Background: Dual specificity protein phosphatases (DSPs) bind MAPKs through specific interaction motifs.
Results: A novel motif (IYT) in Msg5 mediates a common docking domain-independent interaction with the yeast cell integrity kinase Slt2.
Conclusion: Distinct mechanisms allow Msg5 to differentially bind Slt2 and Mlp1 or mating/pseudohyphal MAPKs Fus3 and Kss1.
Significance: Elucidation of the mechanisms by which MAPKs interact with key regulators is vital to understanding cell signaling.
Keywords: Cell Signaling, Dual Specificity Phosphoprotein Phosphatase, MAP Kinases (MAPKs), Protein-Protein Interactions, Yeast, Binding Motif, Mlp1, Slt2
Abstract
MAPK phosphatases (MKPs) are negative regulators of signaling pathways with distinct MAPK substrate specificities. For example, the yeast dual specificity phosphatase Msg5 dephosphorylates the Fus3 and Slt2 MAPKs operating in the mating and cell wall integrity pathways, respectively. Like other MAPK-interacting proteins, most MKPs bind MAPKs through specific docking domains. These include D-motifs, which contain basic residues that interact with acidic residues in the common docking (CD) domain of MAPKs. Here we show that Msg5 interacts not only with Fus3, Kss1, and Slt2 but also with the pseudokinase Slt2 paralog Mlp1. Using yeast two-hybrid and in vitro interaction assays, we have identified distinct regions within the N-terminal domain of Msg5 that differentially bind either the MAPKs Fus3 and Kss1 or Slt2 and Mlp1. Whereas a canonical D-site within Msg5 mediates interaction with the CD domains of Fus3 and Kss1, a novel motif (102IYT104) within Msg5 is involved in binding to Slt2 and Mlp1. Furthermore, mutation of this site prevents the phosphorylation of Msg5 by Slt2. This motif is conserved in Sdp1, another MKP that dephosphorylates Slt2, as well as in Msg5 orthologs from other yeast species. A region spanning amino acids 274–373 within Slt2 and Mlp1 mediates binding to this Msg5 motif in a CD domain-independent manner. In contrast, Slt2 uses its CD domain to bind to its upstream activator Mkk1. This binding flexibility may allow MAPK pathways to exploit additional regulatory controls in order to provide fine modulation of both pathway activity and specificity.
Introduction
Mitogen-activated protein kinase (MAPK) pathways constitute very tightly regulated signaling modules that both transduce and convert extracellular stimuli into appropriate cellular responses. Mammalian MAPKs comprise four main subfamilies: the extracellular signal-regulated kinases 1 and 2 (ERK1/2), the c-Jun N-terminal kinases (JNKs), the p38 MAPKs, and ERK5 (1). These MAPKs contain a highly conserved TXY motif in the kinase activation loop in which both threonine and tyrosine residues must be phosphorylated to achieve kinase activation. In contrast, atypical MAPKs, exemplified by ERK3 and ERK4, contain an SEG motif in which the serine residue is the sole phosphoacceptor (2). Several mechanisms exist to ensure high efficiency and fidelity within each specific pathway. Among these, selective recognition by specific protein-protein interactions between the components of a particular pathway is critical for the maintenance of accurate signal transduction (3, 4).
MAPKs are activated through phosphorylation by MAPK kinases and phosphorylate a wide range of protein substrates, including transcription factors, protein kinases, and other cell response effectors. In turn, MAPK signaling can be down-regulated by dephosphorylation and inactivation by a family of MAPK phosphatases (MKPs).5 These phosphorylation/dephosphorylation events require not only a transient enzyme-substrate interaction involving the active sites of these enzymes but also the formation of a complex between the MAPK and its cognate activator, substrate, or inactivator. This is normally achieved through specific docking interactions between conserved regions within both MAPKs and MAPK-interacting proteins (3). Such docking sites are located outside the catalytic domains of these proteins and promote binding specificity and high affinity interactions.
One well characterized docking motif found in MAPK-interacting proteins is the D motif, which has also been termed the δ-domain or DEJL (docking site for ERK and JNK, LXL) motif. This is closely related to the conserved kinase interaction motif (KIM) found within MAPK phosphatases and comprises a cluster of positively charged amino acids, normally lysines or arginines, followed by a submotif containing two hydrophobic residues leucine, isoleucine, or valine separated by one residue ((K/R)1–3X2–6ϕXϕ) (5, 6). Swapping one D domain for another can completely change the specificity of protein-protein interactions, indicating that these motifs are essential to maintain binding selectivity among MAPK pathway components (7). A second type of docking motif named the DEF motif (for docking site for ERK FXF) has been found only in some subsets of MAPK substrates. This consists of two phenylalanines separated by one amino acid and followed by proline (FXFP) (8).
Docking sites have been also characterized within MAPKs themselves. The region of the MAPK that interacts with D motifs is complex and is formed by the common docking (CD) domain, the ED (for Glu/Asp-containing) motif, and a hydrophobic docking groove (6). The CD domain is shared by all MAPK subfamilies and is an electrostatic surface depression containing a cluster of negatively charged amino acids (aspartic and glutamic acids) located within the C-terminal domain of the kinase (9). Substitution of these residues by charge-neutral amino acids completely abrogates docking, indicating the importance of the electrostatic interaction of the CD domain with the positively charged amino acids within the D domains of MAPK-interacting proteins (10). The ED motif is located close to the CD domain in the tertiary structure of MAPKs and has also been reported to contribute to binding specificity (11). Close to the CD/ED regions resides a hydrophobic docking groove that is involved in binding to the hydrophobic submotif of the D site of MAPK partners (10). In contrast, the DEF motif binds to a separate hydrophobic pocket that only becomes accessible upon MAPK activation (12). In addition, a novel interaction motif (FRIEDE) within the atypical MAPKs ERK3 and ERK4 has been shown to mediate binding to their substrate MAPKAP kinase MK5 (13). Comparison of the MAPK docking interactions from yeast to humans reveals the existence of conserved molecular mechanisms underlying the specificity of these interactions (11, 15).
Dual specificity phosphatases (DSPs) are able to dephosphorylate both threonine and tyrosine residues in their target proteins. A subfamily of these DSPs specifically targets MAPKs. These enzymes are designated as dual specificity MKPs and play an important role in the regulation of MAPK signaling. All MKPs share a common structure comprising an N-terminal non-catalytic regulatory domain, which contains the sequences necessary for MAPK docking, and a C-terminal catalytic domain (16).
Saccharomyces cerevisiae has five MAPK signaling pathways involved in mating, pseudohyphal/invasive growth, cell wall integrity (CWI), osmoregulation, and ascospore formation, mediated by Fus3, Kss1, Slt2, Hog1, and Smk1, respectively (17). Fus3 and Kss1 are orthologous to ERK1/2, and Hog1 is orthologous to p38 (18). The closest mammalian MAPK to Slt2 is ERK5 (19). The CWI-related protein Mlp1 is a pseudokinase paralog of Slt2 that lacks catalytic residues critical for protein kinase activity as well as the conserved Thr residue within the MAPK activation loop dual phosphorylation site.
Whereas an individual MAPK can be inactivated by more than one protein phosphatase, it is also the case that a single protein phosphatase is able to interact with and inactivate several MAPKs (20). For example, Msg5 has been shown to dephosphorylate Fus3 and Slt2, affecting both basal and inducible kinase activities (21–24). Msg5 is expressed in yeast cells as two isoforms that differ in the first 45 amino acids due to the use of alternative translation initiation sites (23). We have recently shown that this MKP is able to interact not only with Fus3 and Slt2 but also with Kss1, a MAPK whose phosphorylation is not down-regulated by Msg5 (25). Moreover, whereas both Msg5 isoforms interact similarly with Slt2, the longer form binds both Fus3 and Kss1 with higher affinity than the shorter isoform. In a study of the docking interactions within and between the Fus3- or Kss1-mediated pathways, Reményi et al. (15) found docking motifs in different proteins known to physically interact with Fus3, such as Ste7, Msg5, and Far1. Mutation of a D domain within Msg5 partially reduced the ability of this MKP to decrease the elevated mating pathway output displayed by an msg5-null mutant, indicating a contribution of the docking interaction mediated by this motif to the ability of Msg5 to inactivate Fus3. But the incomplete effect of D domain mutation also suggests that additional Fus3-docking motifs may be present in Msg5 (15). The interaction with Slt2 has been shown to be mediated by the N-terminal domain of Msg5 (23).
Here we report the characterization of the interactions between Msg5 and a number of yeast MAPKs, showing the involvement of distinct regions located at the N terminus of Msg5 in MAPK binding. D domain-dependent mechanisms participate in the binding to mating (Fus3 and Kss1) MAPKs. In contrast, a novel motif of Msg5 is involved in the interaction with the cell wall integrity (Slt2 and Mlp1) kinases. We also provide evidence indicating that, as is the case in other MAPK pathways, the CD domain of Slt2 mediates binding to its activator Mkk1, whereas a CD-proximal region containing the conserved MAPK L16 loop is involved in binding to the negative regulator Msg5 in a CD domain-independent fashion.
EXPERIMENTAL PROCEDURES
Yeast Strains, Genetic Methods, and Culture Conditions
The S. cerevisiae strains PJ69-4A (MATa, trp1-901, leu2-3112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ) and PJ69-4α (MATα, trp1-901, leu2-3112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ), from Clontech, were used for two-hybrid assays. Strains DD1-2D (msg5-1::LEU2) (21), YMF1 (MATa, ura3-52, his4, trp1-1, leu2-3, 112, canR, MSG5::6MYC::LEU2) (23), BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), and Y07373 (BY4741 isogenic, msg5Δ::KanMX4) from Euroscarf (Frankfurt, Germany) were used for Western blotting analysis experiments. Standard procedures were employed for yeast genetic manipulations (26). Yeast cultures were performed as reported previously (27). When necessary, Congo red (Sigma) or α-factor (Sigma) was added at the indicated concentrations.
