Background: Subtilisin-like proteases from Dichelobacter nodosus are important enzymes required for footrot pathogenesis.
Results: The S1 pockets of virulent and benign basic proteases underpin their differential proteolytic activity.
Conclusion: The virulent BprV protease possesses key properties in S1 pocket required for effective tissue destruction.
Significance: A bacterial pathogen can exploit subtle amino acid changes in proteases to mediate disease pathogenesis.
Keywords: Bacterial Pathogenesis, Crystal Structure, Extracellular Matrix, Serine Protease, Virulence Factors, Footrot, Subtilisin-like Proteases
Abstract
The ovine footrot pathogen, Dichelobacter nodosus, secretes three subtilisin-like proteases that play an important role in the pathogenesis of footrot through their ability to mediate tissue destruction. Virulent and benign strains of D. nodosus secrete the basic proteases BprV and BprB, respectively, with the catalytic domain of these enzymes having 96% sequence identity. At present, it is not known how sequence variation between these two putative virulence factors influences their respective biological activity. We have determined the high resolution crystal structures of BprV and BprB. These data reveal that that the S1 pocket of BprV is more hydrophobic but smaller than that of BprB. We show that BprV is more effective than BprB in degrading extracellular matrix components of the host tissue. Mutation of two residues around the S1 pocket of BprB to the equivalent residues in BprV dramatically enhanced its proteolytic activity against elastin substrates. Application of a novel approach for profiling substrate specificity, the Rapid Endopeptidase Profiling Library (REPLi) method, revealed that both enzymes prefer cleaving after hydrophobic residues (and in particular P1 leucine) but that BprV has more restricted primary substrate specificity than BprB. Furthermore, for P1 Leu-containing substrates we found that BprV is a significantly more efficient enzyme than BprB. Collectively, these data illuminate how subtle changes in D. nodosus proteases may significantly influence tissue destruction as part of the ovine footrot pathogenesis process.
Introduction
Ovine footrot is a highly contagious disease that causes significant economic losses to the worldwide sheep industry, mainly due to its impact on wool and meat production (1). The disease varies in severity, ranging from a mild interdigital dermatitis (benign footrot) to a necrotic separation of the horn of the hoof from the underlying soft tissue (virulent footrot) (2, 3). Footrot is caused by the pathogenic bacterium Dichelobacter nodosus, a Gram-negative anaerobic rod that is classified into virulent and benign strains according to the severity of the disease (2, 4–6).
Both virulent and benign strains of D. nodosus secrete a number of subtilisin-like serine proteases (subtilases) (5, 6). These proteases have recently been shown to play a pivotal role in the pathogenesis of footrot through their ability to degrade components of the extracellular matrix (ECM)5 (7), which is consistent with the extensive tissue damage observed at the infected lesion. Because D. nodosus has a poor capacity to synthesize amino acids it is likely that these proteases function to degrade proteinaceous substrates, enabling an important source of amino acids and energy to be provided to the bacterium in the infected lesion (8). Proteolytic processing, a general feature of the biosynthesis of functional subtilisin-like proteases (9), is required to yield the final mature D. nodosus subtilase enzymes. This involves the removal of a signal sequence, a separate proregion, and the C-terminal domain (10, 11).
In virulent strains of D. nodosus, three subtilases are synthesized and secreted: the acidic proteases AprV2 and AprV5 and the basic protease BprV. Benign strains of D. nodosus secrete the closely related subtilases AprB2, AprB5, and BprB, which differ in sequence from their virulent counterparts by only a few amino acids (10–14). Importantly, these subtilases have been shown to play an important role in the virulence of D. nodosus (7). Specifically, an aprV2 mutant was shown to be avirulent in an ovine footrot model, with virulence restored by complementation with the wild-type aprV2 gene. Similar studies were carried out on AprV5 and BprV (7), and although the respective mutants were avirulent, the complementation studies did not enable definitive conclusions to be drawn regarding the role of these enzymes in disease.
Previous biochemical analysis had shown that subtilases from the virulent strains display higher elastinolytic and caseinolytic activity than those produced by the benign strains (15–17), presumably as a result of the small number of sequence differences between the catalytic domains of virulent and benign subtilases. The most divergent of these enzymes are the basic proteases. There are nine amino acid sequence variations between the catalytic domains of BprV and BprB. At present, the molecular basis for the difference in biochemical activity between these two enzymes and their contribution to footrot pathogenesis have not been established (6).
In this study, we show that ovine hoof keratin can be efficiently degraded by the basic proteases of D. nodosus. Our biochemical characterization of these enzymes further revealed that BprV is more effective than BprB in degrading components of the ECM. Crystal structure determination, along with biochemical analysis of mutants with key amino acid substitutions, established that the S1 pocket of these proteases is the key determinant for their difference in activity. Primary substrate specificity profiling indicated that the two proteases have a similar substrate specificity, but differ in their kinetics for cleavage of peptide substrates. BprV is more effective in cleaving P1 Leu-containing peptides, whereas the benign protease BprB is more effective in cleaving P1 Phe-containing peptides. These data are consistent with differences in the size and hydrophobic nature of their respective S1 pockets. Analysis of the amino acid content of ECM proteins reveals that leucine occurs in a higher abundance compared with phenylalanine, providing one possible explanation for the higher proteolytic activity of BprV against ECM-related substrates. Collectively, this study provides molecular insight into the mechanism of footrot pathogenesis, whereby differences in the properties of the S1 pocket of a bacterially derived protease may underpin the severity of tissue destruction.
