 cells were accomplished using Superfect transfection reagent (Qiagen). Ectopic expression in COS-7 cells was carried out using Fugene6 (Roche).
Immunoprecipitations were performed using anti Flag M2-agarose beads (Sigma-Aldrich) following the manufacturer's protocol. 1 mg of protein of cell lysate was used unless otherwise specified. Immunoprecipitated proteins were eluted from the beads using 3xFlag peptide. In vitro SUMOylation reaction was performed using bacterially purified histidine-tagged CRABP-II and an in vitro SUMOylation kit obtained from Active Motif following the manufacturer's protocol.
Quantitative Real-time PCR (Q-PCR)
Total RNA was isolated from the cells using TRIzol. 1 μg of mRNA was reverse-transcribed into cDNA using the high capacity RNA to cDNA kit from Applied Biosystems (Gaithersburg, MD). Taqman chemistry assays on Demand probes for RARβ (Hs00233407_m1) was purchased from Applied Biosystems. 18 S rRNA (4352930–1007027) was used as an internal control. Detection and data analysis were carried out on an ABI StepOne Plus Real-Time PCR system.
Live Cell Imaging
EGFP-tagged CRABPII in COS-7 cells was imaged using laser-scanning confocal system (Leica). To image live cells, cells were transfected with GFP-CRABP-II in suspension and placed in LabTek glass chambers (Nalge Nunc Iternational). 24 h post-transfection, cells were incubated with 5% charcoal-treated FBS in DMEM for 2 h and then treated with RA (1 μm). A Leica DMI6000 inverted microscope that was motorized for automated fluorescence filter sets for UV, Blue, Green, Red was used. When working with live cells, the microscope was equipped with an environmental chamber for the control of temperature, CO2 concentration, and humidity. Images were collected with excitation at 488 nm and emission at 530 nm. To image cell nuclei in live cells, cells were incubated with Syto 59 (0.5 μm, 10 min) and visualized with excitation at 622 nm and emission at 645 nm.
Colocalization Studies
COS-7 cells stably expressing EGFP-tagged proteins were treated with vehicle or RA (1 μm, 30 min). Cells were fixed with 3% paraformaldehyde in PBS for 2 h, washed with PBS, blocked with 5% FBS in PBS for 1 h, and incubated with primary antibodies and then secondary antibodies (1 h each, room temperature). The primary antibodies were: rabbit polyclonal anti-human calnexin, or rabbit anti-human succinate dehydrogenase (Santa Cruz Biotechnology). The secondary antibody was donkey anti-rabbit IgG, conjugated with Alexa Fluor 594 (Molecular Probes, Invitrogen). Cells were extensively washed with PBS, air dried, and treated with SlowFade preservation reagent from Invitrogen (Carlsbad, CA). Confocal microscopy experiments were performed at the Imaging Core Facility of the Genetics department at CWRU. A Leica DMI6000 inverted microscope was used. The red fluorescence images of Alexa Fluor 594-labeled ER and Mitochondrial markers were obtained with excitation at 594 nm, and emission at 620 nm. Consecutive imaging of the same field of cells was performed for visualization of EGFP-CRABPII, with excitation of EGFP at 488 nm, and emission at 530 nm. Overlay of the two separate images (red and green) of the same cells was performed by using the Visualization program (a Volocity software). Yellow pixels were displayed where both red and green fluorescence was detected. Similar procedures were used to localize endogenous CRABP-II in MCF-7 cells. For colocalization with mitochondria, CRABP-II was visualized using rabbit anti mouse secondary antibodies conjugated with Alexa Fluor 488 and mitochondria were visualized using MitoTracker (200 nm). ER was stained using antibodies against calreteculin and goat anti chicken secondary antibodies labeled with Alexa Fluor 633.
Image Analysis
Confocal fluorescence microscopy images were quantitatively analyzed with Image J. Measurements of fluorescence intensity per surface unit within and outside nuclei, were used to calculate ratios indicative of distribution of EGFP-CRABP-II between nuclei and cytoplasm.
Generation of COS-7 Cells Lines Stably Expressing EGFP-CRABP-II
Cells were transfected with plasmid harboring EGFP-CRABP-II, which encodes for neomycin (G418) resistance and allows for selection of transfected cells. 24 h following transfection, Geneticin (500 μg/ml) was added. Cells were selected for 7–14 days. Culture medium was replaced with fresh G418-containing DMEM supplemented with 10% FBS daily. The survival of EGFP-expressing cells was monitored with an inverted fluorescence microscope. Cells stably expressing the construct were pooled.