DNA Manipulation and Plasmids
General DNA methods were performed using standard techniques. Oligonucleotide primers used for DNA amplification and directed mutagenesis by PCR are indicated in supplemental Tables I and II.
Vectors pGBKT7 (bearing the Gal4 DNA binding domain) and pGADT7 (bearing the Gal4 activation domain) were obtained from Clontech. Yeast MAPK-containing constructs pGADT7-Kss1, pGADT7-Fus3, pGADT7-Smk1, pGADT7-Slt2, and pGADT7-Hog1 were previously described (28), apart from pGADT7-Mlp1, which was constructed by subcloning the PCR-amplified MLP1 coding sequence from S. cerevisiae genomic DNA into pGADT7 as an NdeI-XhoI fragment. Mutated derivatives of these reading frames (pGADT7-Kss1CD, pGADT7-Fus3CD, pGADT7-Slt2CD, pGADT7-Slt2CD3, and pGADT7-Mlp1CD) were modified by overlap extension PCR-mediated site-directed mutagenesis and subcloned as described for the wild-type construct (28). The MSG5 reading frame was amplified from S. cerevisiae genomic DNA; wild-type, mutant, and truncated reading frames derived from this template were subcloned as NdeI-XhoI fragments into the NdeI/SalI sites in pGBKT7: pGBKT7-Msg5, pGBKT7-Msg5(1–245), pGBKT7-Msg5(246–489), pGBKT7-Msg5(126–489), pGBKT7-Msg5(1–123), pGBKT7-Msg5(90–489), pGBKT7-Msg5(1–45), pGBKT7-Msg5(46–489), pGBKT7-Msg5MD1, pGBKT7-Msg5MD2, pGBKT7-Msg5MD1/2, pGBKT7-Msg5MD3, pGBKT7-Msg5(1–123)MD1/2, and pGBKT7-Msg5(1–123)MD3.
For pGBKT7-Msg5MD1/2 and pGBKT7-Msg5(1–123)MD1/2, PCR was performed as for single point mutations but using mutant half-product templates to generate compound double and truncated mutant reading frames following the second round of amplification. YCplac22MSG5MD1, YCplac22MSG5MD2, and YCplac22MSG5MD3 plasmids were generated by site-directed mutagenesis of YCplac22MSG5m (23) following the method described previously (23). YCplac111-Slt2(1–373) was constructed by cloning a ∼3.2-kb EcoRI-EcoRI fragment containing SLT2 into YCpLac111 (CEN4, LEU2) (23) and then introducing a consecutive pair of stop codons in the corresponding protein positions 374 and 375 by site-directed mutagenesis of the obtained plasmid.
In order to express Msg5 fused to GST in yeast, the previously described plasmid pEG(KG)-MSG5 was used (23). pEG(KG)-Slt2(274–373) was constructed by PCR-amplifying the corresponding SLT2 region using YCplac111-Slt2(1–373) as template and then subcloning the BamHI- digested PCR product into pEG-KG (2 μm, GAL1–10UAS-CYC1).
To obtain recombinant GST-Mkk1, plasmid pGEX-MKK1 was used (29). Recombinant Msg5-GST, Msg5MD3-GST, and Msg5(1–123)MD3-GST were produced from plasmids constructed by subcloning PCR products featuring NdeI and XhoI sites at the 5′- and 3′-ends into a modified pGEX-P1 vector (gift of Dr. B. McStay). Recombinant Slt2, Mlp1, and Mkk1 His-tagged proteins or protein fragments were produced from pET15B-derived plasmids after cloning the corresponding PCR-amplified SLT2 and MLP1 gene fragments in NdeI and XhoI sites.
Yeast Two-hybrid Assays
Assays were performed according to the manufacturer's instructions (Matchmaker 3 kit, Clontech). The vector pGBKT7 or its derivatives were transformed into PJ69-4A cells and mated with PJ69-4α cells transformed with plasmid pGADT7 or its derivatives. Yeast diploids carrying both types of plasmids were selected on synthetic dextrose (SD) medium deficient for leucine and tryptophan. In order to perform a qualitative analysis of the two-hybrid interactions, representative colonies from the different diploid strains were cultivated overnight in liquid YPD, and the corresponding cellular suspensions were prepared. 10-μl volumes of inocula (normalized by optical density at 600 nm) were spotted onto two types of media: SD medium lacking leucine and tryptophan and SD medium lacking leucine, tryptophan, histidine, and adenine. Cell growth on the latter medium provides evidence for the occurrence of protein-protein interaction.
For semiquantitative analysis of the two-hybrid interactions, exponentially growing diploid cells bearing the relevant plasmids were collected, and the β-galactosidase activity was determined using o-nitrophenyl-β-galactoside as a substrate, according to the manufacturer's instructions (Clontech).
Preparation of Bacteria and Yeast Extracts
Recombinant GST- or histidine-tagged proteins were expressed in Escherichia coli strain Rosetta DE3 (Novagen). Cells were collected and lysed by sonication in PBS buffer containing 2 mm PMSF, 1 mm DTT, 1 mm EDTA, and 1 mg/ml lysozyme in the presence of protease inhibitor mixture (Roche Applied Science). Extracts were clarified by centrifugation and then stored at −80 °C. For obtaining yeast extracts, the procedures employed have been described previously (27).
Binding Assays
For in vivo binding assays, yeast cells bearing pEG(KG)-derived plasmids were grown in complete synthetic medium lacking uracil with 2% galactose at 24 °C. Cells were collected and lysed as above in lysis buffer lacking SDS and Nonidet P-40. Yeast lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences) for 2 h. Beads were washed extensively with the same buffer and resuspended in SDS loading buffer, and proteins were analyzed by SDS-PAGE and immunoblotting as described (23). In vitro binding assays were performed by mixing E. coli extracts containing GST or GST-fused proteins with E. coli extracts bearing the corresponding His-tagged proteins and then processed as above.
Immunoblot Analysis
Immunodetection of Myc- and HA- tagged proteins was carried out using either monoclonal 9E10 (Covance) or monoclonal 4A6 (Millipore) and monoclonal HA.11 (Covance) antibodies, respectively. Actin was immunodetected by using monoclonal C4 antibodies (MP Biomedicals). Polyclonal anti-phospho-p44/p42 MAPK (Thr-202/Tyr-204) (Cell Signaling), anti-GST (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and monoclonal anti-His (Sigma) antibodies were also used as described previously (27). Immunodetection of Gal4BD fusion proteins was carried out using monoclonal GAL4DNA-BD antibody (Clontech). The primary antibodies were detected using either a horseradish peroxidase-conjugated secondary antibody with the ECL detection system (Amersham Biosciences) or a fluorescently conjugated secondary antibody with an Odyssey Infrared Imaging System (LI-COR Biosciences).
Real-time Quantitative PCR Assays
Real-time quantitative PCR assays were performed as described previously (30), using an ABI 7700 instrument (Applied Biosystems). Each cDNA was assayed in at least duplicate PCR reactions. For quantification, the abundance of each gene was determined relative to the standard transcript of ACT1, and the final data of relative gene expression between the two conditions tested were calculated following the 2−ΔΔCt method, as described (31). The primer sequences are available upon request.
RESULTS
Distinct Regions of Msg5 Are Responsible for Its Interaction with Yeast MAPKs
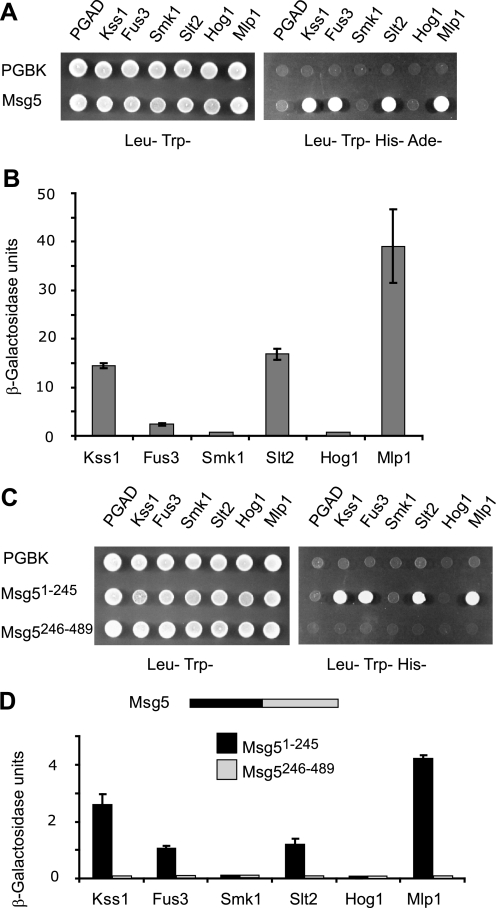
In order to gain insight into the mechanisms that regulate the interaction of Msg5 with its substrate MAPKs, we first wanted to define the MAPKs that physically interact with this phosphatase. To this end, the yeast two-hybrid assay was used. Msg5 was fused to the Gal4-DNA binding domain (Gal4BD), and the five MAPKs (Kss1, Fus3, Smk1, Slt2, and Hog1) as well as the pseudokinase Slt2 paralog Mlp1 (32) were fused to the Gal4 activation domain (Gal4AD). Expression of all of these hybrid proteins was analyzed by Western blotting (supplemental Fig. S1). Formation of a complex between Msg5 and the corresponding MAPK results in reconstitution of the Gal4 transcription factor and expression of the Gal4-dependent reporter genes HIS3, ADE2, and lacZ. These interactions can then be monitored by either growth of the diploid strains on selective medium lacking adenine and histidine or semiquantitative assays of β-galactosidase activity. As shown in Fig. 1A, only the yeast cells expressing Msg5 together with Kss1, Fus3, Slt2, or Mlp1 were able to grow on selective medium lacking adenine and histidine. In addition, expression of lacZ was only observed in these same cells (Fig. 1B). The strongest interaction with Msg5, in terms of increased β-galactosidase activity, was observed with Mlp1. Interactions with Slt2 and Kss1 were also significant, whereas the lowest strength interaction was observed with Fus3. Western blotting analysis revealed that this was not due to low level expression of Gal4AD-Fus3 (supplemental Fig. S1). Despite being expressed to a similar extent as the other kinases (supplemental Fig. S1), neither Hog1 nor Smk1 was able to interact with Msg5 in the two-hybrid assay (Figs. 1, A and B). A catalytically inactive mutant of Msg5C319S (21) also failed to interact with either Hog1 or Smk1 (data not shown), ruling out the possibility that the absence of binding was due to dephosphorylation of these kinases by Msg5.
