Background: Zcchc11 is a uridyltransferase enzyme whose biological functions are poorly understood.
Results: Knockdown of Zcchc11 arrested cells in G1. Conversely, overexpression of full-length, catalytically inactive, or N-terminal Zcchc11 promoted entry into S phase.
Conclusion: Zcchc11 acts through a uridyltransferase-independent mechanism to promote cell cycle progression.
Significance: The N-terminal uridyltransferase-null half of Zcchc11 is biologically active and is a novel regulator of proliferation.
Keywords: Cyclin-dependent Kinase (CDK), Cell Cycle, Cyclins, Retinoblastoma (Rb), RNA-binding Protein, Zcchc11, Uridyltransferase
Abstract
Zcchc11 is a uridyltransferase protein with enzymatic activity directed against diverse RNA species. On the basis of its known uridylation targets, we hypothesized that Zcchc11 might regulate cell proliferation. Confirming this, loss-of-function and complementary gain-of-function experiments consistently revealed that Zcchc11 promotes the transition from G1 to S phase of the cell cycle. This activity takes place through both Rb-dependent and Rb-independent mechanisms by promoting the expression of multiple G1-associated proteins, including cyclins D1 and A and CDK4. Surprisingly, a Zcchc11 construct with point mutations inactivating the uridyltransferase domain enhanced cell proliferation as effectively as wild-type Zcchc11. Furthermore, truncated mutant constructs revealed that the cell cycle effects of Zcchc11 were driven by the N-terminal region of the protein that lacks the RNA-binding domains and uridyltransferase activity of the full protein. Therefore, the N-terminal portion of Zcchc11, which lacks nucleotidyltransferase capabilities, is biologically active and mediates a previously unrecognized role for Zcchc11 in facilitating cell proliferation.
Introduction
Zcchc11 (zinc finger CCHC domain-containing 11; also called TUT4 (terminal uridyltransferase 4)) is an RNA-binding protein implicated in the post-transcriptional regulation of diverse RNA species (1–3). On the basis of two domains homologous to the catalytic core of polymerase β, Zcchc11 has been classified as belonging to a family of non-canonical nucleotidyltransferases characterized by the yeast enzyme Cid1 (caffeine-induced death 1) (4). By homology, Zcchc11 contains two nucleotidyltransferase domains; the more N-terminal of these domains is catalytically inactive, whereas the C-terminal region is an active uridyltransferase enzyme capable of adding free uridine residues to RNA molecules (1, 3, 5, 6). Outside of these domains, Zcchc11 bears little sequence homology to other proteins. Therefore, the functions of Zcchc11 are poorly understood at present but have been inferred from its enzymatic activity. Cid1 has been tied to cell cycle progression in both yeast and Caenorhabditis elegans (7–9). Therefore, one likely role for Zcchc11 is the promotion of cell proliferation.
Further supporting this hypothesis, many RNA targets of Zcchc11 uridylation have important proliferative roles. We found previously that Zcchc11 uridylates microRNA-26a, inhibiting its silencing action and thereby increasing expression of microRNA-26a target genes, including interleukin-6 (1). MicroRNA-26a also targets cell cycle proteins, inhibits cell proliferation, and is decreased in multiple cancers (10, 11). In mouse embryonic stem cells, Zcchc11 uridylates the precursor form of let-7 microRNA family members, which inhibits their processing into mature microRNAs, thereby decreasing let-7 content (3, 5). let-7 family members target multiple cyclins as well as oncogenes and can function as tumor suppressors (2, 12, 13), again implicating Zcchc11 as a potential mediator of the cell cycle. Finally, Zcchc11 can uridylate histone mRNAs (14), contributing to the decreased histone content following completion of DNA replication (15). Given these known uridylation targets and the confirmed function of Cid1, we hypothesized that Zcchc11 may play critical roles in cell proliferation driven by its nucleotidyltransferase activity.
EXPERIMENTAL PROCEDURES
Cell Lines
All cells were passaged in American Type Culture Collection-recommended medium supplemented with 10% FBS and 1× penicillin/streptomycin. H2009 cells were maintained in DMEM/F-12 medium supplemented with 0.005 mg/ml insulin, 0.01 mg/ml transferrin, 30 nm sodium selenite, 10 nm hydrocortisone, 10 nm β-estradiol, and 4.5 mm l-glutamine (16). Human umbilical vein endothelial cells were maintained in endothelial cell growth medium (Lonza). SF-295 cells were obtained from the NCI-Frederick Cancer Tumor/Cell Line Repository; they were maintained in RPMI 1640 medium supplemented with 2 mm l-glutamine. Where indicated, cells were arrested in G2 by treatment with 100 ng/ml nocodazole 24 h prior to collection.