RESULTS
RA Stabilizes Interactions of CRABP-II with the Sumo E2 Ligase Ubc9 and Triggers Its SUMOylation
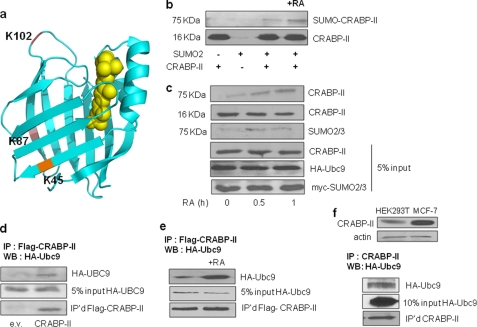
Examination of the primary sequence of CRABP-II using SUMOplot revealed three putative SUMOylation sites centered at K45, K87, and K102 of the protein (Fig. 1). Of these, K45 and K87 are located in the β sheets of the β-clam structure of the protein. K102 is located in the loop that flanks the entrance to the pocket, opposite the helix-loop-helix region of CRABP-II. To examine whether these residues comprise functional SUMOylation sites, bacterially expressed recombinant histidine-tagged CRABP-II (14) was used. The protein was subjected to in vitro SUMOylation using a SUMOylation kit containing SUMO2/3 (Active Motif). Proteins were resolved by SDS-PAGE and CRABP-II visualized by immunoblots (Fig. 1b). Subjecting CRABP-II to in vitro SUMOylation resulted in the appearance of additional band at 75 kDa, reflecting the generation of SUMOylated CRABP-II. Interestingly, SUMOylation was enhanced in the presence of RA. To examine whether CRABP-II is SUMOylated in cells, HEK293T cells were co-transfected with a vector encoding Flag-tagged CRABP-II and an expression vector harboring cDNA for Myc-tagged SUMO2. 24 h post-transfection, cells were placed in retinoid-depleted (charcoal-treated) media overnight and then treated with RA (1 μm) for 30 and 60 min. Cells were lysed in a buffer containing N-ethylmaleimide (NEM) and CRABP-II immunoprecipitated using anti-Flag M2-agarose beads. Precipitated proteins were resolved by SDS-PAGE and CRABP-II and SUMO2 were visualized by immunoblots (Fig. 1c). Similarly to the response of the purified protein in vitro, a higher molecular weight band, which could be immunostained with antibodies against either CRABP-II or SUMO2/3, was observed (Fig. 1c).
FIGURE 1.
CRABP-II associates with Ubc9 and is SUMOylated in response to RA. a, three-dimensional crystal structure of hCRABP-II (PDB ID: 2FR3) (17) drawn using Pymol. The putative SUMOylation residues are highlighted. b, recombinant bacterially expressed CRABP-II was subjected to in vitro SUMOylation in the absence or presence of RA (1 μm). Protein was resolved by SDS-PAGE and visualized by immunoblotting using anti-CRABP-II antibody. In addition to a band corresponding to the molecular mass of CRABP-II (∼16 kDa), a slower migrating band with an apparent molecular mass of 75 kDa was observed. c, HEK293T cells were co-transfected with plasmids encoding Flag-CRABP-II, HA-Ubc9, and Myc-SUMO2. 24 h post-transfection, cells were cultured in charcoal-treated medium overnight and treated with vehicle or RA (1 μm). Cells were lysed in a buffer containing 20 mm NEM. Proteins were immunoprecipitated using anti-Flag M2 antibodies immobilized on agarose beads, resolved by 15% SDS-PAGE, and visualized by immunoblotting using anti-CRABP-II or anti-SUMO2/3 antibodies. d, proteins in lysates of HEK293T cells ectopically expressing Flag-CRABP-II and HA-Ubc9 were precipitated using Flag M2 antibodies immobilized on agarose beads. Proteins were resolved and visualized by immunoblot using anti-HA antibodies. Precipitated CRABP-II was visualized using anti-Flag antibodies. e, HEK293T cells ectopically expressing Flag-CRABP-II and HA-Ubc9 were treated with vehicle or RA (1 μm, 1 h). Cell lysate proteins were immunoprecipitated using Flag M2 antibodies immobilized on agarose beads, resolved, and immunoblotted using anti-HA antibodies. Precipitated CRABP-II was visualized using anti-Flag antibodies. f, top: comparison of the expression levels of CRABP-II in HEK293T and MCF7 cells determined by immunoblots using anti-CRABP-II antibodies. Bottom: endogenously expressed CRABP-II from MCF-7 cells ectopically expressing HA-Ubc9. Proteins were resolved and visualized by immunoblotting using anti-HA antibodies. Precipitated CRABP-II was visualized using anti-CRABP-II antibodies.