EXPERIMENTAL PROCEDURES
Protein Production, Crystallization, and Data Collection
BprV and BprB were purified and crystallized as described previously (18). Expression plasmids and mutagenesis primers used to prepare the protease mutants are reported in supplemental Tables S1 and S2. Data collection statistics for BprV and BprB have been reported previously (18) and are reproduced in supplemental Table S3.
Structure Determination and Refinement and Structural Analysis
Unless stated otherwise, all programs used for structural and crystallographic analysis were located within the CCP4 interface (19) to the CCP4 suite (20). The structures of the BprV and BprB proteases were solved by molecular replacement using PHASER (21). The 1.7-Å crystal structure of the AprB2 protease (3LPC) (7) was used as a search model for BprB, whereas the refined BprB structure was used as a search model for BprV. Chainsaw was used to truncate all nonconserved side chains with the exception of alanine and glycine, to the Cγ (22). Both structures were completed by cycling between manual building and refinement. Manual building and maximum likelihood refinement were carried out using COOT (23) and REFMAC5 (24), respectively. Water molecules were added to all models using ARP/warp (25). Structure validation was carried out using MOLPROBITY (26) and COOT (23). Refinement statistics are presented in Table 1. For calculation of the S1 pocket volumes of the basic proteases, the web tool CASTp was used (27). The coordinates and structure factors are available from the Protein Data Bank (BprV; 3TI7, BprB; 3T19). Raw data and images are available from TARDIS (28).
TABLE 1.
Structure refinement statistics for BprV and BprB
| Parameters | BprV | BprB |
|---|---|---|
| Space group | P21 | P21 |
| Unit cell | ||
| a (Å) | 38.5 | 38.5 |
| b (Å) | 89.6 | 90.5 |
| c (Å) | 47.7 | 44.1 |
| β (°) | 113.6 | 109.0 |
| Resolutiona (Å) | 44.8 − 2.0 | 45.3 − 1.8 |
| Rcryst (%) | 16.3 | 15.6 |
| Rfreefree (%) | 22.6 | 20.7 |
| Nonhydrogen atoms | 2,805 | 2,970 |
| Solvent | 314 | 460 |
| Ramachandran plot | ||
| Favored region (%) | 97.0 | 96.8 |
| Additionally allowed region (%) | 2.7 | 2.9 |
| Generously allowed region (%) | 0 | 0 |
| Disallowed region (%) | 0.3 | 0.3 |
| r.m.s.d. from ideality | ||
| Bond length (Å) | 0.009 | 0.008 |
| Bond angles (º) | 1.153 | 1.054 |
| B factors | ||
| Mean main chain (Å2) | 14.4 | 9.8 |
| Mean side chain (Å2) | 16.3 | 11.7 |
| Mean water molecule (Å2) | 23.4 | 22.1 |
| r.m.s.d. bonded B factor | ||
| Main chain | 0.6 | 0.5 |
| Side chain | 2.0 | 1.7 |
a R = Σ |Fobs − Fcal|/ΣFobs, where Rfree is calculated with the 5% of data omitted from the refinement and Rcryst with the remaining 95% of the data included in the refinement.
Proteolytic Digestion of Hoof Material and Substrate Identification by in-Gel Tryptic Digestion and LC-MS
Hoof material (14% (w/v)) from a disease-free sheep was incubated with 100 μg/ml purified recombinant protease at 37 °C in assay buffer (20 mm Tris-HCl, pH 8, and 5 mm CaCl2) for 1, 2, 4, 6, 8, and 24 h. The degraded products were separated on 12% SDS-PAGE. Major bands from samples obtained at 24 h were excised and subjected to in-gel tryptic digestion by incubating the excised samples with 0.5 μg of trypsin at 37 °C for 24 h. Tryptic digests were analyzed by LC-tandem MS using a HCT ULTRA ion trap mass spectrometer (Bruker Daltonics) coupled online with a 1200 series capillary HPLC (Agilent Technologies). Proteins were identified by searching the LC-tandem MS data against the National Center for Biotechnology Information (NCBI) nonredundant and Swiss-Prot databases using the MASCOT search engine (version 2.1; Matrix Science) with all taxonomy selected.
In Vitro Cleavage of Fibronectin
Recombinant protease (50 nm) was incubated with 1 μm human fibronectin (BD Biosciences) in assay buffer (20 mm Tris, pH 8, 5 mm CaCl2) at 25 °C for 5, 10, 15, 30, 45 min, and 48 h. The cleavage reaction was stopped by incubating the reaction samples in SDS loading buffer at 95 °C for 5 min. The cleavage products of fibronectin were visualized after SDS-PAGE by staining with Coomassie Blue. Densitometry analysis was performed using a GS-800 calibrated densitometer (Bio-Rad) and software, Quantity One 4.6.9.
