Background: Focal adhesion kinase (FAK) and CUB-domain-containing protein 1 (CDCP1) are Src family kinase (SFK) substrates.
Results: SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734 is induced by increased CDCP1 expression and changes in cell attachment.
Conclusion: SFK switching between FAK and CDCP1 may be relevant to malignant transformation.
Significance: Targeting of this switch may be a rational approach to treat diseases such as cancer.
Keywords: Adhesion, Cancer Tumor Promoter, Cell Surface Protein, Membrane Proteins, Src, CDCP1, FAK
Abstract
Reciprocal interactions between Src family kinases (SFKs) and focal adhesion kinase (FAK) are critical during changes in cell attachment. Recently it has been recognized that another SFK substrate, CUB-domain-containing protein 1 (CDCP1), is differentially phosphorylated during these events. However, the molecular processes underlying SFK-mediated phosphorylation of CDCP1 are poorly understood. Here we identify a novel mechanism in which FAK tyrosine 861 and CDCP1-Tyr-734 compete as SFK substrates and demonstrate cellular settings in which SFKs switch between these sites. Our results show that stable CDCP1 expression induces robust SFK-mediated phosphorylation of CDCP1-Tyr-734 with concomitant loss of p-FAK-Tyr-861 in adherent HeLa cells. SFK substrate switching in these cells is dependent on the level of expression of CDCP1 and is also dependent on CDCP1-Tyr-734 but is independent of CDCP1-Tyr-743 and -Tyr-762. In HeLa CDCP1 cells, engagement of SFKs with CDCP1 is accompanied by an increase in phosphorylation of Src-Tyr-416 and a change in cell morphology to a fibroblastic appearance dependent on CDCP1-Tyr-734. SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734 also occurs during changes in adhesion of colorectal cancer cell lines endogenously expressing these two proteins. Consistently, increased p-FAK-Tyr-861 levels and a more epithelial morphology are seen in colon cancer SW480 cells silenced for CDCP1. Unlike protein kinase Cδ, FAK does not appear to form a trimeric complex with Src and CDCP1. These data demonstrate novel aspects of the dynamics of SFK-mediated cell signaling that may be relevant during cancer progression.
Introduction
Src family kinases (SFKs),2 comprising the non-receptor tyrosine kinases Src, Yes, Fyn, Lyn, Lck, Hck, Fgr, Blk, and Yrk (1), are essential mediators of signals required for a range of physiological processes (2). In addition, overexpression and increased activity of Src in human epithelial cancers is common and proposed to play a key role in cancer progression, particularly during later stages when tumor cells acquire metastatic abilities (3–7).
SFK activation can be achieved by a number of mechanisms including phosphorylation, intramolecular protein-protein interactions, and interactions with binding proteins (8). Regulation by phosphorylation is primarily controlled at two tyrosine residues. Phosphorylation of Tyr-527 causes a closed and inactive confirmation, whereas its dephosphorylation induces a conformational change causing activation. Maximal kinase activity requires autophosphorylation of Tyr-416 (2). Another mechanism of Src activation is by displacement of intramolecular interactions at its Src homology 2 and 3 domains by higher affinity ligands such as the non-receptor tyrosine kinase focal adhesion kinase (FAK) (8, 9), another protein up-regulated and/or activated in a range of human malignancies that plays a role in cellular processes such as adhesion, migration, and cell death (10–12). In particular, FAK has a key role in integrin-mediated signaling during cell adhesion (13). Upon binding of integrins, FAK becomes activated through autophosphorylation at Tyr-397, which allows for high affinity binding of Src at this site (14, 15), inducing conformational changes and further phosphorylation of FAK, mediated by SFKs at Tyr-407, -576/577, -861, and -925 (16–18).
Several recent studies have recognized the cell surface glycoprotein CUB-domain-containing protein 1 (CDCP1) as a SFK substrate (19–23) that promotes cancer-associated cellular changes in vitro and in animal models (23–30). Phosphorylation of CDCP1 by SFKs is thought to occur initially at Tyr-734 followed by further SFK-mediated phosphorylation at Tyr-743 and -762 and recruitment of protein kinase C δ (PKCδ) at this last site (21, 23). Formation of a trimeric protein complex of SFKs, CDCP1, and PKCδ plays a critical role in facilitating a CDCP1-mediated anti-apoptotic cell phenotype in vitro (30). The potential functional importance of phosphorylation of CDCP1 by SFKs is further indicated by the observation that it is induced by a number of stimuli including loss of cell adhesion (23, 31, 32), cleavage by trypsin-fold serine proteases (20, 33), cell detachment during mitosis (22, 31, 32), and cell shedding (32).