To examine association between CRABP-II and the SUMO E2 ligase Ubc9 (15), Flag-CRABP-II and HA-tagged Ubc9 were ectopically overexpressed in HEK293T cells. 48 h post-transfection, cells were lysed, Flag-CRABP-II precipitated, and precipitated proteins resolved by SDS-PAGE. Immunoblots revealed that Ubc9 co-precipitated with CRABP-II (Fig. 1d) and that the interaction was enhanced in cells treated with RA (Fig. 1e). Similarly, Ubc9 co-precipitated with endogenous CRABP-II in MCF-7 cells, which express a high level of this protein (Fig. 1f).
Residue K102 Is Required for RA-induced Nuclear Translocation of CRABP-II
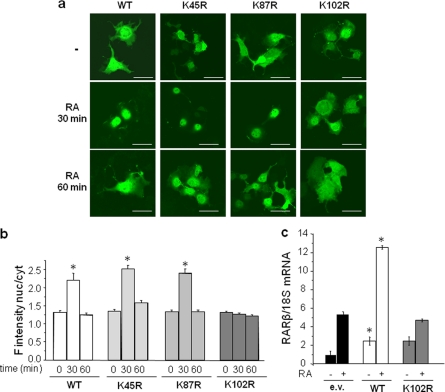
CRABP-II is predominantly cytosolic in the absence of a cognate ligand but, upon binding of RA, it translocates to the nucleus where it delivers the hormone to RAR, thereby activating the receptor to induce the transcription of RAR responsive genes. To examine whether CRABP-II SUMOylation is involved in the RA-triggered nuclear import of the protein and identify the residue(s) that mediates such an involvement, CRABP-II mutants lacking the putative SUMOylation sites were generated. Residues K45, K87, and K102 were individually replaced by arginines in the context of an expression vector encoding CRABP-II tagged with EGFP. EGFP-CRABP-II or the corresponding mutants were ectopically expressed in COS-7 cells. Cells were cultured in retinoid-depleted medium, treated with RA (1 μm) for 30 and 60 min and confocal fluorescence microscopy was used to visualize the proteins (Fig. 2, a and b). As often observed (e.g. Ref. 3, 16), overexpression of the EGFP-tagged proteins resulted in leakage into the nucleus even in the absence of ligand and thus the apo-protein was distributed across the cell. Nevertheless, similarly to the behavior of the endogenous protein (9), addition of RA induced massive mobilization of EGFP-CRABP-II to the nucleus, which was complete within ∼30 min post ligand treatment (Fig. 2, a and b). Interestingly, 60 min following addition of RA, EGFP-CRABP-II regained its pre-treatment localization. Similarly to the WT protein, the CRABP-II-K45R and CRABP-II-K87R mutants translocated to the nucleus within 30 min of RA treatment and redistributed to the cytosol 30 min later. In contrast, the CRABP-II-K102R mutant did not move to the nucleus in response to RA (Fig. 2, a and b).
FIGURE 2.
Mutation of K102 impairs the ability of CRABP-II to mobilize to the nucleus and enhance the activity of RAR. a, EGFP-tagged CRABP-II and corresponding K45R, K87R, and K102R mutant were ectopically expressed in COS-7 cells. Cells were treated with vehicle or RA as denoted and protein image using confocal fluorescence microscopy. Bar, 40 μm. b, quantitation of fluorescence intensity per surface unit in cytosol and nuclei imaged as in a. Ratios of fluorescence intensity in nuclei/cytosol are shown. Values are mean ± S.E., n = 40. *, p < 0.05 versus corresponding untreated cells. c, COS-7 cells were co-transfected with expression vectors for RARα and for Flag-CRABP-II or Flag-CRABP-II-K102R. 24 h post-transfection, cells were cultured in charcoal-treated media overnight and then treated with vehicle or RA (1 μm, 4 h). RNA was isolated, and the expression level of RARβ measured by Q-PCR. *, p < 0.05 versus corresponding e.v. controls.
The effect of the K102R mutation on the ability of CRABP-II to enhance the transcriptional activity of RAR was examined by monitoring the expression of the endogenous RAR target gene RARβ. COS-7 cells were co-transfected with expression vectors for RARα and CRABP-II or its K102R counterpart. Cells were treated with RA (1 μm, 4 h), and the expression level of RARβ mRNA measured by real-time quantitative PCR (Q-PCR). The data (Fig. 2c) show that, while WT-CRABP-II effectively enhanced the transcriptional activity of RAR, CRABP-II-K102R had no effect on receptor activity. It is important to note that it was previously established that mutation of K102 of CRABP-II does not alter the global folding of the protein or its affinity toward RA (11). Hence, K102 is specifically required for mobilization of CRABP-II to the nucleus, suggesting that SUMOylation of this residue is critical for the process.