Congo Red Elastin Digestion Assay
Elastin Congo Red (5 mg; Sigma) was incubated with 5 μm recombinant protease in assay buffer (150 mm NaCl, 25 mm BisTris, pH 6.5, 5 mm CaCl2, 10% glycerol) at 37 °C for 5 h. Quantitative measurement of elastase activity was carried out as described previously (29).
Determination of Kinetic Constants for Cleavage of AAPVn
Recombinant protease (1 μm) was incubated with N-methoxysuccinyl-Ala-Ala-Pro-Val p-Nitroanilide (AAPVn) (0.5, 1, 2, 4, 6, 8, and 10 mm; Sigma) in assay buffer (20 mm Tris, pH 8, 5 mm CaCl2) at 37 °C for 20 h. The increase in absorbance at 405 nm, as a result of free nitroanilide in solution, was monitored at 5-min intervals for 20 h. Initial velocities determined from the linear portion of the curves were plotted against AAPVn concentration and fitted by nonlinear regression to the Michaelis-Menten equation to allow determination of Km and Vmax values using GraphPad Prism 5.
Determination of the Substrate Specificity of Purified Subtilases Using the REPLi Library
The REPLi library (Mimotopes) is a peptide library that allows rapid identification of peptide substrates cleaved by the protease of interest and thus the determination of the specificity of the enzyme (30). It contains 512 pools of peptides with each pool containing up to eight different variable tripeptides with the template layout of MeOC-GGXXXGG-DPA-KK, where each X represents a variable alternative amino acid based on similar physiochemical properties, i.e. A/V, F/Y, I/L, D/E, R/K, D/E, S/T, Q/N, and P. There are no Gly, His, Trp, Cys, or Met residues in the variable tripeptide region. The resulting combinations of variable tripeptides give rise to 3375 different peptides in the library in total. Methoxycarbonyl (MeOC) is the fluorophore, and dipicolinic acid (DPA) is the fluorophore quencher. Protease (BprV or BprB; 10 nm) was incubated with 50 μm peptide substrate in each well in assay buffer (20 mm Tris, pH 8, 5 mm CaCl2) at 37 °C in a microtiter plate. Cleavage of the peptides resulted in the release of the DPA quencher, producing a fluorescence signal that was measured every 30 s for 30 cycles using excitation and emission wavelengths of 320 and 420 nm, respectively. The initial velocity of the reaction was obtained by measuring the slope of the linear region of the curves, allowing comparison of the cleavage rate between peptide pools.
P1 Specificity Determination for BprV and BprB Using Synthesized Peptides Identified from the REPLi Screen
The peptide pool containing peptides with the sequence MeOC-GG-I/L-A//V-F/Y-GG-DPA-KK was identified from the REPLi screen as having the highest rate of cleavage by BprV. Eight peptides representing the potential peptides present in this pool were synthesized individually (Table 2). Protease (70 nm) was incubated with varying concentrations of peptide substrate in assay buffer at 37 °C in a microtiter plate. Cleavage of peptide resulted in the release of the DPA quencher, producing a fluorescence signal. The fluorescence signal was measured as before every 14 s for 200 cycles. Initial velocities were determined from the linear portion of the curves, which were plotted against substrate concentration and fitted by nonlinear regression to the Michaelis-Menten equation to allow determination of Km and Vmax values using GraphPad Prism 5.
TABLE 2.
Activity of BprV, BprB, and mutants against the AAPVn peptide substrate
| Proteases | Km | Kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm·s−1 | |
| BprV | 3.13 ± 0.6 | 8.9 × 10−3 | 2.84 |
| BprB | 3.84 ± 0.5 | 6.4 × 10−3 | 1.67 |
| BprB D180G | 4.28 ± 0.44 | 7.4 × 10−3 | 1.73 |
| BprB D182G | 3.74 ± 0.51 | 7.9 × 10−3 | 2.11 |
| BprB D180G/D182G | 4.02 ± 0.52 | 8.9 × 10−3 | 2.21 |
The protease-treated peptide samples were subjected to mass spectrometry analysis for determination of the P1 specificity of the BprV and BprB proteases. The protease-treated peptide samples were co-spotted onto the MALDI target plate with Matrix solution of 10 mg/ml a-cyano-4-hydroxycinnamic acid (Laser BioLabs, Sophia-Antipolis, France) in 50% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. The samples were analyzed on an Applied Biosystems (Foster City, CA) 4700 Proteomics Analyzer MALDI TOF/TOF in reflectron mode with a mass range of 400–1500 Da, focus mass of 1000 Da at 1500 shots/spectrum using plate model calibration against 4700 tune mix. Peak detection on the spectra was performed using the software 4700 Series Explorer version 3.0.
RESULTS
X-ray Crystal Structure of BprV and BprB
To gain an insight into how the observed sequence variations between the enzymes might influence the proteolytic activity of BprV and BprB, we determined their respective crystal structures to 2-Å and 1.8-Å resolution, respectively (see Table 1 for structure refinement statistics). Both constructs comprised residues 1–344 together with a C-terminal histidine tag. Residues 2–341 (BprV) and 2–342 (BprB) could be modeled into the electron density. Several side chains were also disordered (Asp145 in BprB; Tyr144, Asp145, Gln171, and Arg189 in BprV) and were not included in the final model. Both structures contain three coordinated calcium ions and two disulfide bonds (Cys89-Cys141 and Cys183-Cys220). The overall structure of the two proteases was highly similar, with a r.m.s.d. of 0.58 Å over 329 Cα atoms. The sequence identity between BprV and another structurally characterized D. nodosus secreted subtilase, AprV2 (6), is 73% (r.m.s.d. of 0.56 Å over 339 Cα with 3LPA; supplemental Figs. S1 and S2). We will primarily describe the structures with reference to BprV.