The in vivo importance of CDCP1 phosphorylation has been indicated by reports showing that p-CDCP1-Tyr-734 is expressed by gastric cancer 44As3 cells undergoing peritoneal dissemination in mice and not by surrounding stroma and that p-CDCP1-Tyr-734 levels are markedly up-regulated in ∼30% of human scirrhous-type gastric cancers (30). This residue is also required for CDCP1-mediated experimental metastasis of melanoma cells in mice (25). In addition, another CDCP1 tyrosine, Tyr-743, is phosphorylated in a wide range of cancers but not in normal cells not undergoing mitosis or shedding (32).
To examine the role of tyrosine phosphorylation in CDCP1 biology we have generated HeLa cells stably expressing this protein or a mutant lacking the critical SFK phosphorylation site at Tyr-734. CDCP1 was basally phosphorylated in these cells, and unexpectedly, its expression eliminated SFK-mediated phosphorylation of FAK-Tyr-861. CDCP1 expression was accompanied by a change in HeLa cell morphology that was restored together with phosphorylation of FAK-Tyr-861 in HeLa cells expressing CDCP1-Y734F and also when the activity of SFKs was selectively inhibited. Our data suggest that overexpression of CDCP1 can induce SFK substrate switching from FAK-Tyr-861 to CDCP1-Tyr-734. Importantly, we also observed this switching in colorectal cancer cell lines endogenously expressing FAK and CDCP1. However, switching in these cells was mediated by changes in cell anchorage. These data highlight two settings under which SFKs can switch between FAK-Tyr-861 and CDCP1-Tyr-734. As both settings (increased expression of CDCP1 and changes in cell adhesion) occur during cancer progression, these observations may be useful in understanding SFK·CDCP1-mediated mechanisms occurring during malignant transformation.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies were from the following suppliers: rabbit anti-matrix metalloproteinase-9 (#ab38898) antibody from Abcam (Cambridge, MA); rabbit polyclonal antibody against unspecified C-terminal residues of CDCP1 from Cell Signaling Technology (Danvers, MA; #4115); goat anti-lipocalin2 antibody (#AF1757) and a stem cell array kit (#ARY010) from R&D Systems (Bio-Scientific Pty Ltd, Gymea, Australia); rabbit anti-Src (#2108) and anti-p-Src (#2101) antibodies from Cell Signaling Technology, rabbit anti-p-FAK-Tyr-861 antibody (#44626G) that detects both p-CDCP1-Tyr-734 and p-FAK-Tyr-861 (20), mouse anti-smooth muscle actin (#18-0106) and anti-cytokeratin-8/-18 (#18-0213) antibodies, and goat anti-mouse Alexa Fluor 488 and 647 secondary antibodies from Invitrogen; rabbit anti-FLAG epitope (DYKDDDDK) and mouse anti-tubulin antibodies from Sigma; monoclonal anti-phosphotyrosine antibody PY20 (#525295) from Calbiochem; monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody from Chemicon International (Boronia, Australia); antibodies against FAK (#05-537) and p-FAK-Tyr-397 (#05-1144) from Millipore (North Ryde, Australia); HRP-conjugated secondary antibodies from Thermo Fisher Scientific (Scorseby, Australia). Anti-CDCP1 monoclonal antibodies 41-2 (19, 24, 34) and 10D7 (24) were previously described. Control immunoglobulins (IgGs) were from Sigma and Invitrogen. Protein A/G-agarose and Complete EDTA-free protease inhibitor were from Roche Applied Sciences. G418 and puromycin were from InvivoGen (San Diego, CA), and the SFK selective inhibitor SU6656 (35) was from Invitrogen. Annexin V-conjugated Alexa Fluor 647 was from Biolegend (Australian Biosearch, Karrinyup, Australia). All other reagents were from Sigma. The CDCP1-FLAG-encoding expression construct has been described previously (33). Site-directed mutagenesis, to introduce the CDCP1 mutation Y734F, was performed using Pfu Ultra polymerase (Stratagene, La Jolla, CA). The sequence of constructs was confirmed by DNA sequencing at the Australian Genome Research Facility (St. Lucia, Australia). pLKO.1 lentiviral shRNA constructs targeting CDCP1 were purchased from OpenBiosystems, and the pLKO.1-scramble control was from Addgene (Cambridge, MA).
Cell Culture and Transfections
Cells were purchased from ATCC (Manassas, VA). Untransfected HeLa cells and colon cancer CaCo2, HCT116, SW480, and SW620 cells were cultured in complete media of DMEM supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) in a 5% CO2-humidified atmosphere at 37 °C. All cells were passaged exclusively non-enzymatically using Versene (Invitrogen) or 500 μm EDTA in PBS. HeLa cells were transfected with pcDNA3.1 (vector) or CDCP1-FLAG or CDCP1-FLAG-Y734F expression constructs using Lipofectamine 2000 (Invitrogen). Stably transfected cells were selected in G418 (800 μg/ml)-containing media for 10 days before clonal selection by fluorescence-activated cell sorting using the anti-CDCP1 monoclonal antibody 10D7 and expansion and maintenance in G418-containing media. Cells were examined by transmitted light microscopy analysis using a Nikon Eclipse TE2000-U microscope.