Transfection
Cells were plated at a density sufficient to remain subconfluent throughout the experiment. siRNA delivery was achieved using 4–8 μl of DharmaFECT 1 transfection reagent/ml of medium. Cells were treated with 100 nm non-targeting siCONTROL siRNA 1 (Dharmacon) or Zcchc11-targeting siRNA (5′-UAU CUG UAC AUG UCU GUU GUU-3′) for 24 h before receiving fresh complete medium for another 24–48 h. Two alternative siRNAs were also used target to Zcchc11: Z2, 5′-GGA UUU GGA UUU CGU GAU A-3′; and Z3, 5′-UGU CAU AUG UGU ACG GAU A-3′.
Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen). The creation of GFP-tagged, FLAG-tagged, and DADA mutant (with two aspartic acids mutated to alanines) Zcchc11 constructs has been described previously (1). N- and C-terminal Zcchc11 constructs were PCR-amplified from the original Zcchc11-FLAG plasmid and ligated back into the pEGFP vector (Clontech) between the BsrGI and NotI restriction sites.
Senescence-associated β-Galactosidase (SABG)2 Staining
Cells were plated at very low concentration. 48 h post-transfection, they were fixed and stained using an SABG assay kit (Cell Signaling Technology). For quantification, a minimum of 200 cells/well were counted by individuals blinded to the treatment groups.
Flow Cytometry
Annexin V/7-aminoactinomycin D staining was performed using the BD Pharmingen annexin V/phycoerythrin apoptosis detection kit I. As a positive control for apoptosis, non-transfected cells were treated with 10 μm camptothecin for 24 h. Cell cycle analyses were performed by sequentially treating cells with the following: 1) 4 mm sodium citrate, 0.01% Triton X-100, 0.1 mg/ml RNase A, and 0.5 mg/ml propidium iodide (PI) for 10 min at room temperature and 2) 400 mm NaCl, 0.01% Triton X-100, and 0.5 mg/ml PI for at least 30 min at 4 °C. Cell cycle estimates were performed using an unconstrained Dean-Jett-Fox algorithm from the FlowJo software package (TreeStar Inc.).
Immunofluorescence
Cells were fixed in 10% formaldehyde, permeabilized with methanol, and stained overnight with mouse anti-Ki67 antibody (proliferating cell nuclear antigen; Cell Signaling Technology) followed by anti-mouse IgG conjugated to Alexa Fluor 488 (Cell Signaling Technology) as recommended by the manufacturer. Quantifications were performed for at least 100 cells from each field using overlays of phase-contrast and immunofluorescence images of the same field by researchers blinded to treatment group.
Immunoblotting
For all immunoblotting experiments, 20–50 μg of whole cell lysate was run through 4–12% bis-tris/SDS- or 3–8% Tris acetate/SDS-polyacrylamide gels and transferred onto Immobilon P membranes using the NuPAGE protein blotting system (Invitrogen). All antibodies were obtained from Cell Signaling Technology except goat anti-Zcchc11 (ProSci Inc.), rabbit anti-goat (R&D Biosystems), and goat anti-p21 and mouse anti-cyclin A (Santa Cruz Biotechnology). The anti-phospho-Rb (retinoblasoma protein) antibody was specific to phosphorylation at Ser-807 and Ser-811 (Cell Signaling Technology). Immunoblots were visualized using the ECL Plus chemiluminescence system (Amersham Biosciences).
Northern Blotting
RNA was extracted with phenol/chloroform using TRIzol (Invitrogen). 15 μg of total RNA/well was run on a urea-15% polyacrylamide gel and transferred onto a BrightStar-Plus nylon membrane (Ambion). T4 polynucleotide kinase (New England Biolabs) was used to 32P label the following antisense oligonucleotide probes: U6, 5-GCC ATG CTA ATC TTC TCT GTA TC-3′; and Let-7a, 5′-AAC TAT ACA ACC TAC TAC CTC A-3′.
RT-PCR
RNA was extracted as described above, and RT-PCR was performed using 10 ng of total RNA with a TaqMan RNA-to-CT kit (Applied Biosystems) using the following primers and probes: histone H3, 5′-GCT TGT GAG GCC TAC TTG GTA-3′ (forward primer), 5′-GAG CTG GAT GTC TTT GGG CAT A-3′ (reverse primer), and 5′-AGC ATG GAT GGC GCA AAG GTT TGT GT-3′ (probe); and 18S ribosomal RNA, 5′-ATTCGAACGTCTGCCCTATCA-3′ (forward primer), 5′-GTCACCCGTGGTCACCATG-3′ (reverse primer), and 5′-TCG ATG GTA GTC GCC GTG CCT ACC-3′ (probe).