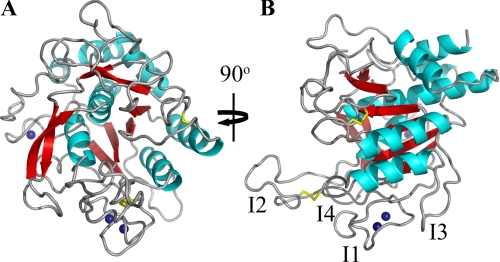
Briefly, BprV adopts the conserved subtilisin-like fold, which consists of a central six-stranded parallel β-sheet sandwiched between two and five α-helices (Fig. 1A). A two-stranded anti-parallel β-sheet runs perpendicular to the central β-sheet. The catalytic triad, which comprises Asp41, His105, and Ser277, is located at the C terminus of the β-sheet. Like AprV2 (7), both contain (Fig. 1B and supplemental Fig. S2) the I2 exosite loop, a region important for mediating enzyme-substrate interactions (7). Tyr92 on the I2 exosite loop is conserved in both BprV and BprB and has previously been shown to be an important determinant for efficient degradation of elastin (7).
FIGURE 1.
1.8 Å crystal structure of BprV. A, view of the BprV structure showing the conserved subtilisin-like fold. Disulfide bonds are shown as yellow sticks, and the three calcium ions are shown as blue spheres. B, view of BprV structure with the I1 to I4 loops labeled. The figure was prepared using PyMOL (31).
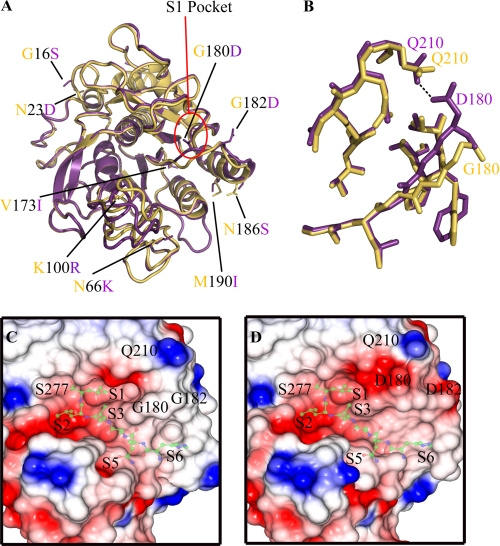
Sequence analysis and superimposition experiments revealed the location of the nine amino acid differences between the two proteases. Eight of these substitutions (GBprV16SBprB, N23D (α1-helix), N66K (I1 loop), K100R (I2 loop), G180D, G182D, N186S, and M190I (α4-helix)) were located on the surface of the proteases and were largely exposed to the solvent (Fig. 2A). The conservative substitution V173I formed part of the hydrophobic core of the protein and was buried between the β3-strand and α3-helix (Fig. 2A). Notably, only the G180D and G182D substitutions caused any significant change in backbone conformation. Furthermore, both of the G180D and G182D substitutions were located within the substrate binding groove of the enzymes.
FIGURE 2.
Structural variations between the BprV and BprB proteases. A, overlay between the BprV and BprB structures showing the location of the nine amino acid substitutions. BprV-specific residues are shown as yellow sticks, and BprB-specific residues are shown as purple sticks. The S1 pockets of the proteases are highlighted by a red circle. B, structural overlay of the residues that form the S1 pockets of BprV and BprB. The S1 pockets of BprV and BprB are shown as yellow and purple sticks, respectively. The S1 pockets of both proteases are formed by residues 176–180, 204–208, and 215–218. C and D, electrostatic potential surfaces of the S1 pockets of BprV (C) and BprB (D) showing the Gly180 and Gly182 residues of BprV and Asp180 and Asp182 residues of BprB. The catalytic residue Ser277 is labeled, as is the Gln210 residue with which Asp180 of BprB makes a hydrogen bond (shown). Neutral regions are colored white, acidic regions are colored red, and basic regions are colored blue. Semitransparent peptide substrates (ball and stick) have been modeled onto the substrate binding clefts of BprV and BprB and are included to help visualize the subsites of the two proteases. A and B were prepared using PyMOL (31), and C and D were prepared using CCP4MG (32, 33).
Comparison of the BprV and BprB structures revealed that the substitutions at residues 180 and 182 caused significant changes to the nature of the S1 substrate binding pocket (formed by residues 176–180, 204–208, and 215–218) (Fig. 2B and supplemental Fig. S3). Notably, Asp180 of BprB formed a hydrogen bond with Gln210, which resulted in a 2-Å shift of the loop relative to its equivalent position in BprV (Fig. 2B). This change resulted in a significantly smaller S1 pocket in BprV compared with BprB (volume of S1 pocket in BprV = 120 Å3, whereas in BprB = 145 Å3). In addition, the S1 pocket of BprV appeared to be more hydrophobic than BprB, which was relatively acidic (Fig. 2, C and D). Finally, the presence of the two glycine residues (Gly180 and Gly182) in the loop that forms the rim of the BrpV S1 site would be anticipated to enhance the flexibility of the pocket and its ability to accommodate substrates.