FIGURE 1.
Interaction of Msg5 with MAPKs. A, qualitative analysis of two-hybrid interaction by growth analysis of diploid yeast cells transformed with plasmids encoding the indicated proteins. Vector pGBKT7 (bearing the Gal4 DNA-binding domain) or pGBKT7-Msg5 was transformed into PJ69-4A S. cerevisiae strain and mated with PJ69-4α S. cerevisiae strain transformed with plasmid pGADT7 (bearing the Gal4 activation domain, pGADT7-Kss1, pGADT7-Fus3, pGADT7-Smk1, pGADT7-Slt2, pGADT7-Mlp1, or pGADT7-Hog1. Yeast diploids were spotted onto SD medium lacking leucine and tryptophan (left) or SD medium lacking leucine, tryptophan, histidine, and adenine (right). Cell growth on the latter medium provides evidence for the occurrence of protein-protein interaction. B, semiquantitative analysis of the two-hybrid interaction between Msg5 and the distinct MAPKs based on the level of induction of β-galactosidase. Experiments were performed in triplicate on the diploid strains described in A, with error bars representing S.D. C, qualitative analysis of the two-hybrid interaction between the N-terminal Msg5 (Msg5(1–245)) or C-terminal Msg5 (Msg5(246–489)) region and the different MAPKs. Before mating, pGBKT7, pGBKT7-Msg5(1–245) or pGBKT7-Msg5(246–489) were transformed into PJ69-4A cells, and pGADT7, or pGADT7-Kss1, pGADT7-Fus3, pGADT7-Smk1, pGADT7-Slt2, pGADT7-Mlp1, or pGADT7-Hog1 were transformed into PJ69-4α cells. Two-hybrid assays were performed as indicated in A. D, semiquantitative analysis of the two-hybrid interaction between the same Msg5 protein fragments as in C and the different MAPKs. A schematic showing both halves of Msg5 is also displayed. Experiments were performed in triplicate on the diploid cells described in C, with error bars representing S.D.
Msg5 is composed of an N-terminal domain (approximately residues 1–245) and a C-terminal catalytic domain (approximately residues 246–489). To determine the involvement of these two regions in interactions with distinct MAPKs, we extended our two-hybrid analysis by using constructs containing either the isolated N-terminal or C-terminal domains of Msg5. As observed in Fig. 1C, the N-terminal domain of Msg5 interacted with all four of the MAPKs that bind full-length Msg5. Although the β-galactosidase levels were significantly reduced when compared with those measured between full-length Msg5 and these MAPKs, the strongest interaction was again observed with Mlp1 (Fig. 1D). The C-terminal domain of Msg5 fused to Gal4BD did not show interaction with any of the MAPKs (Fig. 1D), although it was highly expressed (supplemental Fig. S1). These results indicate that the N-terminal domain of Msg5 is both necessary and sufficient to mediate protein-protein interactions with MAPK substrates.
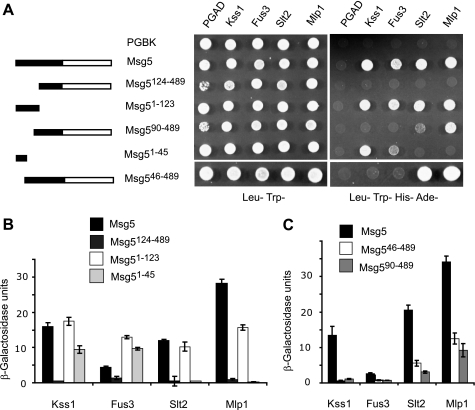
In order to further elucidate the region of Msg5 that is responsible for the interactions with Fus3, Kss1, Slt2, and Mlp1, we next tested a set of Msg5 truncations fused to Gal4BD for binding to the distinct Gal4AD-MAPK fusions. The expression of these hybrid Msg5 proteins was also monitored by Western blotting (supplemental Fig. S1). As shown in Fig. 2, A and B, the region encompassing the first 123 amino acids of Msg5 (Msg5(1–123)) is necessary and sufficient for the interaction with all four MAPKs. In contrast, although highly expressed (supplemental Fig. S1), a construct lacking the first 123 residues (Msg5(124–489)) failed to interact with all four MAPKs. However, although an Msg5 construct lacking the first 45 amino acids interacted with both Slt2 and Mlp1, it could no longer bind to either Fus3 or Kss1 (Fig. 2, A and C). Consistent with these data, an Msg5 fragment containing only the first 45 amino acids interacted with Fus3 and Kss1 but not with Mlp1 and Slt2 (Fig. 2, A and B). These two-hybrid results are also in agreement with our previously reported data showing that the short Msg5 isoform, which lacks the first 45 amino acids, displays a much lower affinity for Fus3 and Kss1 than the full-length Msg5 isoform in co-purification assays, whereas both isoforms bind Slt2 with the same affinity (25). Finally, a mutant of Msg5 lacking the first 89 amino acids did not interact with Fus3 and Kss1 but was still able to interact with Slt2 and Mlp1 (Fig. 2, A and C).
FIGURE 2.
Mapping of Msg5 regions that mediate MAPK interaction. A, schematic of full-length and Msg5 fragments analyzed for MAPK interaction (left) and qualitative analysis of the two-hybrid interaction by growth analysis of diploid cells bearing plasmids that encode the indicated proteins or protein fragments (right). Before mating, pGBKT7-Msg5, pGBKT7, pGBKT7-Msg5(126–489), pGBKT7-Msg5(1–123), pGBKT7-Msg5(90–489), pGBKT7-Msg5(1–45), or pGBKT7-Msg5(46–489) were transformed into PJ69-4A cells, and pGADT7 and either pGADT7-Kss1, pGADT7-Fus3, pGADT7-Slt2, or pGADT7-Mlp1 were transformed into PJ69-4α cells. Two-hybrid assays were performed as indicated in Fig. 1A. B and C, semiquantitative analysis of the two-hybrid interaction between Msg5 or the different Msg5 fragments and the distinct MAPKs based on the level of induction of β-galactosidase. Experiments were performed in triplicate on the diploid cells described in A, with error bars representing S.D.
In summary, these two-hybrid data show that different regions of Msg5 mediate the interaction of this phosphatase with either mating- or cell wall integrity-related kinases. The first 45 amino acids of Msg5 are necessary for interaction with Fus3 and Kss1 and dispensable for binding to Slt2 and Mlp1, whereas the region spanning amino acids 90 to 123 is required for interaction with the latter two kinases.
Docking Site-dependent and -independent Mechanisms Mediate Binding of Msg5 to Distinct MAPKs
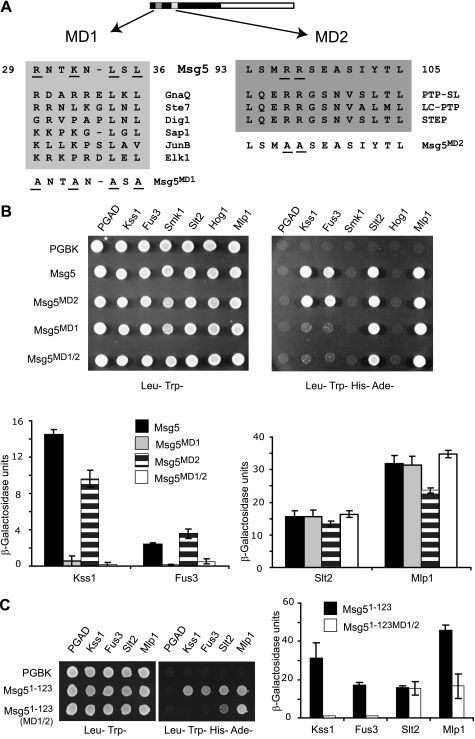
The fact that different regions of Msg5 mediate its binding to distinct MAPKs suggested that this phosphatase might have evolved different interaction mechanisms to selectively act on its MAPK substrates. To gain insight into this issue, we studied the involvement of classical docking interactions on the binding of Msg5 to MAPKs. Msg5 has previously been shown to contain an interaction motif for Fus3 and Kss1, spanning amino acids 27 to 39. This is referred to here as MD1 (Msg5 docking 1) and has previously been found to contribute to the ability of Msg5 to act on Fus3 (15). This MD1 docking motif is similar to the D domains found in MAPK kinases, transcription factors, and other MAPK substrates (Fig. 3A). However, to date, no docking motif has been identified that mediates the interaction of Msg5 with Slt2. Because our results indicate that the first 123 amino acids of Msg5 are involved in binding to Fus3, Kss1, Slt2, and Mlp1, we searched for additional putative MAPK docking domains in this region. This analysis identified an additional amino acid motif, located between residues 93 and 105 in Msg5, that showed sequence homology with kinase interaction motifs previously identified in mammalian tyrosine-specific MAPK phosphatases (33), such as PTP-SL, LC-PTP, or STEP (Fig. 3A). This motif contains two conserved basic residues (arginines 96 and 97), whereas the characteristic LXL submotif is replaced by YXL. To evaluate the role of the MD1 and MD2 motifs in the interactions of Msg5 with the various MAPKs, we generated Msg5 mutants in which key residues were substituted by alanine (Fig. 3A) and analyzed their ability to interact with MAPKs using the two-hybrid assay. Mutations in MD1 (R29A, K32A, L34A, and L36A) clearly abrogated the ability of Msg5 to interact with either Fus3 or Kss1 (Fig. 3B), whereas interactions with either Slt2 or Mlp1 were completely unperturbed. In contrast, mutations within MD2 involving substitution of the two arginines (R96A and R97A) had no effect on interactions with either Fus3 or Slt2 and only led to a slight reduction in the interaction with Kss1 and Mlp1. The corresponding arginine residues of PTP-SL, LC-PTP, and STEP have been shown to be essential for the interaction of these phosphatases with the mammalian MAPKs ERK2 and p38 (34, 35). Thus, despite the resemblance between MD2 and the KIM found in mammalian MAPK-specific tyrosine phosphatases, the conserved arginines do not appear to play a role in mediating Msg5 binding to MAPKs. Accordingly, a mutant of Msg5 containing substitutions in both MD1 and MD2 behaved the same as the MD1 mutant of Msg5 in two-hybrid assays (Fig. 3B). Finally, mutation of both MD1 and MD2 in the Msg5 fragment encompassing the first 123 amino acids abolished its interaction with both Fus3 and Kss1 but not Slt2 and Mlp1 (Fig. 3C). These results confirm the essential role of MD1 in the interaction of Msg5 with Fus3 and Kss1 and its dispensability for the interaction of Msg5 with Slt2 and Mlp1 while indicating that MD2 does not play a functional role in MAPK recognition.