Lentiviral shRNA Gene Silencing
CDCP1 expression was suppressed using a pLKO.1 lentiviral shRNA knockdown system (OpenBiosystems, Millennium Science, Surrey Hills, Australia) with a scramble shRNA construct (Addgene) as a control. To generate lentivirus, the pLKO.1 shRNA construct and packaging plasmids (pCMV-VSVG and pCMV-dR8.2-dvpr) were cotransfected into HEK293T cells with Lipofectamine 2000 (Invitrogen). The conditioned media was collected 48 and 72 h post-transfection and pooled followed by filtration through a 0.45-μm filter. Filtered conditioned media was used to infect target cells in the presence of 8 μg/ml hexadimethrine bromide (Sigma). Cells were initially infected for 3 h, allowed to recover overnight in complete media, and then infected a second time overnight. After washing, cells were cultured for 24 h in complete media before selection in puromycin (2 μg/ml)-containing medium for 1 week. Gene silencing efficiency was assessed by Western blot analysis.
Cell Treatments
To examine the effect of inhibiting SFK activity on cell morphology, cells were detached with Versene then washed with PBS, counted, and seeded at 2 × 105 cells/T25 flask in complete media containing G418 (700 μg/ml) in the presence of either SU6656 (10 μm) or vehicle (0.075% v/v DMSO). After 1 h this medium was replaced with complete medium containing G418 (700 μg/ml). After a total of 24 h, cell morphology was assessed by light microscopy using a Nikon Eclipse TE2000-U microscope as described below. The level of cell death induced by SU6656 treatment was examined by flow cytometry analysis of annexin V-stained cells as described below. To examine the effect of SFK inhibition on phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861, lysates were collected for Western blot analysis (described below) of HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells treated with vehicle (0.37% v/v DMSO) or increasing concentrations of SU6656 (0.1, 0.27, 1, 2.7, and 10 μm) for 60 min. To examine changes in phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861, the adhesion/de-adhesion status of HeLa CDCP1 cell clones 2 and 3 and colon cancer HCT116 and SW480 cells was modulated. For adhesion experiments, cells were cultured in DMEM to 50% confluence then serum-starved for 24 h before detachment with 500 μm EDTA in PBS. After 30 min in suspension cells were allowed to re-adhere to plastic for periods of up to 45 min. Cells in suspension were then collected from media by low speed centrifugation and lysed in buffer containing protease inhibitor mixture, 2 mm sodium vanadate, 10 mm sodium fluoride, Triton X-100 (1%), 50 mm Tris-HCl (pH 7.4), and NaCl (150 mm). Adhered cells were lysed in situ in the same buffer and after clearance by centrifugation were combined with lysates obtained from cells in suspension. For de-adhesion experiments, cells were cultured to 50% confluence then serum-starved for 24 h before incubation with 500 μm EDTA in PBS for periods of up to 45 min. Adhered and suspended cells were collected and lysed as described above.
Quantification of Cell Morphology
Images of fields of cells acquired by light microscopy using a 10× objective on a Nikon Eclipse TE2000-U microscope were analyzed using MetaMorph Imaging Software (Molecular Devices, Bio-Strategy Pty Ltd, Hawthorne East, Australia) to determine the length of the short and long axis of each cell. Based on a previously reported approach (36), the shape factor for each cell was determined as the ratio of short axis to long axis. For epithelial-shaped cells this value approaches 1, and for elongated, fibroblastic cells it approaches 0. Cells were grouped into one of two categories based on shape factor (0–0.49 and 0.5–1), and these categories were graphed against the proportion of cells in each category. For analysis of HeLa cells stably transfected with vector or expression constructs encoding either CDCP1 or CDCP1-Y734F, at least 50 cells were analyzed. For cells treated with SU6656, at least 80 cells were analyzed per experiment, and experiments were performed 3 times. Statistical analysis for change in cell morphology was performed using Student's t test with a 95% confidence interval using SigmaPlot 11 (Systat Software, Inc., San Jose, CA). A p value < 0.05 was considered significant and is indicated by an asterisk in Fig. 1.
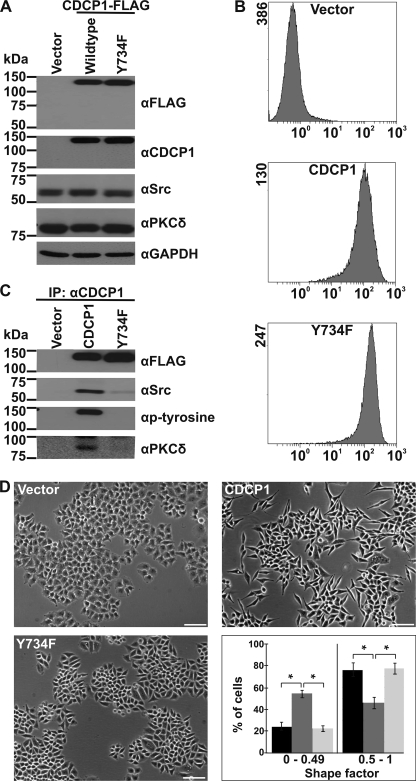
FIGURE 1.