Differential Proteolytic Activity of BprV and BprB against ECM Substrates
Given the changes in and around the S1 pocket we investigated the activity of each enzyme against potential target substrates. Specifically, we tested the ability of each enzyme to degrade fibronectin, insoluble elastin, and hoof material isolated from a disease-free sheep. We also tested the ability of each enzyme to hydrolyze the peptide substrate, AAPVn, which has previously been used to characterize the enzymes.
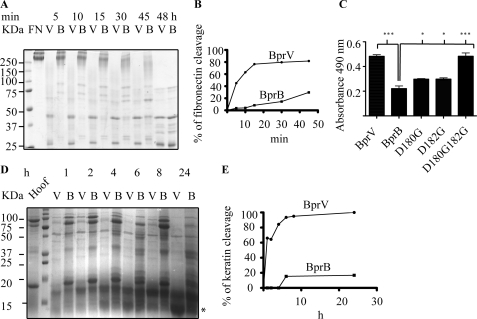
We found that the proteolytic activity of BprV on each substrate tested was significantly higher than BprB (Fig. 3). Notably, BprV was able to mediate the degradation of fibronectin 5-fold faster than BprB at the 30-min reaction time point (Fig. 3,A and B). Interestingly, the fibronectin digestion patterns produced by the two enzymes at the 48-h time point were similar, suggesting that the two proteases cleave the substrate at identical sites, but at different rates (Fig. 3A). The difference in the catalytic efficiencies of the two enzymes was further demonstrated by the ∼2-fold higher efficiency in substrate turnover of AAPVn than BprB (Table 2). The ability of BprV to degrade insoluble elastin was also 2-fold higher than BprB (Fig. 3C). Furthermore, BprV was shown to be more effective in degrading sheep hoof (Fig. 3D). To identify the major 12-kDa band that appeared on the SDS-PAGE as a result of hoof degradation mediated by the basic proteases, in-gel trypsin digestion followed by LC-MS analysis was conducted. The result showed that ovine keratin was the major hoof substrate targeted by both basic proteases for degradation and that BprV degraded keratin 6-fold faster than BprB at the 6-h reaction time point (Fig. 3, D and E). Together, these results show that the virulent protease, BprV, is more efficient at degrading components of the ECM than the benign protease, BprB.
FIGURE 3.
Protease activity of BprV (V) and BprB (B) on components of the extracellular matrix. A, 50 nm purified protease incubated with 1 μm fibronectin at 25 °C. Samples were taken at the indicated time points, and the reaction was stopped by the addition of SDS-PAGE loading buffer and subsequent boiling. Degraded fibronectin products were analyzed using SDS-PAGE. B, quantitative measurement of fibronectin cleavage by densitometry analysis. C, quantitative measurement of elastase activity of the basic proteases. Each purified protease (5 μm) was incubated with 5 mg of Congo Red elastin at 37 °C for 5 h. Elastase activity was measured by reading the absorbance of the recovered supernatant at 490 nm for detection of the proteolytically released Congo Red dye. Mean ± S.E. (error bars) from triplicate experiments are shown, and statistical significance was calculated using one-way ANOVA (p < 0.05 between BprV and BprB, p < 0.05 for all BprB mutants compared with BprB). D, activities of BprV and BprB on ovine hoof material tested by incubating 100 μg/ml of the proteases with a 14% (w/v) insoluble hoof fragment at 37 °C. Samples of supernatant were taken at the indicated time points, and the reaction was stopped by the addition of SDS-PAGE loading buffer, followed by boiling for 5 min. Degraded hoof products were analyzed using SDS-PAGE. The 12-kDa band (asterisk) corresponding to the major degraded product was excised and subjected to in-gel trypsin digestion, followed by LC-MS analysis, to determine the identity of degraded product. E, quantitative measurement of keratin cleavage by densitometry analysis.
S1 Pockets of the Basic Proteases Underpin Their Differential Proteolytic Activity
To determine whether the differences observed between the S1 pockets of the two enzymes form the basis for the difference in their activity, we used site-directed mutagenesis to replace Asp180 and Asp182 of BprB, either alone or in combination, with the equivalent glycine residues in BprV. The activity of these substituted proteins against AAPVn and insoluble elastin was then characterized. Relative to BprB, both of the single substitution derivatives, BprBD180G and BprBD182G, displayed enhanced activity against these substrates (Fig. 3B and Table 2). The elastase-like activity of the two single substitutions was slightly increased, with the kcat/Km values of the BprBD180G and BprBD182G mutants for AAPVn hydrolysis increased by 4 and 27%, respectively. Significantly, the double substitution derivative displayed much higher activity, similar to the level of activity of BprV, against the substrates tested. Compared with BprB, the activity of this enzyme for cleavage of elastin was increased 2-fold (Fig. 3B), and the kcat/Km value for AAPVn hydrolysis was increased by 33% (Table 2).