The SUMO Ligase Ubc9 Delays the Exit of CRABP-II from the Nucleus and Enhances Its Ability to Activate RAR
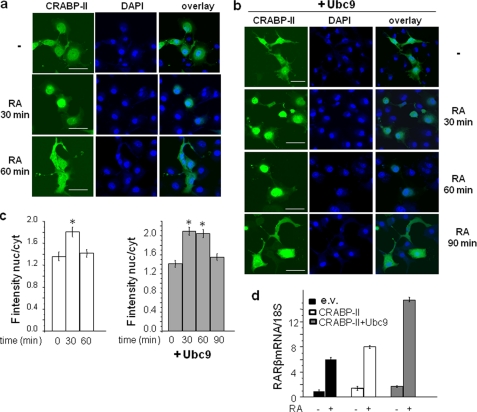
To examine whether SUMOylation is involved in regulating the nucleocytoplasmic shuttling of CRABP-II, the effect of expression of the SUMO conjugating enzyme Ubc9 on the nuclear localization of the protein was examined. COS-7 cells were transfected with EGFP-CRABP-II along with an empty vector or an expression vector encoding Ubc9. Cells were treated with RA and proteins visualized by confocal fluorescence microscopy (Fig. 3, a and b). While CRABP-II moved to the nucleus within 30 min of RA treatment and redistributed to the cytosol 30 min later (Fig. 3, a and c), overexpression of Ubc9 resulted in a longer residency of CRABP-II in the nucleus (Fig. 3, b and c). Ectopic expression of Ubc9 also augmented the ability of CRABP-II to enhance the transcriptional activity of RAR, reflected by induction of the RAR target gene RARβ (Fig. 3d). These observations may be understood to reflect that CRABP-II is deSUMOylated in the nucleus but becomes re-SUMOylated when in the cytosol. Increased expression of Ubc9 will potentiate an additional cycle of CRABP-II through the nucleus and, consequently, lengthen its apparent nuclear residence time and enhance its ability to activate RAR.
FIGURE 3.
Ubc9 delays the exit of CRABP-II from the nucleus and augments the ability of CRABP-II to enhance the activity of RAR. a, EGFP-CRABP-II was ectopically expressed in COS-7 cells. Cells were treated with vehicle or RA as denoted and CRABP-II imaged using confocal fluorescence microscopy. Bar, 40 μm. b, EGFP-CRABP-II and Ubc9 were ectopically co-expressed in COS-7 cells. Cells were treated with vehicle or RA as denoted and CRABP-II imaged using confocal fluorescence microscopy. c, quantitation of fluorescence intensity per surface unit in cytosol and nuclei imaged as in Fig. 2, a and b, measured using Image J. Ratios of fluorescence intensity in nuclei/cytosol are shown. Values are mean ± S.E., n = 40; *, p < 0.05 versus corresponding untreated cells. c, COS-7 cells were co-transfected with expression vectors for RARα and for Flag-CRABP-II or Flag-CRABP-II-K102R. 24 h post-transfection, cells were cultured in charcoal-treated media overnight and then treated with vehicle or RA (1 μm, 4 h). RNA was isolated, and the expression level of RARβ measured by Q-PCR.
The K102R Mutation Impairs Interactions with Ubc9 and SUMOylation of CRABP-II
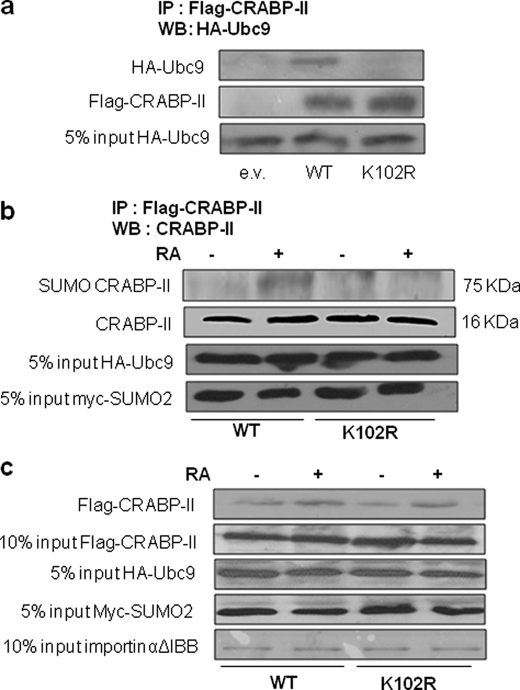
The importance of the K102 residue for CRABP-II SUMOylation was examined by monitoring the effect of mutating this residue on the ability of CRABP-II to interact with Ubc9. COS-7 cells were co-transfected with an expression plasmid for Flag-CRABP-II or its K102R counterpart and with a vector encoding HA-Ubc9. 48 h post-transfection, cells were lysed, and proteins immunoprecipitated using agarose-bead-conjugated anti-Flag antibodies. Precipitated proteins were resolved by SDS-PAGE and immunoblotted using HA antibodies. The data (Fig. 4a) show that the K102R mutation impaired the ability of CRABP-II to associate with Ubc9. To examine the effect of the K102R mutation on SUMOylation of CRABP-II, Flag-CRABP-II or its K102R mutant, HA-Ubc9, and Myc-tagged SUMO2 were ectopically co-expressed in HEK293T cells. 24 h post-transfection, cells were cultured in charcoal-treated medium overnight and then treated with RA (1 μm, 60 min). Cells were lysed, and Flag-tagged proteins precipitated. Proteins were resolved by SDS-PAGE and immunoblotted for CRABP-II. The data (Fig. 4b) show that the K102R mutation abolished the ability of RA to induce SUMOylation of CRABP-II. Hence, of the three putative SUMOylation sites of CRABP-II (Fig. 1a), K102 appears to be the sole site to be SUMOylated in response to RA.