FIGURE 3.
Involvement of distinct docking domains of Msg5 in MAPK binding. A, scheme of Msg5 showing the location of two putative docking domains (MD1 and MD2), which contain conserved amino acid residues (underlined) found in MAPK docking sites of different MAPK-interacting proteins, including yeast Ste7 and Dig1 and mammalian GnaQ, Sap1, JunB, Elk1, PTP-SL, LC-PTP, and STEP. The amino acid substitutions introduced into Msg5 to generate the mutated versions Msg5MD1, Msg5MD2, and Msg5MD1/2 are underlined. B, qualitative (top) and semiquantitative (bottom) analysis of the two-hybrid interaction of diploid cells containing plasmids that encode the indicated proteins. Before mating, pGBKT7, pGBKT7-MSG5, pGBKT7-Msg5MD1, pGBKT7-Msg5MD2, or pGBKT7-Msg5MD1/2 were transformed into PJ69-4A cells, and pGADT7 or pGADT7-Kss1, pGADT7-Fus3, pGADT7-Smk1, pGADT7-Slt2, pGADT7-Mlp1, and pGADT7-Hog1 were transformed into PJ69-4α cells. Two-hybrid assays were performed as indicated in Fig. 1, A and B. C, semi qualitative (left) or semiquantitative (right) analysis of the two-hybrid interaction of diploid cells bearing plasmids that encode the indicated proteins. Before mating, pGBKT7, pGBKT7-Msg5(1–123) or pGBKT7-Msg5(1–123)MD1/2, expressing the first 123 amino acids of Msg5 without or with both MD1 and MD2 substitutions, respectively, were transformed into PJ69-4A cells, and pGADT7, pGADT7-Kss1, pGADT7-Fus3, pGADT7-Slt2, or pGADT7-Mlp1 were transformed into PJ69-4α cells. Two-hybrid assays were performed as indicated in the legend to Fig. 1, A and B.
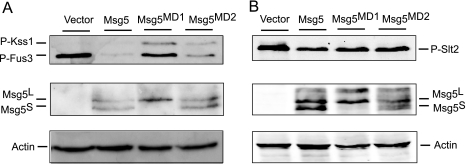
Deletion of MSG5 results in increased phosphorylation of both Fus3 and Slt2, under both basal and induced conditions (25). To study the importance of docking interactions in the negative regulation of these MAPKs, we analyzed the amount of dually phosphorylated Fus3 and Slt2 in msg5Δ cells carrying a centromeric plasmid encoding either wild type Msg5, a MD1 mutant in the two leucines of the LXL submotif (Msg5L34A,L36A), or the MD2 mutant (Msg5R96A,R97A). As shown in Fig. 4A, the MD1 mutant protein was unable to reduce the increased levels of phospho-Fus3 in pheromone-stimulated msg5Δ cells to the extent promoted by either wild type Msg5 or the MD2 mutant. Therefore, disruption of the docking interaction greatly decreased the efficiency of the action of Msg5 on Fus3. In contrast, expression of Msg5 mutated in either MD1 or MD2 reduced the increased Slt2 phosphorylation exhibited by msg5Δ mutants under CWI-stimulating conditions to a similar extent as wild type Msg5 (Fig. 4B).
FIGURE 4.
Effect of mutation of the putative Msg5 docking domains on MAPK phosphorylation. A, Western blot analysis of cell extracts from the DD1-2D (msg5Δ) strain transformed with the empty vector YCplac22, YCplac22MSG5m, YCplac22MSG5MD1m, or YCplac22MSG5MD2m expressing Msg5–6Myc, Msg5MD1-6Myc, or Msg5MD2-6Myc, respectively. Cells were grown to mid-log phase in YPD medium at 24 °C and then treated with 50 nm α-factor for 10 min. B, Western blot analysis of cell extracts from the same strains as in A, after treatment with 30 μg/ml Congo red for 2 h. Analyses were performed using anti-phospho-p44/p42 MAPK (Thr-202/Tyr-204) antibody for immunodetection of phosphorylated Fus3, Kss1, and Slt2 (top), anti-Myc for detecting the two Msg5 isoforms Msg5S and Msg5L (23) (middle), and anti-actin antibodies for actin detection as loading control (bottom). Reproducible results were obtained in different experiments, and selected images correspond to representative blots.
All of these data reinforce the importance of the docking interaction through MD1 between Msg5 and Fus3 for the regulatory role of this phosphatase on the mating pathway and suggest that the interaction of Msg5 with Slt2 and Mlp1 is not mediated by canonical MAPK docking domains. Because this MD1 mutant form of Msg5 only carries the substitutions of leucines 34 and 36 by alanine, these results indicate that these two amino acids are essential for the functionality of this docking motif. The tagging of these Msg5 constructs with the Myc epitope allowed us to analyze the relative amount of each Msg5 isoform in the blots. Interestingly, Msg5MD1 is mainly present as the long isoform (Fig. 4). Although other possibilities cannot be ruled out, this result suggests that the mRNA region of MSG5 corresponding to the MD1-coding sequence is critical for the translation of the short form of this phosphatase.
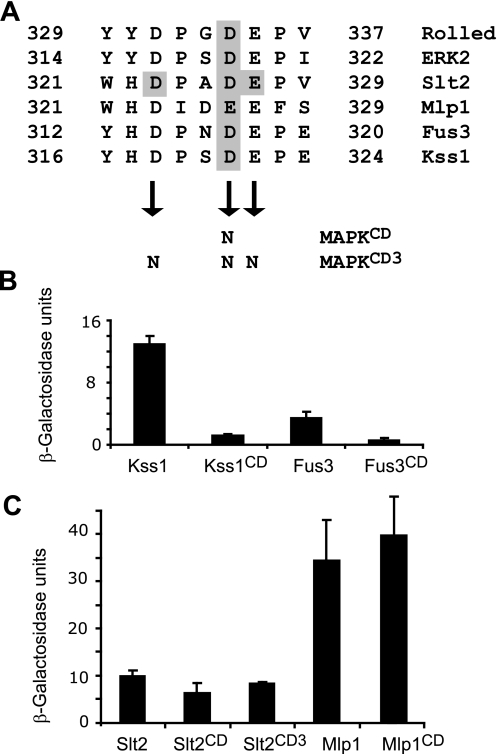
A complementary approach to analyze the importance of docking interactions between MAPKs and their partners is to mutate the MAPK CD domain. Alignment of the sequences corresponding to the CD domain of mammalian Erk2 and Drosophila Rolled with those of Slt2, Mlp1, Fus3, and Kss1 revealed conserved aspartic and glutamic acids in the yeast proteins (Fig. 5A). Mutation of aspartic acid 319 to asparagine in Erk2 gives rise to a hyperactive protein due to a deficient binding and reduced sensitivity to the DSP MKP-3 (36). D317N and D321N CD domain mutations in Fus3 and Kss1, respectively, did not affect protein levels (supplemental Fig. S1) but greatly reduced the interaction of both MAPKs with Msg5 in the two-hybrid assay (Fig. 5, A and B). These results are in agreement with the involvement of the Msg5 canonical docking domain MD1 in its interaction with Fus3 and Kss1 as well as with the importance of the CD domain of these MAPKs in binding to Ste7, Ste5, Dig1, and Dig2 (37) or Far1 (15). In contrast, the Slt2 D326N and Mlp1 E326N CD domain mutations did not decrease the association with Msg5 (Fig. 5, A and C). Moreover, a combination of the D326N mutation with two other changes in conserved residues within the CD of Slt2, namely aspartic acid 323 and glutamic acid 327 (Fig. 5A), did not result in a reduction in the binding of this Slt2 mutant (Slt2CD3) to Msg5 (Fig. 5C). The two-hybrid results reported so far show that binding between Msg5 and Slt2 or Mlp1 is not mediated by classical docking interactions involving D and CD domains, in contrast to that occurring with Fus3 or Kss1.