P1 Substrate Specificity of the Basic Proteases
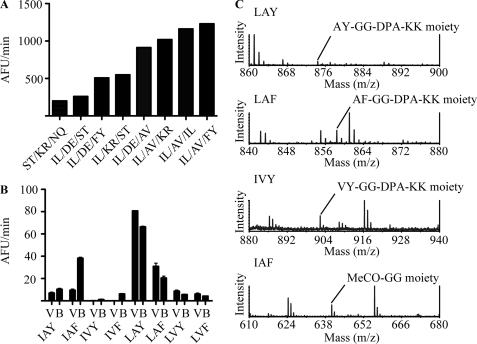
We hypothesized that the changes around the S1 pocket may have altered the primary substrate specificity of each enzyme. To test this hypothesis, the primary substrate specificity of BprV was first characterized using screening of the REPLi (30). Screening of the library using BprB was unsuccessful due to the lack of activity for the protease against the library, even at concentrations considerably higher than those used for BprV. The peptides identified as being most effectively cleaved by BprV were then selected for more detailed kinetic analysis compared with BprB. Of the 512 pools of peptides tested, eight pools of peptides from the library were cleaved efficiently by BprV (Fig. 4A). The majority of the peptides cleaved contained an I/L at the first variable core position, followed by either a small hydrophobic amino acid (A/V in the top three pools) or a charged residue (in four of eight pools) at the second variable core position. However, there were three pools that contained I/L and A/V at the first and second positions that were not cleaved by BprV. These pools had a D/E, N/Q, or proline at the third position, indicating that these amino acids inhibited the cleavage of these peptides when presented at the third variable position. Furthermore, BprV was also able to cleave peptides from a pool that had hydrophilic residues at all three variable core positions (the (S/T)-(K/R)-(N/Q) pool) although to a much lesser extent. This result indicated that BprV could accept P1 side chains other than hydrophobic residues.
FIGURE 4.
Identification of REPLi peptides cleaved by the BprV and BprB proteases. A, rate of cleavage of eight REPLi peptide pools best cleaved by the BprV protease. The peptide pools (50 μm) were incubated with 10 nm BprV at 37 °C. Fluorescence intensities were measured at 30-s intervals for 30 min. The initial velocities of cleavage for the pools are shown. B, rate of cleavage of eight synthesized peptides based on the (I/L)-(A/V)-(F/Y) pool of the REPLi library by BprV and BprB. The proteases (70 nm) were incubated with each peptide substrate (50 μm) at 37 °C, and fluorescence intensities were measured at 30-s intervals for 30 min. The initial velocities of cleavage of each peptide are shown. Mean ± S.E. (error bars) from a triplicate experiment are shown. C, representative mass spectra identifying cleaved products of the peptides, LAY, LAF, LVY, and IAF after cleavage with BprV and BprB.
To determine the P1 specificity of BprV, eight peptides representing all possible sequence combinations from the peptide pool that returned the highest relative rate ((I/L)-(A/V)-(F/Y)) were synthesized (Table 3), and the relative rate of cleavage was determined for each peptide (Fig. 4B). The relative rate of cleavage by the benign protease, BprB, was also determined for each peptide. Subsequently, the four peptides that returned the highest relative rate against BprV (IAF, LAY, LAF, LVY) were chosen for more detailed kinetic studies. Increasing concentrations of peptide were incubated with 70 nm BprV or BprB, and the kinetics of cleavage for these peptides were determined (Table 4). Mass spectrometry was also performed on the cleavage products to determine the P1 specificity of the proteases. Only one cleavage point was detected for all four peptides. The results showed that both proteases cleaved the LAY, LAF, and LVY peptides after leucine (Fig. 4C). By contrast, both proteases cleaved the IAF peptide after phenylalanine (Fig. 4C), indicating that leucine was preferred over isoleucine at the P1 position. Because both proteases cleaved the LAF peptide after leucine, not phenylalanine, it was concluded that they prefer leucine over phenylalanine at the P1 position. In addition, the kinetics data indicated that BprV was more efficient at substrate turnover on the peptides LAY, LAF, and LVY with 64%, 26 and 87% higher kcat/Km than BprB, respectively (Table 4). By contrast, BprB was more efficient at substrate turnover for the peptide IAF, which cleaved after phenylalanine, with a 57% higher kcat/Km value than BprV (Table 4).
TABLE 3.
Eight peptides synthesized for determining the P1 specificity of BprV and BprB proteases
| Peptide 1 | MeOC-GGIAYGG-DPA-KK |
| Peptide 2 | MeOC-GGIAFGG-DPA-KK |
| Peptide 3 | MeOC-GGIVYGG-DPA-KK |
| Peptide 4 | MeOC-GGIVFGG-DPA-KK |
| Peptide 5 | MeOC-GGLAYGG-DPA-KK |
| Peptide 6 | MeOC-GGLAFGG-DPA-KK |
| Peptide 7 | MeOC-GGLVYGG-DPA-KK |
| Peptide 8 | MeOC-GGLVFGG-DPA-KK |
TABLE 4.