CDCP1 expression alters HeLa cell morphology dependent on the SFK binding site. Analyses of HeLa cells stably transfected with vector or expression constructs encoding either CDCP1 or CDCP1-Y734F are shown. A, anti (α)-FLAG, -CDCP1 (10D7), -Src, - PKCδ, and -GAPDH Western blot analysis. B, flow cytometry analysis using the anti-CDCP1 monoclonal antibody 10D7. C, anti-FLAG, -Src, -phosphotyrosine, and -PKCδ Western blot analysis of proteins immunoprecipitated (IP) from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells using the monoclonal anti-CDCP1 antibody 41–2. D, transmitted light microscopy analysis. Cells were imaged using a Nikon Eclipse TE2000-U microscope (bar, 100 μm). To quantify the effect of CDCP1 expression on HeLa cell morphology, a shape factor was determined for each cell using MetaMorph software as described under “Experimental Procedures.” At least 50 cells (for each of three independent cell platings) were analyzed for each cell type, and cells were grouped into the two shape factor categories 0–0.49 and 0.5–1. These categories were graphed against the proportion of cells in each category. Black, HeLa vector cells; dark gray, HeLa CDCP1 cells; light gray, HeLa CDCP1-Y734F cells. *, p < 0.05.
Flow Cytometry
To assess CDCP1 expression levels, cells grown to 70–90% confluence were de-adhered with Versene then stained for 30 min on ice with anti-CDCP1 antibody 10D7 (3 μg/ml) in PBS. After washes with ice-cold PBS, cells stained with 10D7 were incubated for 30 min on ice with a goat anti-mouse Alexa Fluor 488 secondary antibody (1:750 dilution). After washes with ice-cold PBS, a minimum of 10,000 cells were analyzed using a Cytomics FC-500 MPL flow cytometer (Beckman Coulter; Gladesville, Australia). To assess the level of cell death caused by SU6656 treatment, treated and untreated cells were stained with annexin V-conjugated Alexa Fluor 647 as previously described (37) and analyzed using an LSR II flow cytometer (BD Biosciences). Data plots generated from flow cytometry analyses are representative of three independent experiments performed in triplicate.
Cell Lysis, Immunoprecipitation, Western Blot, and Stem Cell Array Analysis
For experiments other than adhesion/de-adhesion assays, whole cell lysates were collected in a buffer containing protease inhibitor mixture, 2 mm sodium vanadate, 10 mm sodium fluoride, and either Triton X-100 (1%), 50 mm Tris-HCl (pH 7.4), and NaCl (150 mm) or CHAPS (1%), PBS (pH 7.4), and MgCl2 (2 mm). Protein concentration was determined by microbicinchoninic acid assay (Thermo Scientific). For immunoprecipitations, total protein (500 μg) was precleared for 2 h at 4 °C by end-to-end rotation with immobilized protein A/G beads. After centrifugation, the supernatant was mixed with the immunoprecipitating antibody (5 μg) and incubated overnight at 4 °C by end-to-end rotation. Fresh aliquots of protein A/G beads were then added, and the mixture was incubated for 4 h at 4 °C by end-to-end rotation. The beads were then washed three times in cell lysis buffer, then associated proteins were eluted into Laemmli sample buffer. Cell lysates and immunoprecipitated proteins were separated by SDS-PAGE under either non-reducing (for Western blot analysis using 10D7 as primary antibody) or reducing conditions and transferred to nitrocellulose membranes, which were blocked in 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). Membranes were incubated with primary antibodies diluted in blocking buffer for 1–3 h at room temperature, washed with TBS-T, and then incubated with species-appropriate HRP-conjugated secondary antibodies for 60 min. After washes, membranes were incubated with SuperSignal West Pico Substrate (Thermo Fisher Scientific) then exposed to film. Consistent protein loading and transfer was determined by reprobing membranes stripped in Restore Western blot stripping buffer (Pierce) with either an anti-GAPDH or anti-tubulin antibody. Densitometry analysis was performed using Image J. p values were determined from 4 experiments using one-tailed Student's t test with a 95% confidence interval using GraphPad Prism. A p value < 0.05 was considered significant and is indicated by an asterisk in Fig. 4.
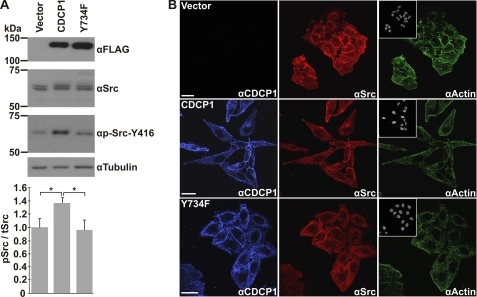
FIGURE 4.