FIGURE 5.
Involvement of the common docking domain of MAPKs on their interaction with Msg5. A, amino acid sequence of the CD domains found in yeast Slt2, Mlp1, Fus3, and Kss1, Drosophila melanogaster Rolled, and mammalian Erk2. Shaded characters indicate the amino acids substituted by asparagines in the different MAPKs (CD and CD3 mutations). B and C, quantitative analysis as indicated in Fig. 1B of the two-hybrid interaction of diploid cells bearing plasmids that encode the indicated proteins. Before mating, pGBKT7, pGBKT7-Msg5 were transformed into PJ69-4A cells, and pGADT7, pGADT7-Kss1, pGADT7-Kss1CD, pGADT7-Fus3, pGADT7-Fus3CD (B), pGADT7-Slt2, pGADT7-Slt2CD, pGADT7-Slt2CD3, pGADT7-Mlp1, or pGADT7-Mlp1CD (C) were transformed into PJ69-4α cells. Error bars, S.D.
Msg5 Regulates the Phosphorylation of the N-terminal Kinase Domain of Slt2 by Binding the Region Spanning Amino Acids 274 to 373
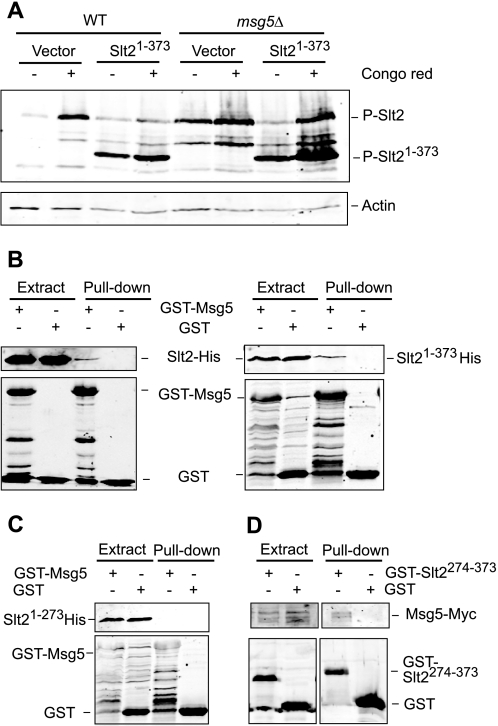
It has previously been shown that removal of the extreme C terminus of Slt2 (amino acids 394–484) increases the level of phosphorylation of the TXY motif within the activation loop (38). One possible explanation for this observation is that this region mediates binding to a negative regulator. Therefore, we studied the effect of Msg5 on the phosphorylation of a C-terminal truncation of Slt2 (Slt2(1–373)). As shown in Fig. 6A, the Slt2(1–373) mutant showed higher levels of activation loop phosphorylation than wild-type Slt2, both in unstimulated conditions and after exposure of cells to the Slt2 activator Congo red in wild type cells. Interestingly, levels of Slt2(1–373) phosphorylation were further increased under both basal and inducing conditions in cells lacking Msg5 (msg5Δ). This result indicates that Msg5 is still able to regulate the phosphorylation of the truncated kinase, suggesting that the C terminus of Slt2 is dispensable for binding to Msg5.
FIGURE 6.
Mapping of the Slt2 region that mediates binding to Msg5. A, Western blot analysis of cell extracts from the BY4741 strain (WT) and the isogenic mutant strain Y07373 (msg5Δ) transformed with the vector YCpLac111 or plasmid YCplac111-Slt2(1–373). Cells were grown to mid-log phase in YPD medium at 24 °C (−) and then treated with 30 μg/ml Congo red (CR) for 2 h as indicated. Immunodetection was performed with anti-phospho-p42/44 MAPK (top) and anti-actin (bottom) antibodies as loading control. B and C, Western blot analysis of the in vitro co-purification of recombinant Msg5 with Slt2 and different truncated Slt2 versions. E. coli extracts containing GST or GST-Msg5 were incubated with E. coli extracts containing Slt2-His, Slt2(1–373)-His (B), or Slt2(1–274)-His (C) and glutathione-Sepharose to pull down GST-complexes. Immunodetection was performed using anti-poly-His (top) and anti-GST (bottom) antibodies. D, Western blot analysis of co-purification of Msg5 with Slt2(274–373) fragment. Yeast extracts of the Msg5–6myc-tagged strain YMF1 transformed with the indicated plasmid, pEG(KG) (GST) or pEG(KG)-SLT2(274–373) (GST-SLT2274–373), were incubated with glutathione-Sepharose to pull down GST complexes. Immunodetection was performed with anti-Myc (top) and anti-GST antibodies (bottom). Reproducible results were obtained in different experiments, and selected images correspond to representative blots.
Because Slt2 has been shown to dimerize (39), we wanted to rule out the possibility that the effect of Msg5 on Slt2(1–373) phosphorylation was facilitated by binding to endogenous full-length Slt2 protein. To this end, recombinant GST-Msg5 and both full-length Slt2 and Slt2(1–373) tagged in the N terminus with a poly-His epitope were produced in E. coli and tested for in vitro interaction by a pull-down assays. As shown in Fig. 6B, both Slt2 and Slt2(1–373) showed similar binding to GST-Msg5, suggesting that Slt2 does not bind Msg5 through the MAPK C-terminal tail.
In an attempt to localize the Msg5-binding region within Slt2, we carried out pull-down experiments using a mutant of Slt2 (Slt2(1–273)) lacking a further 100 amino acids from the C terminus of the protein. Unlike Slt2(1–373), Slt2(1–273) did not bind GST-Msg5 (Fig. 6C), suggesting that the Slt2 region spanning amino acids 274 to 373 mediates binding to Msg5. In support of this, we showed that the Slt2 region 274–373 fused to GST (GST-Slt2(274–373)) bound Msg5-Myc when expressed in yeast (Fig. 6D). Binding between this Slt2 fragment and Msg5 was also confirmed by in vitro pull-down experiments using poly-His-tagged Slt2(274–373) and GST-Msg5, both produced in E. coli (Fig. 7A).
FIGURE 7.
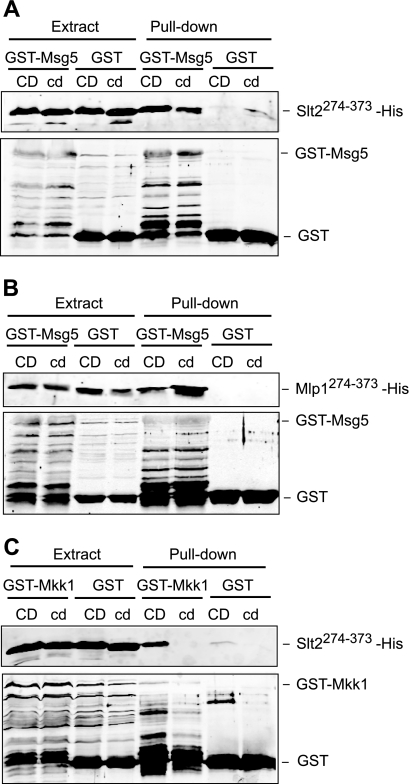
Involvement of the common docking domain of Slt2 and Mlp1 on binding to distinct interactors. A, Western blot analysis of the in vitro co-purification of recombinant Msg5 with the indicated Slt2 fragments. E. coli extracts containing GST or GST-Msg5 were incubated with E. coli extracts containing Slt2(274–373)-His (CD) or Slt2(274–373)(323N,326N,327N)-His (cd) and glutathione-Sepharose to pull down GST complexes. Immunodetection was performed using anti-poly-His (top) and anti-GST (bottom) antibodies. B, Western blot analysis of the in vitro co-purification of recombinant Msg5 with the indicated Mlp1 fragments. E. coli extracts containing GST or GST-Msg5 were incubated with E. coli extracts containing Mlp1(274–373)-His (CD) or Mlp1(274–373)(326N)-His (cd) and glutathione-Sepharose to pull down GST complexes. Immunodetection was performed as in A. C, Western blot analysis of the in vitro co-purification of recombinant Mkk1 with the indicated Slt2 fragments. E. coli extracts containing GST or GST-Mkk1 were incubated with E. coli extracts containing Slt2(274–373)-His (CD) or Slt2(274–373)(323N,326N,327N)-His (cd) and glutathione-Sepharose to pull down GST complexes. Immunodetection was performed as in A. In all cases, reproducible results were obtained in different experiments, and selected images correspond to representative blots.
The Same Region of Slt2 Mediates Binding to Its Activator Mkk1 and Msg5 through CD-dependent and -independent Binding Mechanisms, Respectively
The Msg5-binding region of Slt2, spanning amino acids 274 to 373, includes the CD domain (Fig. 5A). We have previously shown by hybrid assays that the Slt2 CD domain is not required to mediate the interaction of Slt2 with Msg5. In order to confirm the dispensability of this motif in Slt2-Msg5 binding, in vitro interaction assays with recombinant proteins individually produced in E. coli were performed. A poly-His-tagged version of Slt2(274–373) mutated in three of the critical CD domain residues (Slt2(274–373)(323N,326N,327N)) was produced in E. coli and assayed for binding to recombinant GST-Msg5. As observed in Fig. 7A, the CD mutant Slt2 fragment bound to the phosphatase similarly to the wild type Slt2 fragment, suggesting that the Slt2 CD domain is not involved in this interaction. GST-Msg5 was also able to bind an equivalent poly-His-tagged fragment of Mlp1 (Mlp1(274–373)) as well as the corresponding CD-mutated version of this fragment (Mlp1(274–373)(326N)) (Fig. 7B). Altogether, these results confirm the dispensability of the CD domain within Slt2 for binding to Msg5 and suggest that a similar binding mechanism is likely to operate in the interaction of Mlp1 with Msg5.