Activity of BprV and BprB against synthesized peptide substrates identified from REPLi screen
| Bpr | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | mm·s−1 | |
| BprV P3P2P1↓P1′P2′ | |||
| IAF↓ | 50.9 ± 14.6 | 5.7 × 10−3 | 112 |
| L↓AY | 97.8 ± 23.6 | 32.8 × 10−3 | 335 |
| L↓AF | 87.7 ± 20.7 | 13.5 × 10−3 | 154 |
| L↓VY | 115.4 ± 35.9 | 11.7 × 10−3 | 101 |
| BprB P3P2P1↓P1′P2′ | |||
| IAF↓ | 44.0 ± 11.3 | 11.5 × 10−3 | 261 |
| L↓AY | 77.1 ± 24.2 | 15.7 × 10−3 | 204 |
| L↓AF | 121.4 ± 33.8 | 14.8 × 10−3 | 122 |
| L↓VY | 91.1 ± 31.5 | 4.9 × 10−3 | 54 |
DISCUSSION
In this study, we have structurally and biochemically characterized the basic proteases BprV and BprB from virulent and benign strains of D. nodosus, respectively. We further investigated the biochemical activities of the two enzymes against components of the ECM (fibronectin and insoluble elastin), the peptide substrate, AAPVn, as well as keratin that was derived from sheep hoof. Broadly, these analyses provided evidence that the proteolytic activity of BprV and BprB was significantly different and that BprV was more efficient at degrading potential host-derived substrates.
Sequence-based comparisons reveal that the catalytic domains of BprV and BprB differ by nine amino acids. The crystal structures of both enzymes revealed that only two nonconservative substitutions, GBprV180DBprB and GBprV182DBprB, would be likely to influence catalytic function. These positions are located around the rim of the S1 binding pocket and significantly influence its size and chemical properties. Specifically, in BprV the S1 pocket is generally hydrophobic in nature, whereas in BprB the presence of the two Asp residues confers more acidic properties upon this region. Furthermore, movements in the position of Asp180 and Asp182 in BprB results in a larger S1 pocket compared with that of BprV (145 Å3 and 120 Å3 for BprB and BprV, respectively). Importantly, we have demonstrated via site-directed mutagenesis that the S1 pockets of the two enzymes underpin their differential biochemical activity; most notably, substitution of Asp180 and Asp182 of BprB with Gly residues resulted in a BprB derivative with enhanced proteolytic activity against insoluble elastin and AAPVn.
To understand further how changes in the properties of the S1 pockets of the two basic proteases mediate differences in their biochemical activity, we comprehensively analyzed the primary substrate specificity of the two enzymes using the REPLi combinatorial peptide substrate library and mass spectrometry. Consistent with previous studies (10), these data reveal that the two proteases have similar specificity for the P1 residue in the peptide substrates tested and prefer cleaving after Leu over Phe. We also observed that BprV was more efficient than BprB at cleaving leucine-containing substrates (LAY, LAF, and LVY). By contrast, and consistent with its larger S1 pocket, BprB demonstrated better substrate turnover on the peptide IAF. Compared with BprB, we reasoned that enhanced hydrophobicity of the BprV S1 pocket, as well as a greater flexibility in this region, may confer a higher efficiency for BprV cleavage after Leu.
Because the top three REPLi pools cleaved by BprV contained I/L and A/V at the first and second variable core positions, respectively (Fig. 4A), it can be concluded that small hydrophobic residues are preferred to bind the S1′ subsite of the basic proteases. In addition, we observed that pools that contained D/E, N/Q, or proline at the P2′ position were not cleaved even when the first and second variable core positions of these pools were occupied by I/L and A/V, respectively. These data suggest that acidic, large polar, or proline amino acids at the P2′ position are less preferred for enzyme-substrate interactions.
To correlate the primary substrate specificity data with the differential biochemical activity of the two basic proteases against fibronectin, elastin, and keratin (NCBI accession P02751, AAA30498, and NP001108235, respectively), the amino acid compositions of these three proteins were analyzed. The results indicated that all three proteins have a higher Leu than Phe content (Table 5). Furthermore, the motifs L/A or L/V appear more frequently in all three substrates than the A/F motif (Table 5). Collectively, these observations are consistent with our observations that BprV is more effective than BprB at cleaving ECM and fibrous protein substrates.
TABLE 5.
Leucine and phenylalanine content of components of the extracellular matrix
| ECM proteins | Leu | Phe | Frequency of LA motif | Frequency of LV motif | Frequency of AF motif | Accession |
|---|---|---|---|---|---|---|
| % | % | |||||
| Fibronectin | 5.4 | 2.1 | 5 | 9 | 1 | P02751 |
| Elastin | 5.7 | 3 | 5 | 0 | 2 | AAA30498 |
| Keratin | 10.4 | 2.4 | 1 | 3 | 2 | NP001108235 |
In summary, the data presented in this study suggest that two substitutions around the S1 pocket of the virulent basic protease BprV result in a more efficient enzyme that possesses more focused substrate specificity. These changes appear to favor degradation of proteins that form the skin-horn junction in the ovine hoof. Taken together, these data are consistent with the hypothesis that D. nodosus-secreted subtilisin-like proteases mediate effective tissue destruction and play a significant role in the development of ovine footrot.