Altered HeLa CDCP1 cell morphology is accompanied by increased phosphorylation of Src-Tyr-416 without altering cellular localization of CDCP1 or Src. A, anti (α)-FLAG, -Src, -p-Src-Tyr-416, and -tubulin Western blot analysis of lysates from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells. This analysis is representative of four independent experiments. Data from densitometric analysis of p-Src-Tyr-416 signal relative to total Src signal from four independent experiments are graphed at the bottom. *, p < 0.05. B, confocal microscopy analysis. HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were incubated with anti-CDCP1 and -Src antibodies followed by species-specific secondary antibodies then stained with phalloidin-conjugated Alexa Fluor 488 and DAPI to visualize actin and cell nuclei, respectively. Images were acquired using a Leica SP5 confocal microscope. Bar, 25 μm.
To assess the expression of stem cell markers, HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were seeded in T75 flasks 48 h before lysates were prepared. Cells were harvested at a confluence of 70% using an array kit lysis buffer containing 10 μg/ml aprotinin and 1× protease inhibitor mixture. Stem cell arrays were hybridized with 175 μg of total protein according to the instructions of the manufacturer and then exposed to film.
Confocal Microscopy
Cells grown on sterile coverslips were washed in PBS, fixed in 4% formaldehyde in PBS for 15 min at room temperature, rinsed twice with PBS then permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. After blocking in 0.5% BSA for 30 min, cells were incubated with the mouse anti-CDCP1 antibody 41–2 (5 μg/ml) for 45 min at room temperature followed by incubation with a rabbit anti-Src antibody (1:100 dilution; Cell Signaling Technology #2108). The cells were then washed twice with PBS and incubated with species-specific Alexa Fluor-conjugated secondary antibodies (1:750 dilution) for 30 min at room temperature. Actin and cell nuclei were stained by incubation for 10 min at room temperature with a PBS solution containing Alexa Fluor 488 phalloidin and DAPI (1:1000 dilution). After washes with PBS, coverslips were mounted on slides, and cells were imaged with a Leica TCS SP5 confocal microscope (Leica Microsystems, Sydney, Australia). Images were processed and displayed using Corel Draw (Corel Pty Ltd; Sydney, Australia).
RESULTS
CDCP1 Expression Alters HeLa Cell Morphology-dependent on CDCP1-Tyr-734, the SFK Binding Site
To examine the function of Tyr-734 of CDCP1, HeLa cells were stably transfected with vector (HeLa vector) or expression constructs encoding CDCP1-FLAG (HeLa CDCP1) or CDCP1-FLAG mutated at Tyr-734 (HeLa CDCP1-Y734F), which is the primary site for SFK binding (21). Western blot and flow cytometric analysis, shown in Fig. 1, A and B, respectively, indicated high and uniform expression of CDCP1 and CDCP1-Y734F. These data were obtained from combination of three clones for each stable cell type; however, the same data were obtained from analysis of the individual clones (not shown). In these cells CDCP1 is produced exclusively with a molecular mass of 135 kDa with no evidence of a lower molecular mass CDCP1 species variously reported at 70–85 kDa for some cell lines (19–20, 22, 23). It has previously been shown that the difference in the predicted molecular mass of CDCP1 (∼92 kDa) and its apparent molecular mass of 135 kDa is due in large part to 35–40 kDa of N-linked glycans (19). Consistent with previous reports, Src, CDCP1, and PKCδ form a complex that is dependent on CDCP1-Tyr-734, as mutation of this site completely abolished PKCδ binding and markedly reduced Src binding (Fig. 1C). In line with the decreased binding of Src to CDCP1-Y734F, we found that tyrosine phosphorylation of CDCP1 was only detectable in HeLa CDCP1 but not in HeLa CDCP1-Y734F cells (Fig. 1C). Of note, microscopy analysis revealed that CDCP1 expression altered the morphology of HeLa cells from an epithelial appearance, characteristic of parental and vector-transfected cells, to an elongated, spindle-like fibroblastic morphology (Fig. 1D). Quantitative analysis of the morphology of these cells indicated that there was a statistically significant difference between CDCP1-expressing HeLa cells and HeLa vector and HeLa CDCP1-Y734F cells; ∼55% of HeLa CDCP1 cells had an elongated morphology in comparison with ∼20% of HeLa vector and HeLa CDCP1-Y734F cells (Fig. 1D, graph). CDCP1-Tyr-734 was required for this change as the morphology of HeLa CDCP1-Y734F cells was reminiscent of parental and vector cells (Fig. 1D, images and graph). Significantly, mutation of two other SFKs phosphorylation sites, CDCP1-Tyr-743 and -Tyr-762 (21, 23), did not cause a reversion to an epithelial morphology (supplemental Fig. 1).