FIGURE 4.
Mutation of K102 impairs the ability of CRABP-II to interact with Ubc9 and to undergo SUMOylation but not its RA-induced association with importin α. a, HEK293T cells were transfected with expression vectors for Flag-CRABP-II or Flag-CRABP-II-K102R and for HA-Ubc9. 48 h post-transfection, cells were lysed in a buffer containing 20 mm NEM and a protease inhibitor mixture. Proteins were immunoprecipitated using anti FlagM2-agarose beads, resolved by SDS-PAGE and visualized by immunoblots using antibodies against HA or Flag. b, HEK293T cells were co-transfected with expression vectors for Flag-CRABP-II or its K102R mutant, for HA-Ubc9, and for Myc-SUMO2. 24 h post-transfection, cells were cultured in charcoal-treated medium overnight and treated with vehicle or RA (1 μm, 1 h). Cells were lysed, and proteins immunoprecipitated using anti-Flag M2-agarose beads, resolved by SDS-PAGE, and visualized by immunoblots using antibodies against CRABP-II. c, HEK293T cells were co-transfected with expression vectors for Flag-CRABP-II or Flag CRABP-II-K102R and for HA-Ubc9 and Myc-SUMO2. 24 h post-transfection, cells were cultured in charcoal-treated medium for 16 h and then treated with vehicle or RA (1 μm, 60 min). Cells were lysed and proteins immunoprecipitated using anti-FlagM2-agarose beads. Flag-tagged proteins were eluted from the beads and incubated with bacterially expressed purified GST-importin αΔIBB immobilized on glutathione beads in the presence or absence of RA. Beads were washed band bead-associated proteins resolved by SDS-PAGE and immunoblotted using antibodies against CRABPII. Input controls for Ubc9 and SUMO2 were visualized by immunoblots using antibodies against HA and Myc, respectively. GST-importin αΔIBB was visualized by Coomassie Blue staining.
K102R Mutation of CRABP-II Does Not Abrogate Its Ability to Interact with Importin α in a RA-responsive Manner
RA triggers the nuclear translocation of CRABP-II by stabilizing its interactions with the nuclear import protein importin α (9). The inability of CRABP-II-K102R to mobilize to the nucleus thus raises the possibility that SUMOylation at this residue is required for the interactions of the protein with importin α. The effect of the mutation on the ability of CRABP-II to bind importin α was thus examined. GST-importin αΔIBB, lacking the importinβ-binding self-inhibitory domain, was expressed in Escherichia coli, purified and attached to glutathione beads. HEK293T cells were co-transfected with expression vectors for Flag-CRABP-II or Flag-CRABP-II-K102R and with plasmids encoding HA-Ubc9 and Myc-SUMO2. Cells were treated with RA (1 μm, 60 min) to allow for SUMOylation. Flag-CRABP-II was precipitated from cell lysates, eluted from the agarose beads, and incubated with immobilized GST-importin α in the presence or absence of RA. Bead-associated proteins were pelleted by centrifugation and resolved by SDS-PAGE, followed by immunoblotting with anti CRABP-II antibodies. The data (Fig. 4c) indicate that, similarly to CRABP-II, the K102R mutant associates with importin α and did so in a RA-sensitive manner. Hence, SUMOylation of CRABP-II-K102 is not necessary for the interactions of the protein with the nuclear import machinery.