To establish whether the CD domain present in the Slt2 fragment could mediate binding to other known interactors of this MAPK, we carried out similar pull-down experiments using recombinant GST-Mkk1. Mkk1 is one of the two redundant MAPK kinases acting as Slt2 activators in the CWI pathway (29). As shown in Fig. 7C, GST-Mkk1 bound Slt2(274–373) in a CD-dependent manner, in contrast to that observed with GST-Msg5. These results indicate first that the CD domain present in this fragment is fully functional for binding cognate interactors and, second, that both Mkk1 and Msg5 interact with Slt2 through CD-dependent and independent binding mechanisms, respectively.
Identification of a Novel Motif within Msg5 Involved in Binding to Slt2 and Mlp1
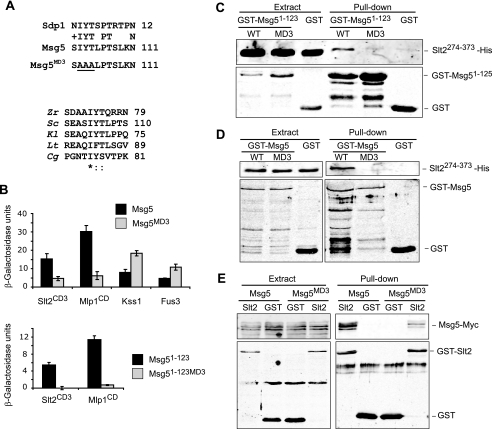
Because Sdp1, another DSP acting on Slt2, lacks the characteristic N-terminal regulatory domain where typical D domains reside, we hypothesized that its binding to this MAPK could be mediated by a similar mechanism as that operating in Msg5. Therefore, we searched for motifs in the first 123 amino acids of Msg5 that were also present in Sdp1. BLAST analysis revealed a short region of homology spanning residues 2–12 in Sdp1 and residues 101–111 in Msg5 (Fig. 8A). The IYT motif within this region is also conserved in Msg5 orthologs from other fungal species, suggesting that it may play a relevant role in the interaction with Slt2.
FIGURE 8.
The MD3 motif mediates binding of Msg5 to Slt2 and Mlp1. A, top, sequence comparison by BLAST analysis between Msg5 and Sdp1 revealed a similar region in the N-terminal part of these two proteins. The area corresponding to the putative Msg5 motif MD3 is shown here. Amino acidic changes introduced in the mutated version Msg5MD3 are underlined. Bottom, multiple alignment of the Msg5 amino acid sequence stretch bearing the MD3 motif with several Msg5 orthologs from different fungi. Alignment was performed by ClustalW (available on the World Wide Web). Identical residues are indicated by asterisks. Dots mark similar residues. Zr, Zygosaccharomyces rouxii; Sc, S. cerevisiae; Kl, Kluyveromyces lactis; Lt, Lachancea thermotolerans; Cg, Candida glabrata. B, semiquantitative analysis of the two-hybrid interaction of diploid cells containing plasmids that encode the indicated proteins. Before mating, pGBKT7, pGBKT7-Msg5, pGBKT7-Msg5MD3, pGBKT7-Msg5(1–123), or pGBKT7-Msg5(1–123)MD3 were transformed into PJ69-4A cells, and pGADT7, pGADT7-Kss1, pGADT7-Fus3, pGADT7-Slt2CD3, or pGADT7-Mlp1CD were transformed into PJ69-4α cells. Two-hybrid assays were performed as indicated in Fig. 1B. C, Western blot analysis of the in vitro co-purification of recombinant GST-Msg5(1–123) (WT) and GST-Msg5(1–123)MD3 (MD3) with Slt2(274–373)-His. E. coli extracts containing GST fusions were incubated with E. coli extracts containing Slt2(274–373)-His and glutathione-Sepharose to pull down GST complexes. Immunodetection was performed using anti-poly-His (top) and anti-GST (bottom) antibodies. D, Western blot analysis of the in vitro co-purification of recombinant GST-Msg5 (WT) and GST-Msg5MD3 (MD3) with Slt2(274–373)-His. E. coli extracts containing GST fusions were incubated with E. coli extracts containing Slt2(274–373)-His and glutathione-Sepharose to pull down GST complexes. Immunodetection was performed as in C. E, Western blot analysis of Msg5–6Myc pulled down by GST-Slt2. Transformants of the yeast DD1-2D strain with YCplac22MSG5m and pEG(KG) (GST) or pEG(KG)-SLT2 (Slt2) were grown to mid-log phase in selective medium at 24 °C, cells were collected, cell extracts were prepared, and GST-Slt2 complexes obtained by purification with glutathione-Sepharose were analyzed by immunoblotting with anti-Myc (top) and anti-GST (bottom) antibodies. In all cases, reproducible results were obtained in different experiments, and selected images correspond to representative blots.
In order to address the involvement of this putative motif (MD3) of Msg5 in Slt2 binding, the conserved Ile102, Tyr103, and Thr104 were changed to alanine in both full-length Msg5 and Msg5(1–123), and the interaction with Slt2, Fus3, Kss1, and Mlp1 was assayed by two-hybrid experiments. We confirmed that mutant hybrid proteins were expressed to a similar extent as the wild type versions (supplemental Fig. S1). As shown in Fig. 8B, in contrast to that observed for Fus3 and Kss1, loss of MD3 reduced the ability of full-length Msg5 to bind both Slt2 and Mlp1 in the two-hybrid system. Moreover, mutation of MD3 within Msg5(1–123) completely abrogated two-hybrid interaction of this fragment with these two kinases. For these assays, we used the CD-mutated versions of Slt2 (Slt2CD3) and Mlp1 (Mlp1CD) to confirm that no CD-dependent mechanism was operating in MD3-mediated binding.
The role of MD3 as a binding motif was confirmed by both in vitro and in vivo co-purification experiments. As shown in Fig. 8, C and D, E. coli-produced MD3-mutated forms of both GST-Msg5 and GST-Msg5(1–125) lost the ability to interact in vitro with the E. coli-produced Slt2(274–373) fragment. Moreover, compared with Msg5 wild type, a significant reduction in the amount of Msg5MD3 pulled down by GST-Slt2 from yeast extracts was also observed (Fig. 8E). All of these results indicate the involvement of this novel Msg5 MD3 motif in the interaction with Slt2 and Mlp1.
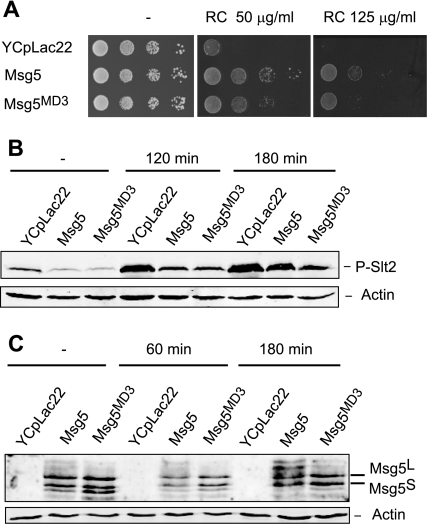
To analyze the physiological consequences of the binding mediated by the MD3 motif, we tested different CWI pathway- associated phenotypes in msg5Δ cells expressing either the Msg5 or the Msg5MD3 mutant proteins. As shown in Fig. 9A, cells expressing Msg5MD3 showed an intermediate Congo red sensitivity between the msg5 deletant and the wild type strain, indicating that this mutant protein is not able to fully complement the lack of Msg5.
FIGURE 9.
Effect of the lack of Msg5 MD3 motif on the CWI pathway. A, sensitivity to Congo red of msg5Δ (DD1-2D) cells transformed with the vector YCplac22m or plasmid YCplac22MSG5m (Msg5) or YCplac22MSG5MD3m (Msg5MD3). Cells were grown in liquid selective medium at 24 °C, and a 10-fold dilution series of this culture was spotted onto YPD agar medium in the absence (−) or presence of the indicated concentration of Congo red and incubated for 2 days at 24 °C. B, Western blotting analysis of the effect of the Msg5 MD3 mutation on Slt2 phosphorylation. The same transformants as in A were grown to mid-log phase in selective medium at 24 °C, and then aliquots were treated or not with Congo red (30 μg/ml) for 120 and 180 min. Protein extracts were prepared, and phospho-Slt2 and actin (as a loading control) were detected by immunoblot analysis with anti-phospho-p42/44 and anti-actin antibodies, respectively. C, Western blotting analysis of the effect of the Msg5 MD3 mutation on Msg5 phosphorylation. The same transformants as in A were grown to mid-log phase in selective medium at 24 °C, and then aliquots were either left untreated or exposed to Congo red (30 μg/ml) for 60 and 180 min. Proteins extracts were prepared, and Msg5–6Myc or Msg5MD3-6Myc and actin (as a loading control) were detected by immunoblotting analysis with anti-Myc and anti-actin antibodies, respectively. In all cases, reproducible results were obtained in different experiments, and selected images correspond to representative blots.