As the morphological change of HeLa CDCP1 cells is reminiscent of an epithelial to mesenchymal transition (EMT), we analyzed the expression of markers of epithelial and mesenchymal cells in the generated cell lines. Western blot analysis using antibodies against matrix metalloproteinase-9, smooth muscle actin and lipocalin2 (mesenchymal markers), and cytokeratin-8 and -18 (epithelial markers) indicated that CDCP1 expression did not alter expression of these markers of EMT (supplemental Fig. 2A). As it has previously been noted that EMT can generate cells with stem cell properties (38), we also probed an array containing antibodies against 15 pluripotent stem cell markers with lysates from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells. No changes in any of the markers were observed (supplemental Fig. 2B). These data suggest that the altered morphology of HeLa cells induced by CDCP1 expression is not due to an EMT.
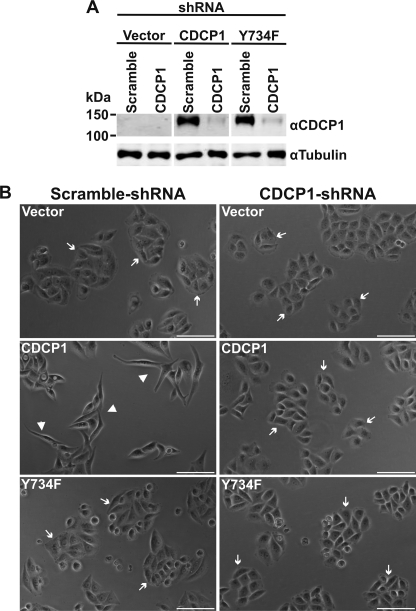
Knockdown of CDCP1 Reverts HeLa CDCP1 Cells to an Epithelial Morphology
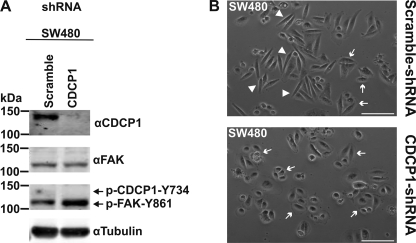
To further address the role of CDCP1 in the observed change in cell morphology, we reduced expression of CDCP1 using an shRNA knockdown approach. Using a virus-mediated protocol, we generated stable polyclonal cells. Anti-CDCP1 Western blot analysis indicated that CDCP1 and CDCP1-Y734F had been specifically reduced by greater than 95% (Fig. 2A). As shown in Fig. 2B and supplemental Fig. 3, light microscopy analysis demonstrated that knockdown of CDCP1 caused reversion of HeLa CDCP1 cell shape to an epithelial morphology. Cell morphology was unaffected by the scrambled control and knockdown of CDCP1-Y734F (Fig. 2B and supplemental Fig. 3).
FIGURE 2.
Knockdown of CDCP1 reverts HeLa CDCP1 cells to an epithelial morphology. A, HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were stably infected with either a CDCP1 shRNA knockdown construct or a scramble control construct. Lysates of these cells were examined by anti-CDCP1 (#4115) and anti-tubulin Western blot analysis. To facilitate comparison, the anti-CDCP1 and -tubulin panels are also displayed in Fig. 5B with contemporaneously performed anti-p-FAK-Tyr-861 and -FAK Western blot analyses. B, transmitted light microscopy analysis. Cells were imaged using a Nikon Eclipse TE2000-U microscope. Arrow, epithelial morphology; arrowhead, elongated, fibroblastic morphology. Bar, 100 μm.
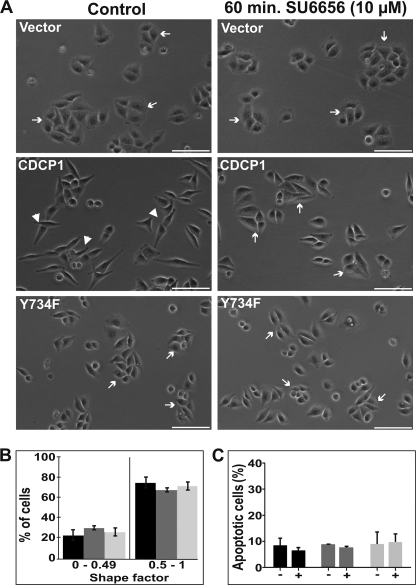
CDCP1-induced HeLa Cell Morphology Change Requires SFK Activity
To directly examine whether SFK activity is required for the observed change in HeLa cell morphology, HeLa CDCP1 cells were either untreated or treated with the SFK-selective inhibitor SU6656 (35) at a concentration of 10 μm. In these experiments cells were plated in the presence of SU6656 (or vehicle for controls). Treatment continued for 1 h, then the medium was changed to normal growth media. After a recovery period of 23 h, cells were photographed. As shown in Fig. 3A and supplemental Fig. 4, SU6656 treatment had no impact on HeLa vector and HeLa CDCP1-Tyr-734 cells but caused the reversion of HeLa CDCP1 cell morphology from an elongated, spindle-like morphology to an epithelial appearance. Quantitative analysis of cell morphology indicated that HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells shared common morphological features after SU6656 treatment (Fig. 3B; compare with the graph in Fig. 1D). In addition, flow cytometry analysis of annexin V-stained cells indicated that SU6656 treatment did not alter the proportion of cells undergoing apoptosis, indicating that the SU6656-mediated altered morphology of HeLa CDCP1 cells was not due to a cytotoxic effect (Fig. 3C). These data indicate that altered HeLa CDCP1 cell morphology requires SFK activity.