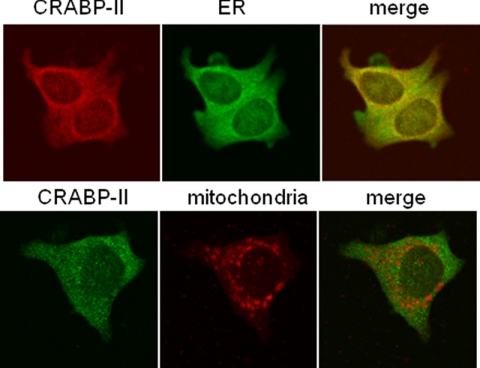
Apo-CRABP-II Is Associated with ER and Leaves This Location upon RA Binding
The K102R mutant is retained in the ER even in the presence of RA. Another possibility for understanding why SUMOylation is required for the nuclear mobilization of CRABP-II is that the apo-protein is associated with a particular extranuclear location, and that RA enables its dissociation from that niche by triggering SUMOylation. We thus set out to determine the subcellular localization of apo-CRABP-II. MCF-7 cells were cultured in charcoal-treated medium to deplete retinoid stores and confocal fluorescence microscopy was used to examine colocalization of endogenous apo-CRABP-II with the endoplasmic reticulum (ER) marker calreticulin or with the mitochondrial-specific dye MitoTracker. The observations (Fig. 5) indicate that apo-CRABP-II displayed marked co-localization with the ER marker. No significant colocalization with the mitochondrial marker was observed.
FIGURE 5.
Apo-CRABP-II endogenously expressed in MCF-7 cells is associated with ER. MCF-7 cells were fixed and immunolabeled for CRABP-II and the endoplasmic reticulum marker calreticulin. Mitochondria were visualized with Mitotracker (see “Experimental Procedures”). Images were obtained using confocal fluorescence microscopy. In the upper panels, yellow pixels in overlays of CRABP-II (red) with the ERE marker calreticulin (green) indicate colocalization.
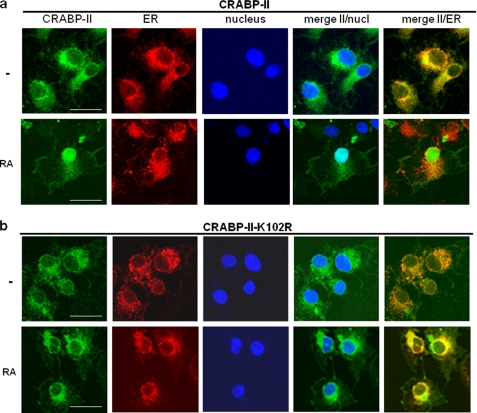
To enable mutagenesis analyses, COS-7 cells, which express a low level of CRABP-II, were used. Cells were transfected with expression constructs for EGFP-tagged CRABP-II or its K102R mutant. As the high expression level of EGFP-CRABP-II obtained by transient expression of the protein masks its exact localization (Figs. 2 and 3), COS-7 cell lines that stably express EGFP-CRABP-II and the corresponding K102R mutant were generated. Cells were stained with antibodies toward the ER marker calnexin or the mitochondrial marker succinate dehydrogenase (SDH, see “Experimental Procedures” for details). The markers and EGFP-CRABP-II were visualized by confocal fluorescence microscopy. The data (Fig. 6) indicate that, similarly to the localization of endogenous CRABP-II in MCF-7 cells, ectopically expressed apo-CRABP-II in COS-7 cells displayed marked co-localization with the ER marker. Upon treatment with RA (1 μm, 30 min), CRABP-II dissociated from the ER and mobilized to nuclei (Fig. 6). Similarly to the WT protein, CRABP-II-K102R co-localized with calnexin, indicating that it is associated with ER. However, unlike WT CRABP-II, the mutant was retained at the ER even in the presence of RA (Fig. 6). These observations thus suggest that RA-induced SUMOylation of CRABP-II at K102 is necessary for triggering the dissociation of the protein from the ER and thus for enabling its RA-induced nuclear translocation.
FIGURE 6.
Apo-CRABP-II ectopically expressed in COS-7 cells is associated with ER; CRABP-II but not its K102R mutant dissociates from ER in response to RA. COS-7 cells stably expressing EGFP-CRABP-II, or EGFP-CRABP-II-K102R were treated with vehicle or RA (1 μm, 30 min). Cells were fixed and immunolabeled for the endoplasmic reticulum marker calnexin. Nuclei were counterstained with DAPI. Images were obtained using confocal fluorescence microscopy (see “Experimental Procedures”). Images of CRABP-II (green), calnexin (red), and nuclei (blue) are shown separately and overlaid. Yellow pixels in overlays of green (EGFP-CRABP-II) and red (calnexin) fluorescence images indicate colocalization. Bar, 40 μm.