However, when we analyzed Slt2 phosphorylation, we observed that both mutant and wild type Msg5 reduced to the same extent the phosphorylation of Slt2 displayed by the msg5 deletant both in basal and stimulating conditions (Fig. 9B). No differences were detected in the expression of several CWI-regulated genes as determined by quantitative PCR in the absence of stimulus. This was not an unexpected result, because we have previously shown that although loss of Msg5 in such conditions causes a significant increase in the phosphorylation of Slt2, this results in only a weak transcriptional response (25). Nevertheless, no significant transcriptional differences between the MD3 mutant and the wild type were detected upon stimulation of the pathway (supplemental Fig. S2). Interestingly, a significant reduction in the phosphorylation of Msg5 by Slt2 upon pathway activation was clearly observed in the Msg5MD3 mutant protein as compared with the wild type (Fig. 9C). This indicates that the interaction mediated by this motif is critical for this Slt2-mediated post-translational modification of Msg5 in response to stimuli that activate the CWI pathway.
DISCUSSION
Many studies have demonstrated that MAPK phosphatases can act as negative regulators of multiple distinct MAPK pathways (20). How they manage to act specifically on the appropriate substrate under given conditions is one of the key issues in understanding their role on signaling. In this context, it is essential to know the mechanisms that mediate the interaction between MKPs and MAPKs. With this goal in mind, we have studied the physical interaction of the MKP Msg5 with distinct yeast MAPKs using yeast two-hybrid and in vitro binding analyses. Whereas certain mammalian MKPs exemplified by DUSP1(MKP-1) have been shown to bind and down-regulate MAPKs from all three major subfamilies (40), we have found that Msg5 only interacts with ERK-type MAPKs, showing high substrate specificity for the ERK1/2 and ERK5 homologs Fus3/Kss1 and Slt2, respectively. Interestingly, the yeast DSP Sdp1 also down-regulates Slt2 (28, 41), but no yeast DSP has so far been shown to act on the p38 homolog Hog1. Therefore, a different evolutionary outcome for p38 kinase regulation seems to have occurred in mammalian cells and yeast, in which Hog1 seems to be exclusively dephosphorylated by serine-threonine-specific and tyrosine-specific phosphatases.
The phosphatase catalytic domain recognizes phosphorylated sites in the target protein. However, this recognition is usually insufficient for providing specificity toward the substrate. Binding of classical MKPs to a target MAPK is mainly mediated through critical interaction motifs known as docking domains, which confer substrate specificity (40). Our work provides novel insights into the mechanisms that mediate the interaction of Msg5, a phosphatase that plays an essential role in controlling signaling through mating and CWI pathways with their target MAPKs.
Our two-hybrid data confirm the results obtained by Remenyi et al. (15), indicating the existence of a D domain in the N terminus of Msg5 that mediates binding to Fus3 and Kss1. This D domain presents the two major elements found in nearly all MAPK docking motifs: a cluster of basic residues at the N terminus, interacting with the negatively charged surface on MAPKs at the CD motif, and a hydrophobic motif near the C terminus that binds a series of shallow hydrophobic pockets on the kinase surface. These authors showed that a version of Msg5 carrying alanine substitutions in both basic and hydrophobic residues had reduced activity in attenuating Fus3-mediated mating pathway output. Our results show that mutation of arginine 29, lysine 32, and leucines 34 and 36 abolishes the interaction with Fus3 and Kss1 but also that mutation of leucines 34 and 36 alone decreases the activity of Msg5 toward Fus3, highlighting the importance of these hydrophobic residues for binding between these two proteins. The importance of hydrophobic interactions mediated by D sites has been described previously for other MAPK interactions (42).
We have also shown that Msg5 uses a D domain-independent mechanism to bind Slt2 and Mlp1 kinases. It is important to note that not all mammalian DSPs bind to MAPKs through D domains and therefore via the CD site of MAPKs. For example, it has been shown that the CD domain of p38 and JNK is not involved in binding and deactivation by the DSP LMW-DSP2 (43). However, in contrast to Msg5, this mammalian protein phosphatase does not contain a canonical D domain or even the regulatory N-terminal domain found in typical MKPs. In addition, it has been shown that a DEF motif can contribute to binding of the mammalian DUSP1/MKP-1 to active ERK2 (44). However, Msg5 does not contain a DEF consensus FX(F/Y)P sequence within the N-terminal region. Here we show that Msg5 binding to Slt2 and Mlp1 is not mediated by a second putative D domain lacking the consensus LXL submotif found in the N-terminal region of Msg5. Therefore, despite having a fully functional D domain that interacts with Fus3 and Kss1 MAPKs, an alternative mechanism for binding the two CWI-related kinases is used by this MKP. Interestingly, mutation of the KIM domain of DUSP1/MKP-1 prevents the interaction of this phosphatase with both ERK1/2 and p38 MAPKs in mammalian cells. In contrast, both the binding of DUSP1/MKP-1 to JNK and its ability to inactivate this kinase are totally unaffected by loss of the KIM, indicating that a different site within DUSP1/MKP-1 is involved in binding to JNK (45).
Our findings demonstrate that a motif consisting of 102IYT104 is involved in Msg5 interaction with the CWI kinases Slt2 and Mlp1. Remarkably, this motif is located close to the D domain that mediates binding to Fus3 and Kss1. This reinforces the importance of the N-terminal domain of Msg5 and generally of MKPs for binding to their substrates, regardless of the interaction mechanism. Although the Msg5 IYT motif is situated next to the two arginines 96RR97, it is not conserved in mammalian phosphatases that possess a consensus D domain, like PTP-SL, LC-PTP, and STEP (see Fig. 3A). In contrast, this motif is present in Sdp1, the other MKP known to act on Slt2, as well as in Msg5 orthologs from other yeast species. This suggests that this motif participates in a novel conserved binding mechanism. It is noteworthy that Sdp1, which possesses a short N-terminal region lacking the characteristic D domains, has been shown to interact with Slt2 but not with the other yeast MAPKs (28).
The use of different binding modes or determinants of MAPK recognition might allow Msg5 to differentially modulate the activities of “mating” or “cell integrity” kinases, depending on specific cellular situations. In fact, post-translational modifications, such as phosphorylation, have been described for Msg5 as a consequence of CWI pathway activation. This phosphorylation is likely to differentially modulate Msg5 binding to CWI or mating-related MAPKs. In support of this, the interaction of Msg5 with Slt2 is reduced under conditions that activate the CWI pathway (23), which is consistent with the characteristic absence of adaptation showed by this pathway. This does not seem to be the case for the mating pathway, in which the action of Msg5 is important for adaptation after pheromone stimulation (21). Our results indicate that binding between Msg5 and Slt2 through the IYT motif can provide specificity in Msg5 phosphorylation by Slt2, because a version of Msg5 that lacks this motif shows a deficient phosphorylation upon CWI pathway stimulation.
In addition, as in the case of Sdp1, Msg5 can be subjected to redox regulation. Formation of an intramolecular disulfide bond under oxidative conditions leads to both activation and selective recognition of tyrosine-phosphorylated MAPK substrates (46). The existence of distinct binding mechanisms could also allow differential modulation of Msg5 action toward distinct MAPKs following oxidative stress.
We have also studied the molecular mechanisms mediating the interaction of Slt2 with different regulators. We show that Slt2 binds its activator Mkk1 through the MAPK CD domain. Bearing in mind that D domains mediate interaction of Slt2 with other CWI pathway components, including activators like the Mkk1-redundant MAPK kinase Mkk2 (29) and substrates like Rlm1 (47) or Swi4 (48), this indicates that Slt2 preferentially uses this prototypic interaction mechanism for binding to activators and substrates. However, our work also shows that this kinase is able to exploit other mechanisms for binding to regulators, such as Msg5. The fact that peptide versions of D domains from different MAPK-interacting proteins inhibit MEK-mediated phosphorylation or MKP-mediated dephosphorylation (49) suggests that activators and down-regulators compete for binding to the same CD sites of cognate MAPKs. Interestingly, the binding determinants within Slt2 for Mkk1 and Msg5 are different but proximally located, allowing possible steric hindrance in the interactions of these proteins with the MAPK. In this way, Slt2 would benefit from the existence of a specific Msg5 interaction mechanism without losing the possibility of competition among distinct MAPK partners. It has been recently shown that MAPK activation triggers dynamic changes within docking sites (50). Therefore, the presence of distinct binding sites in Slt2 could be a means to differentially regulate interaction with Msg5 and Mkk1/2 following Slt2 phosphorylation.
In addition to substrate recruitment, docking interactions have been implicated in allosteric control of MAPK activity. Binding of MAPKs to docking motifs of their partners may cause MAPKs to change their conformation and expose their activation loop (51). For example, binding of Ste5 to Fus3, which occurs through two sites of the kinase, including the D-site-interacting groove, induces partial autophosphorylation and activation of Fus3 (52). It is interesting to note that the Slt2 region mediating binding to Msg5 includes the MAPK L16 loop. This loop, as well as mediating MAPK homodimerization (53), has been regarded as a molecular switch that promotes MAPK activation in response to conformational change (14). Therefore, the possibility that binding of Msg5 to Slt2 could affect its activity by an allosteric change cannot be ruled out. In addition, the novel FRIEDE interaction motif identified in the atypical ERK3 and ERK4 MAPKs as essential for binding to its substrate MK5 is also located within the L16 loop (13). These findings highlight the importance of this region in MAPK function. Fungal MAPKs operating in conserved CWI pathways possess an autoinhibitory extension, located C-terminal to this region (38). It is plausible that non-canonical binding could affect the function of this C-terminal extension in a way different from binding through classical CD domain-mediated docking interactions. Therefore, this may represent a novel binding mechanism shared by fungal CWI MAPKs bearing C-terminal extensions.