RESULTS
Zcchc11 Loss of Function Decreases Cell Proliferation
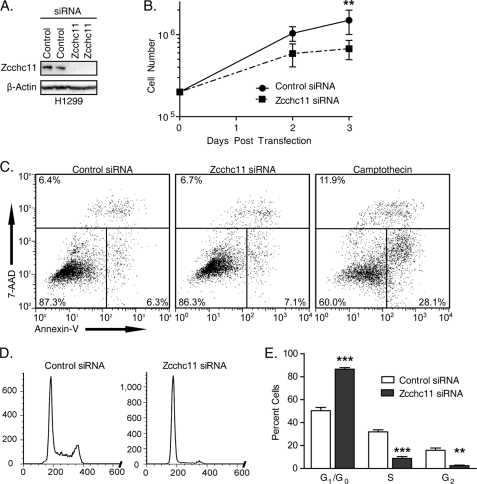
To assess the importance of Zcchc11 in proliferation, we treated the H1299 lung epithelial cell line with Zcchc11-targeting or non-targeting control siRNA and quantified cell numbers over time. Transfection with targeting siRNA reduced Zcchc11 expression to levels nearly undetectable by immunoblotting (Fig. 1A). After 72 h, there were an average of 1.5 × 106 cells/control well compared with only 6.7 × 105 cells/well treated with Zcchc11-targeting siRNA (Fig. 1B). To rule out apoptosis as a cause for this decrease, cells were stained with annexin V and 7-aminoactinomycin D. Similar percentages of control (87%) and Zcchc11 knockdown (86%) cells were viable as indicated by staining negative for both markers (Fig. 1C). Analogous results were observed using the A549 lung epithelial cell line (supplemental Fig. 1).
FIGURE 1.
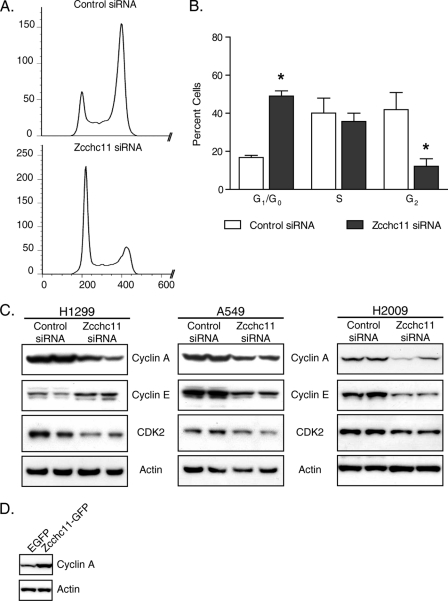
Zcchc11 knockdown inhibits cell proliferation in lung epithelial lines. A, immunoblot of cell lysates 48 h after transfection with Zcchc11-targeting or control siRNA. B, cell counts of H1299 cells transfected with the indicated siRNAs. Error bars represent S.E. from three experiments. **, p < 0.01 by matched-pair two-way analysis of variance with the Bonferroni post hoc analysis. C, staining with annexin V and 7-aminoactinomycin D (7-AAD) following treatment with control or Zcchc11-targeting siRNA or with camptothecin (positive control). Numbers indicate the percentage of cells in each section. D, genomic DNA content, as measured by PI staining, of H1299 cells following siRNA treatment. E, estimates of the percentage of H1299 cells in each phase of the cell cycle measured by PI staining. Error bars represent S.E. from six experiments. **, p < 0.01; ***, p < 0.001.
These results support the hypothesis that Zcchc11 enhances proliferation. To assess the impact of Zcchc11 knockdown on the cell cycle, adherent cells were collected 48 h post-transfection, and DNA was stained with PI. Flow cytometric analysis revealed that ∼50% of treated control cells were in G1 stage, with an additional 32% in S phase and 16% in G2 (Fig. 1, D and E). Treatment with Zcchc11-targeting siRNA induced an accumulation of 87% of cells in G1, leaving just 9 and 3% in S phase and G2, respectively (Fig. 1E). Two siRNAs complementary to different regions of the Zcchc11 transcript also induced an accumulation of cells in G1 (supplemental Fig. 2), arguing against off-target effects of the siRNA. Similar inhibition was observed in A549 cells following Zcchc11 knockdown (supplemental Fig. 3).
Zcchc11 Gain of Function Increases Cell Proliferation
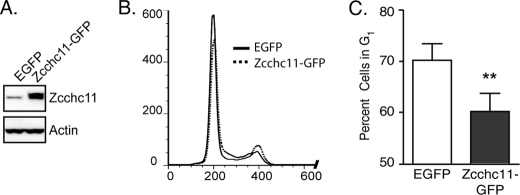
The above results indicated that Zcchc11 expression is necessary for maintaining the base-line proliferation rates of lung epithelial cells in vitro. To test whether Zcchc11 is sufficient to increase G1 progression, H1299 cells were transfected with plasmids encoding either enhanced green fluorescent protein (EGFP) or mouse Zcchc11 bearing a C-terminal GFP tag (Zcchc11-GFP). 24 h after transfection, Zcchc11-GFP appreciably increased the amount of enzyme detected by immunoblotting using an anti-Zcchc11 antibody (Fig. 2A) and produced a significant decrease in the percentage of cells in G1 (Fig. 2, B and C), indicating enhanced passage into S phase.