DISCUSSION
CRABP-II cooperates with the nuclear receptor RAR in mediating the transcriptional activity of their shared ligand, all-trans-RA. Apo-CRABP-II resides in the extranuclear milieu but, following binding of RA. We previously reported that the side chains of the CRABP-II residues that comprise the protein nuclear localization signal, K20, R29, and K30, undergo a conformational change upon ligand binding, enabling recognition by importin α, which mobilizes the protein to the nucleus (9). In the nucleus, CRABP-II directly delivers RA to RAR thereby markedly enhancing the transcriptional activity of the receptor (3, 4, 6–8, 10). The observations described here reveal that RA controls the nuclear translocation of CRABP-II not only by activating the protein NLS (9) but also by triggering SUMOylation at residue K102. The data demonstrate that RA stabilizes association of CRABP-II with the SUMO ligase Ubc9 and triggers SUMOylation of the protein both in vitro and in cultured cells (Fig. 1). The observations further show that RA-induced SUMOylation of CRABP-II specifically targets a single residue, K102 (Fig. 3a). SUMO2 was found to be more efficient than SUMO 1 or SUMO3 in modifying CRABP-II (data not shown). While the molecular weight (M.W.) of SUMO2 is ∼12 kDa, SUMOylated CRABP-II, whose M.W. is ∼15 kDa, migrated on SDS-PAGE gels with an apparent size of ∼75 kDa (Fig. 1). The observations thus suggest that, as often is the case (13), CRABP-II is modified by a SUMO polymer. The M.W. increase of ∼60 kDa further suggests that the modification results in attachment of a polymer containing 5 SUMO moieties.
Mutation of K102 abolished the ability of CRABP-II to mobilize to the nucleus in response to RA and to enhance the transcriptional activity of RAR (Figs. 2 and 4, a and b). Taken together with the observations that overexpression of Ubc9 prolonged the nuclear residence time of CRABP-II (Fig. 3), these data demonstrate that SUMOylation is critical for the RA-induced nuclear mobilization of CRABP-II as well as for CRABP-II-mediated delivery of RA to RAR. Interestingly, the SUMOylated residue, K102, is also an integral component of the RAR interaction domain of CRABP-II (11). It should be noted that it has been reported that bacterially expressed CRABP-II efficiently associates with bacterially expressed RAR (6), indicating that post-translational modifications are not necessary for the formation of CRABP-II-RAR complex. These considerations suggest that removal of the SUMO moieties from CRABP-II may be required for enabling the association of the binding protein with RAR. Overall, the observations suggest that, upon RA binding, CRABP-II is SUMOylated in the cytosol and that the modification is reversed in the nucleus, likely prior to interactions of the protein with RAR (Fig. 7).
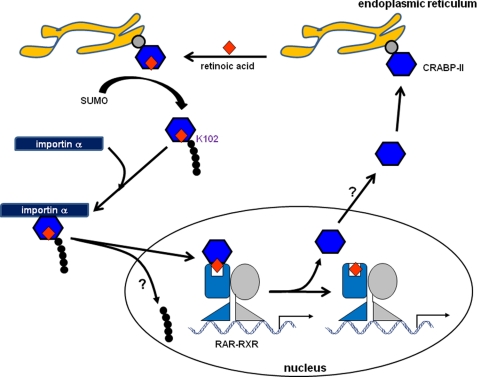
FIGURE 7.
A model for the cytoplasmic-nuclear cycle of CRABP-II. Apo-CRABP-II is associated with ER, likely through recruitment by a yet to be identified protein in the ER membrane. Binding of RA triggers SUMOylation of CRABP-II at residue K102, leading to its dissociation from ER. Free holo-CRABP-II is targeted by importin α which transports it to the nucleus. In the nucleus, CRABP-II directly binds to RAR to form a complex through which RA is channeled to the receptor. Following ligand transfer, the CRABP-II-RA complex rapidly dissociates, allowing the ligated receptor to regulate transcription. CRABP-II is then exported from the nucleus. The mechanisms that mediate the nuclear export of the protein are unknown but may involve intranuclear de-SUMOylation.
In search for the specific step in the in nuclear mobilization process that requires SUMOylation, the effect of mutating the SUMOylated residue K102 on the interactions of CRABP-II with importin α was examined. The data (Fig. 4c) indicated that the mutation did not impair the ability of CRABP-II to bind importin α or to do so in a RA-dependent manner. An alternative possibility is that SUMOylation is required to allow CRABP-II to dissociate from a subcellular compartment with which it is associated in the absence of RA. Co-localization experiments surprisingly revealed that apo-CRABP-II is associated with the endoplasmic reticulum (Figs. 5 and 6). As CRABP-II is a soluble protein and does not contain an identifiable ER localization signal, its association with the ER is likely mediated by binding to a protein(s) in the cytoplasmic surface of the ER. The identity of such a binding partner and the functional significance of its association with CRABP-II remain to be clarified. Regardless of the nature of the partner, the data demonstrate that its association with CRABP-II is negated upon binding of RA, and that the effect requires RA-induced SUMOylation of residue K102 (Fig. 6b). Hence, RA-controlled SUMOylation of CRABP-II releases CRABP-II from the ER, enabling its association with importin α and consequently, its nuclear mobilization (Fig. 7).