FIGURE 3.
CDCP1-induced HeLa cell morphology change requires SFK activity. A, transmitted light microscopy analysis. HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were treated with vehicle (DMSO 0.075% v/v; control) or the SFK selective inhibitor SU6656 (10 μm) for 1 h. The media were then changed to normal growth media and after another 23 h cells were imaged using a Nikon Eclipse TE2000-U microscope. Arrow, epithelial morphology; arrowhead, elongated, fibroblastic morphology. Images are representative of three independent experiments. B, graph of the percentage of cells versus cell shape factor, determined using MetaMorph software analysis as described under “Experimental Procedures.” Black, HeLa vector cells; dark gray, HeLa CDCP1 cells; light gray, HeLa CDCP1-Y734F cells. C, graphic representation of flow cytometry analysis of cells after SU6656 treatment (10 μm 1 h) and re-equilibration for a further 23 h. Cells were lifted, and the percentage of cells undergoing apoptosis was assessed by flow cytometry analysis of cells stained with Alexa Fluor 647-conjugated annexin V. The data are from three experiments performed in triplicate. Black, HeLa vector cells; dark gray, HeLa CDCP1 cells; light gray, HeLa CDCP1-Y734F cells.
Altered HeLa CDCP1 Cell Morphology Is Accompanied by Increased Phosphorylation of Src-Tyr-416 without Altering Cellular Localization of Src
Binding of Src to substrates can reciprocally increase Src activation (39–41). To examine whether CDCP1 expression increased Src activation, we performed anti-pSrc-Tyr-416 Western blot analysis of lysates from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells. As shown in Fig. 4A, this analysis indicated that CDCP1 expression in HeLa cells results in an ∼40% increase in phosphorylation of Src-Tyr-416 and that this statistically significant increase is dependent on Tyr-734 of CDCP1. We also used confocal microscopy to determine whether changes in cell morphology were associated with differences in the localization of Src or CDCP1. As shown in Fig. 4B (blue), CDCP1 localization was unaffected by mutation of CDCP1-Tyr-734, with this protein being localized primarily at the cell surface. Similarly, Src staining was not altered by CDCP1 or CDCP1-Y734F expression, with this protein present throughout the cytoplasm of HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells with evidence of membrane accentuation in each cell type (Fig. 4B, red). Phalloidin staining indicated that actin localization was similar in unstimulated HeLa vector and HeLa CDCP1-Y734F cells, whereas HeLa CDCP1 cells exhibited some evidence of juxta-plasma membrane accentuation of actin localization, although this was heterogeneous among cell populations (Fig. 4B, green). DAPI staining indicated that cell nuclei were not altered by expression of CDCP1 or CDCP1-Y734F (Fig. 4B, inset).
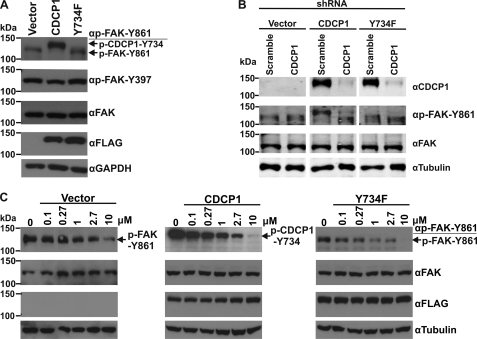
CDCP1-Tyr-734 Is Phosphorylated in Preference to FAK-Tyr-861 in HeLa CDCP1 Cells
To directly examine the phosphorylation of CDCP1-Tyr-734, we performed Western blot analysis with an anti-p-FAK-Tyr-861 antibody that also efficiently recognizes CDCP1-Tyr-734; this antibody detects p-FAK-Tyr-861 at ∼125 kDa and p-CDCP1-Tyr-734 at ∼135 kDa (20, 33). As expected, analysis of lysates from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells detected p-CDCP1-Tyr-734 exclusively in HeLa CDCP1 cells (Fig. 5A). However, whereas p-FAK-Tyr-861 was detected in lysates of HeLa vector and HeLa CDCP1-Y734F cells, it was not present in HeLa CDCP1 cells even though Western blot analysis indicated that total FAK levels were the same in each cell type (Fig. 5A, compare the top panel with the panel second from the top). Interestingly, HeLa cells stably expressing CDCP1-Tyr-743F or CDCP1-Tyr-762F also had high levels of p-CDCP1-Tyr-734 with no evidence of p-FAK-Tyr-861 (supplemental Fig. 5). Of further interest, stable expression of CDCP1 had no significant effect on phosphorylation of FAK-Tyr-397 (Fig. 5A), an auto-phosphorylation site, phosphorylation of which creates a high affinity binding site for Src homology 2 domain-containing proteins including Src (14). These data suggest that CDCP1-Tyr-734 is phosphorylated in preference to FAK-Tyr-861 in HeLa CDCP1 cells. This proposal is supported by analysis of cells silenced for CDCP1 (characterized in Fig. 2). Western blot analysis using the anti-p-FAK-Tyr-861 antibody indicated that silencing of CDCP1 in HeLa CDCP1 cells resulted in phosphorylation of FAK-Tyr-861 to levels seen in HeLa vector and HeLa CDCP1-Y734F cells (Fig. 5B). To confirm the role of SFKs in these tyrosine phosphorylation events, we treated HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells with increasing concentrations of the SFK-selective inhibitor SU6656 for 1 h (the same treatment period used for cells shown in Fig. 3). As shown in Fig. 5C, Western blot analysis of lysates from these cells using the anti-p-FAK-Tyr-861 antibody demonstrated that SU6656 inhibited phosphorylation of both FAK-Tyr-861 and CDCP1-Tyr-734 in a dose-dependent manner.