FIGURE 2.
Overexpression of Zcchc11 promotes G1 exit in lung epithelial cells. A, representative immunoblots for Zcchc11 and actin in cell lysates from H1299 cells following transfection with plasmids encoding either EGFP or Zcchc11-GFP. B, overlay of PI staining of H1299 cells transfected with the indicated plasmids. C, estimates of the percentage of cells in G1 following treatment with the same plasmids. Error bars indicate S.E. from four experiments. **, p < 0.01 by Student's matched-pair t test.
Decreased Replication Induced by Zcchc11 Is the Result of a Lower Proliferation Rate
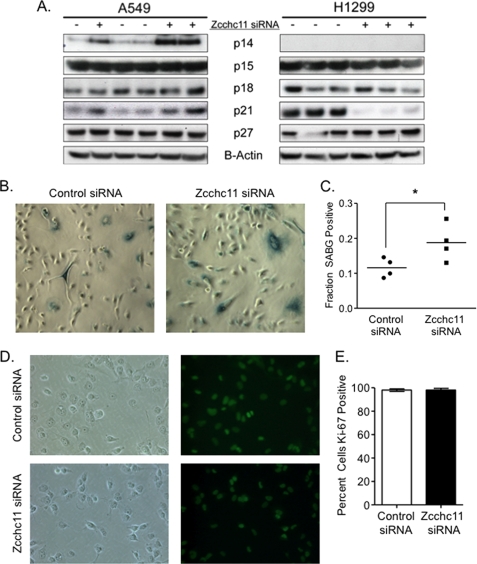
Slower proliferation may be caused by decreased passage through G1, by temporary arrest in G0 (quiescence), or by permanent replicative arrest (senescence). Whether temporary or permanent, replicative arrest is initiated by induction of one or more inhibitory proteins (17). Immunoblots of the more common of these factors, including p21CIP, showed no consistent changes across cell lines following Zcchc11 knockdown (Fig. 3A), arguing that these cells are not consistently undergoing replicative arrest.
FIGURE 3.
Knockdown of Zcchc11 delays passage through G1 arrest but does not induce replicative arrest. A, immunoblotting of whole cell lysates from H1299 and A549 cells treated with control or Zcchc11-targeting siRNA was used to assess the impact of Zcchc11 expression on the levels of common G1 inhibitors. p16INK4 was not detectable in either cell line. B, SABG staining of A549 cells following treatment with control or Zcchc11-targeting siRNA. C, quantification of the percentage of A549 cells staining SABG-positive. *, p < 0.05 by Student's matched-pair t test (n = 3). D, immunofluorescence of Ki67 expression in H1299 cells following siRNA treatment. E, quantification of the percentage of cells staining positive for Ki67 expression in H1299 cells (as in D). Error bars represent S.E. from three separate experiments.
To confirm that Zcchc11 knockdown does not induce senescence, H1299 and A549 cells were stained for SABG activity 48 h after transfection with control or Zcchc11-targeting siRNA. In H1299 cells, we observed no detectable SABG activity in cells treated with either control or Zcchc11-targeting siRNA (data not shown). However, in A549 cells, we found a basal SABG activity in ∼10% of control cells (Fig. 3B) and a small but significant increase in SABG activity following knockdown of Zcchc11 (Fig. 3C). These data match the immunoblotting results that showed p21 induction in A549 but not H1299 cells (Fig. 3A). Although the A549 results suggest that Zcchc11 is capable of furthering senescence in a setting in which it already occurs, the immunoblotting and SABG results with H1299 cells demonstrate that changes in senescence are not an essential feature of the influence of Zcchc11 on the cell cycle.
The resting G0 state is characterized by a loss of Ki67 staining. Immunofluorescence staining revealed that virtually all H1299 cells were cycling in these cultures, and Zcchc11 siRNA did not induce a subpopulation of cells to become negative for Ki67 (Fig. 3, D and E). Taken together, these data indicate that Zcchc11 knockdown causes cells to be blocked in G1 without entering a state of arrest in G0.
Rb-dependent and Rb-independent Regulation of G1 Exit by Zcchc11
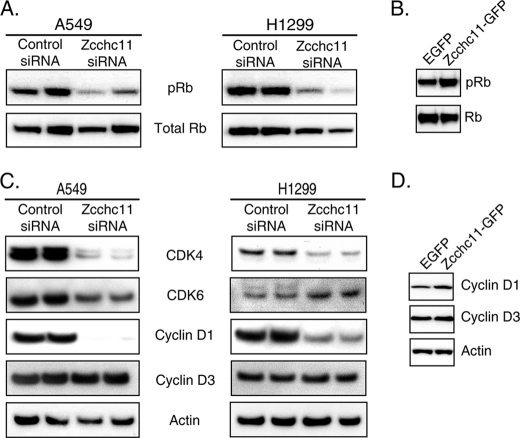
The Rb protein is a central regulator of S phase entry, with the inactivation of Rb by phosphorylation critical to G1 exit (18). To determine whether Zcchc11 levels modulate Rb phosphorylation, cell lysates from H1299 and A549 cells treated with control or Zcchc11-targeting siRNA were analyzed by immunoblotting. In both cell lines, Rb phosphorylation was decreased following Zcchc11 knockdown (Fig. 4A). Complementing these results, the levels of phosphorylated (but not total) Rb were increased following transfection with Zcchc11-GFP (Fig. 4B).