Whereas these and previous observations (9) delineated the molecular mechanisms that regulate the nuclear import of CRABP-II, the process that drives the export of the protein from the nucleus following delivery of RA to RAR remains poorly understood. The data revealed that RA-induced mobilization of CRABP-II to the nucleus is complete within ∼30 min, and that the protein exits the nucleus ∼30 min later (Figs. 2a and 3a). These observations suggest the existence of a clock that, following a set residence time in the nucleus, affects nuclear export of CRABP-II. The observations that overexpression of Ubc9 delays the protein's nuclear export (Fig. 3b) likely reflects a higher nuclear influx of CRABP-II upon enhancement of SUMOylation by the ligase. These observations support the possibility that the timed export process involves intranuclear reversal of the SUMO modification. It is also worth noting that, while a nuclear export signal (NES) was found to exist in other intracellular lipid-binding proteins, e.g. fatty acid-binding protein 4, CRABP-II lacks the Leu residues that comprise this NES (16). Nuclear export of CRABP-II may thus be mediated by another protein. The network of interactions of CRABP-II in the nucleus and the determinants of its transient stay in this location remain to be clarified.
Acknowledgments
We thank Daniel C. Berry for help in plotting the CRABP-II structure. We are very grateful to Cecile Rochette-Egly (IGBMC, Strasbourg) for CRABP-II antibodies, Hung-Ying Kao (Case Western Reserve University, Cleveland), Jun-Lin Guan (University of Michigan, Ann Arbor), and Karsten Weis (University of California, Berkeley) for plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK060684 (to N. N.) and by NIH Training Grant T32 CA059366 (to A. D. P.).
- RA
- retinoic acid
- CRABP
- cellular retinoic acid-binding protein
- PPAR
- peroxisome proliferator-activated receptor
- NEM
- N-ethylmaleimide
- NLS
- nuclear localization signal.
REFERENCES
- 1. Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. (2006) Pharmacol. Rev. 58, 712–725 [DOI] [PubMed] [Google Scholar]
- 2. Berry D. C., Noy N. (2009) Mol. Cell. Biol. 29, 3286–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schug T. T., Berry D. C., Shaw N. S., Travis S. N., Noy N. (2007) Cell 129, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schug T. T., Berry D. C., Toshkov I. A., Cheng L., Nikitin A. Y., Noy N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaw N., Elholm M., Noy N. (2003) J. Biol. Chem. 278, 41589–41592 [DOI] [PubMed] [Google Scholar]
- 6. Dong D., Ruuska S. E., Levinthal D. J., Noy N. (1999) J. Biol. Chem. 274, 23695–23698 [DOI] [PubMed] [Google Scholar]
- 7. Budhu A. S., Noy N. (2002) Mol. Cell. Biol. 22, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manor D., Shmidt E. N., Budhu A., Flesken-Nikitin A., Zgola M., Page R., Nikitin A. Y., Noy N. (2003) Cancer Res. 63, 4426–4433 [PubMed] [Google Scholar]
- 9. Sessler R. J., Noy N. (2005) Mol. Cell. 18, 343–353 [DOI] [PubMed] [Google Scholar]
- 10. Tan N. S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. (2002) Mol. Cell. Biol. 22, 5114–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budhu A., Gillilan R., Noy N. (2001) J. Mol. Biol. 305, 939–949 [DOI] [PubMed] [Google Scholar]
- 12. Schwartz D. C., Hochstrasser M. (2003) Trends Biochem. Sci 28, 321–328 [DOI] [PubMed] [Google Scholar]
- 13. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 14. Donato L. J., Noy N. (2006) Anal. Biochem. 357, 249–256 [DOI] [PubMed] [Google Scholar]
- 15. Müller S., Hoege C., Pyrowolakis G., Jentsch S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202–210 [DOI] [PubMed] [Google Scholar]
- 16. Ayers S. D., Nedrow K. L., Gillilan R. E., Noy N. (2007) Biochemistry 46, 6744–6752 [DOI] [PubMed] [Google Scholar]
- 17. Vaezeslami S., Mathes E., Vasileiou C., Borhan B., Geiger J. H. (2006) J. Mol. Biol. 363, 687–701 [DOI] [PubMed] [Google Scholar]