These data, together with the existence of two isoforms of Msg5 and MAPK-dependent post-translational modifications (23, 25), emphasize the complexity of Msg5 regulation and illustrate different modes used by phosphatases to interact with the MAPKs they inactivate. In this case, the same protein phosphatase is able to bind to four different MAPKs. It uses either classical docking-mediated interactions (with Fus3 and Kss1) or the novel IYT motif-mediated binding mechanism (with Slt2 and Mlp1) here described, in which no canonical docking domains are involved. It is also remarkable that Mlp1, a pseudokinase bearing the tyrosine but not the threonine at the activation loop, is able to interact with Msg5 using the same mechanism as Slt2. This suggests the existence of competition between these related kinases to bind Msg5 that could modulate CWI pathway activation. Elucidation of the molecular mechanisms mediating protein-protein interactions between key signaling regulators and their targets in MAPK pathways may facilitate the design of substrate-selective therapeutic drugs.
Supplementary Material
Acknowledgments
We thank Dr. McStay Marta Flández, Eva Tapia, and Inmaculada Cosano for materials used in this study. We also thank people from Unit 3 of the Departamento de Microbiología II at the Universidad Complutense de Madrid for useful comments and discussion throughout the work. We acknowledge the Unidad de Genómica y Proteómica (PTM/UCM, Madrid, Spain) for DNA sequencing and especially Jesús G. Cantalejo for the quantitative PCR.
This work was supported by Ministerio de Ciencia e Innovación (Spain) Grants BIO2007-67299 and BIO2010-22369-C02-01 and Program for UCM Research Groups Grant 920628 from BSCH-UCM (to M. M.). Research in the CR-UK Stress Response Laboratory (to R. J. D., M. P. D., and S. M. K.) is supported by Cancer Research UK Program Grant C8227/A12053 (to S. M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I and II and Figs. S1 and S2.
- MKP
- MAPK phosphatase
- KIM
- kinase interaction motif
- CD
- common docking
- DSP
- dual specificity phosphatase
- CWI
- cell wall integrity
- SD
- synthetic dextrose.
REFERENCES
- 1. Turjanski A. G., Vaqué J. P., Gutkind J. S. (2007) Oncogene 26, 3240–3253 [DOI] [PubMed] [Google Scholar]
- 2. Coulombe P., Meloche S. (2007) Biochim. Biophys. Acta 1773, 1376–1387 [DOI] [PubMed] [Google Scholar]
- 3. Tanoue T., Nishida E. (2002) Pharmacol. Ther. 93, 193–202 [DOI] [PubMed] [Google Scholar]
- 4. Grewal S., Molina D. M., Bardwell L. (2006) Cell. Signal. 18, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bardwell L., Shah K. (2006) Methods 40, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akella R., Moon T. M., Goldsmith E. J. (2008) Biochim. Biophys. Acta 1784, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayor F., Jr., Jurado-Pueyo M., Campos P. M., Murga C. (2007) Cell Cycle 6, 528–533 [DOI] [PubMed] [Google Scholar]
- 8. Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 9. Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 10. Tanoue T., Yamamoto T., Nishida E. (2002) J. Biol. Chem. 277, 22942–22949 [DOI] [PubMed] [Google Scholar]
- 11. Tanoue T., Nishida E. (2003) Cell. Signal. 15, 455–462 [DOI] [PubMed] [Google Scholar]
- 12. Lee T., Hoofnagle A. N., Kabuyama Y., Stroud J., Min X., Goldsmith E. J., Chen L., Resing K. A., Ahn N. G. (2004) Mol. Cell 14, 43–55 [DOI] [PubMed] [Google Scholar]
- 13. Aberg E., Torgersen K. M., Johansen B., Keyse S. M., Perander M., Seternes O. M. (2009) J. Biol. Chem. 284, 19392–19401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diskin R., Lebendiker M., Engelberg D., Livnah O. (2007) J. Mol. Biol. 365, 66–76 [DOI] [PubMed] [Google Scholar]
- 15. Reményi A., Good M. C., Bhattacharyya R. P., Lim W. A. (2005) Mol. Cell 20, 951–962 [DOI] [PubMed] [Google Scholar]
- 16. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 17. Chen R. E., Thorner J. (2007) Biochim. Biophys. Acta 1773, 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caffrey D. R., O'Neill L. A., Shields D. C. (1999) J. Mol. Evol. 49, 567–582 [DOI] [PubMed] [Google Scholar]
- 19. Truman A. W., Millson S. H., Nuttall J. M., King V., Mollapour M., Prodromou C., Pearl L. H., Piper P. W. (2006) Eukaryot. Cell 5, 1914–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martín H., Flández M., Nombela C., Molina M. (2005) Mol. Microbiol. 58, 6–16 [DOI] [PubMed] [Google Scholar]
- 21. Doi K., Gartner A., Ammerer G., Errede B., Shinkawa H., Sugimoto K., Matsumoto K. (1994) EMBO J. 13, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhan X. L., Deschenes R. J., Guan K. L. (1997) Genes Dev. 11, 1690–1702 [DOI] [PubMed] [Google Scholar]
- 23. Flández M., Cosano I. C., Nombela C., Martín H., Molina M. (2004) J. Biol. Chem. 279, 11027–11034 [DOI] [PubMed] [Google Scholar]
- 24. Andersson J., Simpson D. M., Qi M., Wang Y., Elion E. A. (2004) EMBO J. 23, 2564–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marín M. J., Flández M., Bermejo C., Arroyo J., Martín H., Molina M. (2009) Mol. Genet. Genomics 281, 345–359 [DOI] [PubMed] [Google Scholar]
- 26. Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 27. Martín H., Rodríguez-Pachón J. M., Ruiz C., Nombela C., Molina M. (2000) J. Biol. Chem. 275, 1511–1519 [DOI] [PubMed] [Google Scholar]
- 28. Collister M., Didmon M. P., MacIsaac F., Stark M. J., MacDonald N. Q., Keyse S. M. (2002) FEBS Lett. 527, 186–192 [DOI] [PubMed] [Google Scholar]
- 29. Jiménez-Sánchez M., Cid V. J., Molina M. (2007) J. Biol. Chem. 282, 31174–31185 [DOI] [PubMed] [Google Scholar]
- 30. García R., Bermejo C., Grau C., Pérez R., Rodríguez-Peña J. M., Francois J., Nombela C., Arroyo J. (2004) J. Biol. Chem. 279, 15183–15195 [DOI] [PubMed] [Google Scholar]
- 31. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32. Kim K. Y., Truman A. W., Levin D. E. (2008) Mol. Cell Biol. 28, 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pulido R., Zúñiga A., Ullrich A. (1998) EMBO J. 17, 7337–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zúñiga A., Torres J., Ubeda J., Pulido R. (1999) J. Biol. Chem. 274, 21900–21907 [DOI] [PubMed] [Google Scholar]
- 35. Muñoz J. J., Tárrega C., Blanco-Aparicio C., Pulido R. (2003) Biochem. J. 372, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camps M., Nichols A., Gillieron C., Antonsson B., Muda M., Chabert C., Boschert U., Arkinstall S. (1998) Science 280, 1262–1265 [DOI] [PubMed] [Google Scholar]
- 37. Kusari A. B., Molina D. M., Sabbagh W., Jr., Lau C. S., Bardwell L. (2004) J. Cell Biol. 164, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levin-Salomon V., Maayan I., Avrahami-Moyal L., Marbach I., Livnah O., Engelberg D. (2009) Biochem. J. 417, 331–340 [DOI] [PubMed] [Google Scholar]
- 39. Kim K. Y., Cosano I. C., Levin D. E., Molina M., Martín H. (2007) Yeast 24, 335–342 [DOI] [PubMed] [Google Scholar]
- 40. Patterson K. I., Brummer T., O'Brien P. M., Daly R. J. (2009) Biochem. J. 418, 475–489 [DOI] [PubMed] [Google Scholar]
- 41. Hahn J. S., Thiele D. J. (2002) J. Biol. Chem. 277, 21278–21284 [DOI] [PubMed] [Google Scholar]
- 42. Chang C. I., Xu B. E., Akella R., Cobb M. H., Goldsmith E. J. (2002) Mol. Cell 9, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 43. Aoyama K., Nagata M., Oshima K., Matsuda T., Aoki N. (2001) J. Biol. Chem. 276, 27575–27583 [DOI] [PubMed] [Google Scholar]
- 44. Lin Y. W., Yang J. L. (2006) J. Biol. Chem. 281, 915–926 [DOI] [PubMed] [Google Scholar]
- 45. Slack D. N., Seternes O. M., Gabrielsen M., Keyse S. M. (2001) J. Biol. Chem. 276, 16491–16500 [DOI] [PubMed] [Google Scholar]
- 46. Fox G. C., Shafiq M., Briggs D. C., Knowles P. P., Collister M., Didmon M. J., Makrantoni V., Dickinson R. J., Hanrahan S., Totty N., Stark M. J., Keyse S. M., McDonald N. Q. (2007) Nature 447, 487–492 [DOI] [PubMed] [Google Scholar]
- 47. Jung U. S., Sobering A. K., Romeo M. J., Levin D. E. (2002) Mol. Microbiol. 46, 781–789 [DOI] [PubMed] [Google Scholar]
- 48. Truman A. W., Kim K. Y., Levin D. E. (2009) Mol. Cell Biol. 29, 6449–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bardwell L. (2006) Biochem. Soc. Trans. 34, 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodriguez Limardo R. G., Ferreiro D. N., Roitberg A. E., Marti M. A., Turjanski A. G. (2011) Biochemistry 50, 1384–1395 [DOI] [PubMed] [Google Scholar]
- 51. Zhou T., Sun L., Humphreys J., Goldsmith E. J. (2006) Structure 14, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 52. Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M., Lim W. A. (2006) Science 311, 822–826 [DOI] [PubMed] [Google Scholar]
- 53. Khokhlatchev A. V., Canagarajah B., Wilsbacher J., Robinson M., Atkinson M., Goldsmith E., Cobb M. H. (1998) Cell 93, 605–615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.