FIGURE 4.
Phosphorylation of Rb is regulated by Zcchc11 in lung epithelial cells. A and C, immunoblots of whole cell lysates from H1299 and A549 cells 48 h after transfection with Zcchc11-targeting or control siRNA. B and D, immunoblots from H1299 cells 24 h after transfection with plasmids encoding either EGFP or mouse Zcchc11-GFP.
In the earliest stages of G1-to-S phase progression, Rb is phosphorylated by heterodimers between D-type cyclins and cyclin-dependent kinase CDK4 or CDK6 (18). Further immunoblotting was used to determine whether Zcchc11 influences these early G1 proteins. As shown in Fig. 3C, the quantities of cyclin D1 and CDK4 were markedly decreased by Zcchc11 siRNA, whereas other cell cycle proteins such as cyclin D3 remained unchanged. Conversely, Zcchc11 overexpression induced higher levels of cyclin D1 but not cyclin D3 (Fig. 4D). These complementary gain- and loss-of-function approaches reveal a consistent role of Zcchc11 in enhancing the signaling pathways upstream of Rb phosphorylation.
Unlike D-type cyclins and their corresponding cyclin-dependent kinases, which primarily regulate proliferation by phosphorylating Rb, heterodimers between cyclin A or E and CDK2 are critical regulators at later stages of G1 (18). These complexes regulate S phase entry downstream of or in the absence of Rb (19, 20). To determine whether the effects of Zcchc11 on Rb phosphorylation are solely responsible for the influence of Zcchc11 on the cell cycle, we manipulated Zcchc11 levels in the H2009 lung epithelial cell line, which lacks Rb (21). Initial PI staining experiments showed small differences in the cell cycle profile after Zcchc11 knockdown. To improve the sensitivity of these analyses, cells were treated with nocodazole, which arrests proliferating cells in G2 (22). Using this technique, only 17% of cells remained in G1 48 h after transfection with control siRNA (Fig. 5, A and B). In contrast, nearly 3-fold more cells (49%) remained in G1 when Zcchc11 levels were decreased by siRNA (Fig. 5B). Immunoblots from all three cell lines showed that cyclin A content was consistently dependent on Zcchc11 expression (Fig. 5C). As before, Zcchc11-GFP overexpression had effects complementary to Zcchc11 knockdown and increased cyclin A expression (Fig. 5D).
FIGURE 5.
Rb-independent regulation of passage through the G1 checkpoint by Zcchc11. A, genomic DNA content of H2009 cells following treatment with control or Zcchc11-targeting siRNA and nocodazole to arrest proliferating cells in G2. B, estimated percentage of H2009 cells in each stage of the cell cycle following siRNA and nocodazole treatment as in A. Error bars represent S.E. from three experiments. *, p < 0.05 by two-way analysis of variance with the Bonferroni post hoc test. C, immunoblots of late G1 proteins from H1299, A549, and H2009 cells treated with control or Zcchc11-targeting siRNA. D, representative immunoblot of H1299 cells transfected with either EGFP or Zcchc11-GFP.
Effects of Zcchc11 on Proliferation Are Independent of Uridyltransferase Activity
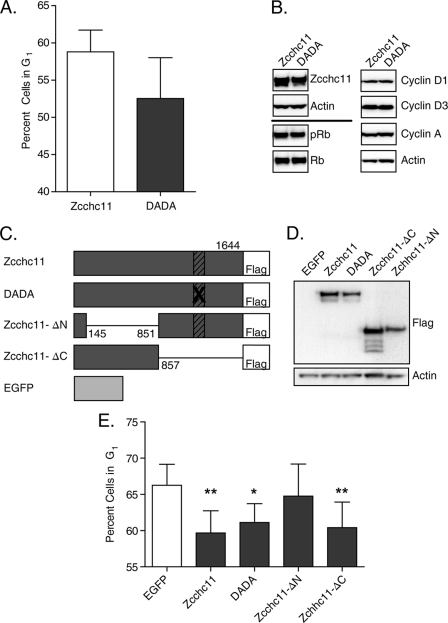
The above studies support a role for Zcchc11 in enhancing cell proliferation, a hypothesis that stemmed from its known functions as a nucleotidyltransferase enzyme (1, 3, 5, 14). The mutation of two aspartic acids to alanines (DADA) in the C-terminal nucleotidyltransferase domain of Zcchc11 is sufficient to ablate its uridyltransferase activity (1, 3, 5). To test whether its nucleotidyltransferase activity dictates the effects of Zcchc11 on cell proliferation, we transfected cells with plasmids encoding either WT mouse Zcchc11 or the DADA mutant form of the protein to compare the effects of each construct on the cell cycle. Surprisingly, transfection of H1299 cells with the DADA mutant did not increase the percentage of cells in G1 compared with wild-type Zcchc11 (Fig. 6A), as would have been expected if the nucleotidyltransferase activity of Zcchc11 were responsible for effects on cell proliferation. Similarly, immunoblots from these cells showed no difference in Rb phosphorylation or in expression of either cyclin A or D1 (Fig. 6B).