Supplementary Material
Acknowledgments
We thank the protein production unit at Monash University for high throughput purification of the recombinant proteases, Dr. John Price at the Department of Biochemistry and Molecular Biology, Monash University, for purified fibronectin, and Professor Richard Whittington at the Faculty of Veterinary Science, University of Sydney, for sheep hoof.
This research was supported by the Australian Research Council through funding to the Australian Research Council Centre of Excellence in Structural and Functional Microbial Genomics.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S3.
The atomic coordinates and structure factors (codes 3TI7 and 3TI9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ECM
- extracellular matrix
- AAPVn
- N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- DPA
- dipicolinic acid
- MeOC
- methoxycarbonyl
- REPLi
- Rapid Endopeptidase Profiling Library
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Roberts D. S., Egerton J. R. (1969) J. Comp. Pathol. 79, 217–227 [DOI] [PubMed] [Google Scholar]
- 2. Green L. E., George T. R. (2008) Vet. J. 175, 173–180 [DOI] [PubMed] [Google Scholar]
- 3. Stewart D. J. (1989) in Footrot and Foot Abscess of Ruminants (Egerton J. R., Yong W. K., Riffkin G. G. eds) pp. 5–45, CRC Press, Boca Raton, FL [Google Scholar]
- 4. Wani S. A., Samanta I. (2006) Vet. J. 171, 421–428 [DOI] [PubMed] [Google Scholar]
- 5. Billington S. J., Johnston J. L., Rood J. I. (1996) FEMS Microbiol. Lett. 145, 147–156 [DOI] [PubMed] [Google Scholar]
- 6. Kennan R. M., Han X., Porter C. J., Rood J. I. (2011) Vet. Microbiol. 153, 59–66 [DOI] [PubMed] [Google Scholar]
- 7. Kennan R. M., Wong W., Dhungyel O. P., Han X., Wong D., Parker D., Rosado C. J., Law R. H., McGowan S., Reeve S. B., Levina V., Powers G. A., Pike R. N., Bottomley S. P., Smith A. I., Marsh I., Whittington R. J., Whisstock J. C., Porter C. J., Rood J. I. (2010) PLoS Pathog. 6, e1001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Myers G. S., Parker D., Al-Hasani K., Kennan R. M., Seemann T., Ren Q., Badger J. H., Selengut J. D., Deboy R. T., Tettelin H., Boyce J. D., McCarl V. P., Han X., Nelson W. C., Madupu R., Mohamoud Y., Holley T., Fedorova N., Khouri H., Bottomley S. P., Whittington R. J., Adler B., Songer J. G., Rood J. I., Paulsen I. T. (2007) Nat. Biotechnol. 25, 569–575 [DOI] [PubMed] [Google Scholar]
- 9. Siezen R. J., Leunissen J. A. (1997) Protein Sci. 6, 501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kortt A. A., Caldwell J. B., Lilley G. G., Edwards R., Vaughan J., Stewart D. J. (1994) Biochem. J. 299, 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riffkin M. C., Focareta A., Edwards R. D., Stewart D. J., Kortt A. A. (1993) Gene 137, 259–264 [DOI] [PubMed] [Google Scholar]
- 12. Lilley G. G., Stewart D. J., Kortt A. A. (1992) Eur. J. Biochem. 210, 13–21 [DOI] [PubMed] [Google Scholar]
- 13. Lilley G. G., Riffkin M. C., Stewart D. J., Kortt A. A. (1995) Biochem. Mol. Biol. Int. 36, 101–111 [PubMed] [Google Scholar]
- 14. Riffkin M. C., Wang L. F., Kortt A. A., Stewart D. J. (1995) Gene 167, 279–283 [DOI] [PubMed] [Google Scholar]
- 15. Depiazzi L. J., Richards R. B. (1979) Aust. Vet. J. 55, 25–28 [DOI] [PubMed] [Google Scholar]
- 16. Liu D., Yong W. K. (1993) Res. Vet. Sci. 55, 124–129 [DOI] [PubMed] [Google Scholar]
- 17. Stewart D. J. (1979) Res. Vet. Sci. 27, 99–105 [PubMed] [Google Scholar]
- 18. Wong W., Kennan R. M., Rosado C. J., Rood J. I., Whisstock J. C., Porter C. J. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 20. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 22. Stein N. (2008) J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Morris R. J., Perrakis A., Lamzin V. S. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 968–975 [DOI] [PubMed] [Google Scholar]
- 26. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Androulakis S., Schmidberger J., Bate M. A., DeGori R., Beitz A., Keong C., Cameron B., McGowan S., Porter C. J., Harrison A., Hunter J., Martin J. L., Kobe B., Dobson R. C., Parker M. W., Whisstock J. C., Gray J., Treloar A., Groenewegen D., Dickson N., Buckle A. M. (2008) Acta Crystallogr. D Biol. Crystallogr. D64, 810–814 [DOI] [PubMed] [Google Scholar]
- 29. Ohman D. E., Cryz S. J., Iglewski B. H. (1980) J. Bacteriol. 142, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thomas D. A., Francis P., Smith C., Ratcliffe S., Ede N. J., Kay C., Wayne G., Martin S. L., Moore K., Amour A., Hooper N. M. (2006) Proteomics 6, 2112–2120 [DOI] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2010) The PyMOL Molecular Graphics System version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 32. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 33. Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1955–1957 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.