FIGURE 6.
Zcchc11-mediated regulation of the cell cycle is independent of its uridyltransferase activity. A, the percentage of H1299 cells in G1 following transfection with either wild-type or DADA mutant Zcchc11 was estimated by staining genomic DNA with PI. Error bars represent S.E. from four experiments. There was no significant difference between groups by Student's matched-pair t test. B, representative immunoblots of H1299 cell lysates following transfection with FLAG-tagged wild-type or DADA mutant Zcchc11 plasmids. C, schematic representation of the mutant Zcchc11 constructs used to assess the impact of specific regions on G1 exit. Hatched boxes indicate the uridyltransferase domain. D, immunoblot of H1299 cells transfected with the indicated mutant Zcchc11 proteins. E, estimated percentage of H1299 cells in G1 following transfection with the indicated plasmids. Error bars represent S.E. from four experiments. *, p < 0.05; **, p < 0.01 by one-way matched-pair analysis of variance with a Bonferroni correction for multiple comparisons.
These results suggested the unexpected conclusion that Zcchc11-mediated regulation of proliferation is independent of its uridyltransferase activity. Consistent with this idea, two previously documented targets of Zcchc11 uridyltransferase activity were interrogated, neither of which was changed as predicted by extrapolation from how Zcchc11 influences these substrates in other settings. Northern blots for Let-7a, which increases in embryonic stem cells after Zcchc11 knockdown (3, 5), showed no change due to knockdown of Zcchc11 in H1299 cells (supplemental Fig. 4). Similarly, histone H3 mRNA, which can be destabilized by Zcchc11-mediated uridylation in HEK293T or HeLa cells (14), was strongly decreased rather than increased following Zcchc11 knockdown in H1299 cells (supplemental Fig. 5). This change is likely downstream of the cell cycle activity of Zcchc11 activity. Taken together, these results indicate that Zcchc11 can impact the cell cycle independent of its uridylation activity or its previously implicated biological effects.
To determine the region of the protein responsible for the cell cycle regulatory functions of Zcchc11, H1299 cells were transfected with plasmids encoding EGFP, Zcchc11, DADA mutant Zcchc11, or one of two truncated Zcchc11 mutants: Zcchc11-ΔN (missing amino acids 145–851) and Zcchc11-ΔC (lacking the last 787 amino acids, including its active uridyltransferase domain) (Fig. 6C). Immunoblotting showed equal expression of all four Zcchc11 constructs (Fig. 6D). As predicted from the previous studies, transfection with either the wild-type or DADA mutant form of Zcchc11 resulted in significant decreases in the fraction of cells in G1 compared with the control EGFP-transfected cells (Fig. 6E). In contrast to the full-length proteins, overexpression of Zcchc11-ΔN, which contained the nucleotidyltransferase domain, was insufficient to alter the fraction of cells in G1 (Fig. 6E). However, overexpression of Zcchc11-ΔC, which contained only the N-terminal region, conveyed a change in the cell cycle profile equal in magnitude to that of the full-length protein (Fig. 6E).
Zcchc11 Controls Cell Proliferation in Diverse Cell Types
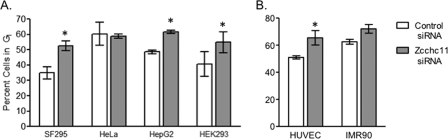
The above data strongly indicate that Zcchc11 contributes to cell proliferation in the limited setting of non-small cell lung carcinoma cells. We have observed Zcchc11 protein expression in a large number of cultured cell lines (data not shown), even in the absence of its expression in corresponding tissues (1). We therefore hypothesized that Zcchc11 may contribute to proliferation in multiple lines of cultured cells. To test this hypothesis, Zcchc11-targeting siRNA was used to knockdown Zcchc11 expression in the following cell lines: HEK293 embryonic kidney cells, HepG2 hepatocellular carcinoma cells, SF-295 neuroblastoma cells, and HeLa cervical carcinoma cells. In three of these cell lines, Zcchc11 knockdown induced a significant accumulation in G1 (Fig. 7A). Conversely, HeLa cells proved insensitive to treatment with Zcchc11 siRNA (Fig. 7A), although immunoblotting revealed substantial knockdown of Zcchc11 expression in these cells (data not shown). These data indicate that Zcchc11 can promote proliferation in multiple different cell lines.
FIGURE 7.
Modulation of Zcchc11 affects cell cycle progression in diverse cell types. The percentage of cells remaining in G1 following Zcchc11 knockdown with siRNA was estimated for a diverse set of immortalized cell lines (A) and normal cells (B). *, p < 0.05 versus treated control cells by two-way analysis of variance with Bonferroni adjustment for multiple comparisons. Data shown for each cell type are representative of three separate experiments.
The experiments were all performed using immortalized cell lines representing, in some way, cancerous cells. To further assess the impact of Zcchc11 expression in more normal cells, we used two cell models of normal cell proliferation in vitro: human umbilical vein endothelial cells and IMR-90 lung fibroblasts. As before, inhibition of Zcchc11 resulted in a significant accumulation of cells in G1 for the human umbilical vein endothelial cells and a similar trend in the IMR-90 cells (Fig. 7B). Thus, we conclude that Zcchc11 expression contributes to proliferation in a diverse set of both normal and cancerous cell types.
DISCUSSION
Using complementary knockdown and overexpression studies, we have identified a novel role for Zcchc11 in regulating the G1-to-S phase transition of the cell cycle. The knockdown results indicate that Zcchc11 is necessary for optimal proliferation in vitro, whereas the overexpression data indicate that Zcchc11 is sufficient to increase proliferation in an already rapidly cycling population of cells. Detectable induction of S phase entry by transient transfections is remarkable, as the estimated transfection efficiency was only ∼67% (data not shown). Therefore, it is likely that these observations understate the impact of Zcchc11 on the cell cycle.
Protein analysis by immunoblotting suggests that Zcchc11 expression contributes to certain early and late G1-associated proteins but not others. Cyclins D1 and A and CDK4 are particularly sensitive to modulation of Zcchc11 levels. The H2009 cell line is sensitive to Zcchc11 knockdown, demonstrating that Zcchc11 is capable of promoting proliferation independent of Rb activity, at least in part by facilitating cyclin A expression. However, in Rb-competent cells, the observed changes in Rb phosphorylation and in the expression of cyclin D1 and CDK4 indicate that Rb-dependent proliferation is also enhanced by Zcchc11. This conclusion is further supported by the smaller magnitude of arrest in H2009 cells compared with the Rb-competent H1299 and A549 cells. We conclude that Zcchc11 promotes proliferation by influencing multiple cell cycle regulatory proteins capable of acting both upstream and downstream of Rb.
Overexpression of mutant Zcchc11 constructs indicated that the cell cycle regulatory function of Zcchc11 is independent of its uridyltransferase activity and is instead located in the N-terminal portion of the protein. This is only the second reported function for the N-terminal region of Zcchc11. A prior study suggested that this portion of Zcchc11 physically interacts with the protein TIFA in the TLR4 complex elicited by LPS stimulation, interfering with the signaling pathway leading to NF-κB activation (23). It is unlikely that TIFA, TLR4 signaling, or NF-κB activity is related to the cell cycle effects observed here. Nothing further is known about the structure-function of this region of Zcchc11. Bioinformatic modeling using the conserved domain search tool from NCBI (24, 25) and the InterProScan algorithm from SWISS-MODEL (26, 27) revealed only two recognized domains in this region other than the nonfunctional nucleotidyltransferase domain and its associated poly(A) polymerase region: a single C2H2 zinc finger (28) and a ribosome-like RNA-binding domain (5). The paucity of known biologically relevant domains suggests that Zcchc11 accelerates the cell cycle through a novel mechanistic pathway, yet to be determined and an important focus for further studies.
Zcchc11 was originally identified as a cytoplasmic nucleotidyltransferase on the basis of sequence homology to the protein Cid1 found in fission yeast (4). Like Zcchc11, Cid1 is a cytoplasmic uridyltransferase (6, 29) that contributes to cell proliferation (7–9). However, substantial differences do exist. Here, we have shown that Zcchc11 promotes G1-to-S phase progression, whereas both the yeast and C. elegans Cid1 proteins act primarily in stressed cells to control the G2-to-M checkpoint (7–9). Furthermore, in yeast, the cell cycle regulatory activity of Cid1 is dependent on its uridyltransferase activity, although its uridylation targets have not been identified (8, 9). Conversely, we have shown here that the proliferative activity of Zcchc11 is independent of its uridyltransferase domain.
In conclusion, we have identified Zcchc11 as a previously unrecognized regulator of cell proliferation in mammalian cells. Zcchc11 contributes to the cell cycle by facilitating the expression of multiple cyclins and cyclin-dependent kinases to enhance steps of G1 progression both upstream and downstream of Rb phosphorylation. Unexpectedly, the effect of Zcchc11 on cell cycling is not mediated by its only known catalytic activity as a uridyltransferase and instead is localized in the N-terminal region of the protein. These results highlight the importance of assessing nucleotidyltransferase-independent activities of this intriguing group of proteins.
Supplementary Material
Acknowledgments
We thank Peter Sicinski, Lester Kobzik, and William Cruikshank for helpful comments and suggestions and Yan Geng for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL079392 and R01 HL068153 (to J. P. M.), R01 HL104053 (to M. R. J.), R00 HL092956 (to L. J. Q.), and T32 HL007035 and T90 DK070078 (to M. T. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- SABG
- senescence-associated β-galactosidase
- PI
- propidium iodide
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Jones M. R., Quinton L. J., Blahna M. T., Neilson J. R., Fu S., Ivanov A. R., Wolf D. A., Mizgerd J. P. (2009) Nat. Cell Biol. 11, 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schultz J., Lorenz P., Gross G., Ibrahim S., Kunz M. (2008) Cell Res. 18, 549–557 [DOI] [PubMed] [Google Scholar]
- 3. Heo I., Joo C., Kim Y. K., Ha M., Yoon M. J., Cho J., Yeom K. H., Han J., Kim V. N. (2009) Cell 138, 696–708 [DOI] [PubMed] [Google Scholar]
- 4. Stevenson A. L., Norbury C. J. (2006) Yeast 23, 991–1000 [DOI] [PubMed] [Google Scholar]
- 5. Hagan J. P., Piskounova E., Gregory R. I. (2009) Nat. Struct. Mol. Biol. 16, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rissland O. S., Mikulasova A., Norbury C. J. (2007) Mol. Cell. Biol. 27, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsen A., Vantipalli M. C., Lithgow G. J. (2006) Science 312, 1381–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Read R. L., Martinho R. G., Wang S. W., Carr A. M., Norbury C. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12079–12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang S. W., Toda T., MacCallum R., Harris A. L., Norbury C. (2000) Mol. Cell. Biol. 20, 3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu J., He M. L., Wang L., Chen Y., Liu X., Dong Q., Chen Y. C., Peng Y., Yao K. T., Kung H. F., Li X. P. (2011) Cancer Res. 71, 225–233 [DOI] [PubMed] [Google Scholar]
- 11. Kota J., Chivukula R. R., O'Donnell K. A., Wentzel E. A., Montgomery C. L., Hwang H. W., Chang T. C., Vivekanandan P., Torbenson M., Clark K. R., Mendell J. R., Mendell J. T. (2009) Cell 137, 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 13. Kumar M. S., Erkeland S. J., Pester R. E., Chen C. Y., Ebert M. S., Sharp P. A., Jacks T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt M. J., West S., Norbury C. J. (2011) RNA 17, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mullen T. E., Marzluff W. F. (2008) Genes Dev. 22, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carney D. N., Bunn P. A., Jr., Gazdar A. F., Pagan J. A., Minna J. D. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3185–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 18. Zheng L., Lee W. H. (2001) Exp. Cell Res. 264, 2–18 [DOI] [PubMed] [Google Scholar]
- 19. Lukas J., Herzinger T., Hansen K., Moroni M. C., Resnitzky D., Helin K., Reed S. I., Bartek J. (1997) Genes Dev. 11, 1479–1492 [DOI] [PubMed] [Google Scholar]
- 20. Qi Y., Tu Y., Yang D., Chen Q., Xiao J., Chen Y., Fu J., Xiao X., Zhou Z. (2007) J. Cell Physiol. 210, 63–71 [DOI] [PubMed] [Google Scholar]
- 21. Bandi N., Zbinden S., Gugger M., Arnold M., Kocher V., Hasan L., Kappeler A., Brunner T., Vassella E. (2009) Cancer Res. 69, 5553–5559 [DOI] [PubMed] [Google Scholar]
- 22. Blajeski A. L., Phan V. A., Kottke T. J., Kaufmann S. H. (2002) J. Clin. Invest. 110, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minoda Y., Saeki K., Aki D., Takaki H., Sanada T., Koga K., Kobayashi T., Takaesu G., Yoshimura A. (2006) Biochem. Biophys. Res. Commun. 344, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 24. Marchler-Bauer A., Bryant S. H. (2004) Nucleic Acids Res. 32, W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zdobnov E. M., Apweiler R. (2001) Bioinformatics 17, 847–848 [DOI] [PubMed] [Google Scholar]
- 27. Arnold K., Kiefer F., Kopp J., Battey J. N., Podvinec M., Westbrook J. D., Berman H. M., Bordoli L., Schwede T. (2009) J. Struct. Funct. Genomics 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aravind L., Koonin E. V. (1999) Nucleic Acids Res. 27, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwak J. E., Wickens M. (2007) RNA 13, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.