FIGURE 5.
CDCP1-Tyr-734 is phosphorylated in preference to FAK-Tyr-861 in HeLa CDCP1 cells. A, anti (α)-p-FAK-Tyr-861 (which also recognizes p-CDCP1-Tyr-734 (20)), p-FAK-Tyr-397, -FAK, -FLAG, and GAPDH Western blot analysis of lysates from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells. B, HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were stably transfected with either a CDCP1 shRNA knockdown construct or a scramble control construct. Lysates of these cells were examined by Western blot analysis using antibodies against CDCP1 (#4115), p-FAK-Tyr-861, FAK, and tubulin. These analyses were performed contemporaneously, and anti-CDCP1 and -tubulin panels are also shown in Fig. 2A to facilitate comparison. C, HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F cells were treated with vehicle (DMSO 0.37% v/v; 0) or with the indicated concentrations of the SFK inhibitor SU6656 for 60 min. Lysates were analyzed by Western blot analysis using antibodies against p-FAK-Tyr-861, FAK, FLAG, and tubulin.
SFK Phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 in HeLa CDCP1 Cells Is Inversely Related and Depends on the Level of Expression of CDCP1, but CDCP1 and FAK Do Not Interact
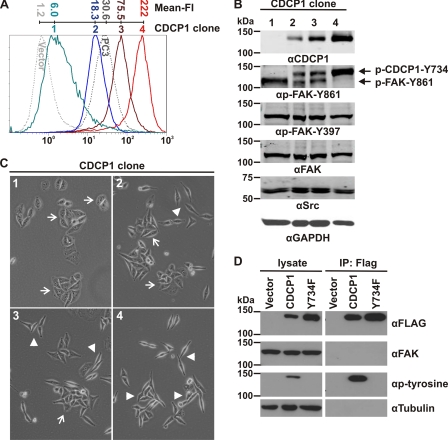
During the generation of HeLa CDCP1 cells, we obtained clones expressing this protein at varying levels. These were used to further investigate the relationship between phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861. As shown in Fig. 6A by flow cytometry analysis using the anti-CDCP1 monoclonal antibody 10D7, four HeLa CDCP1 cell clones (numbered 1–4) expressed CDCP1 at increasing levels on the cell surface. Expression levels ranged from barely detectable (clone 1 in comparison with HeLa vector cells) to approximating the levels seen in endogenous-expressing prostate cancer PC3 cells (clone 2), with two other clones (numbers 3 and 4) expressing CDCP1 at progressively higher levels. To examine the relationship between phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861, we examined these cells by Western blot analysis using the anti-p-FAK-Tyr-861 antibody that cross-reacts with p-CDCP1-Tyr-734 (20). This analysis demonstrated that the level of phosphorylation of CDCP1-Tyr-734 in these cells directly correlated with the level of expression of CDCP1, whereas the level of phosphorylation of FAK-Tyr-861 was inversely correlated with the level of expression of CDCP1 (Fig. 6B). Phosphorylation of FAK-Tyr-397 was unaffected by CDCP1 expression levels. These data suggest that SFKs switch between CDCP1-Tyr-734 and FAK-Tyr-861 as phosphorylation substrates, depending on the level of CDCP1 expressed by HeLa CDCP1 cells. As shown in Fig. 6C, the degree of a fibroblastic morphology present in HeLa CDCP1 clones 1–4 was consistent with the level of expression of CDCP1; the lower the level of CDCP1 expression, the more epithelial the cell morphology and the higher the level of CDCP1 expression the more mesenchymal the cell morphology.
FIGURE 6.
Phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 in HeLa CDCP1 cells is inversely related and depends on the level of expression of CDCP1, but CDCP1 and FAK do not interact. Analyses of four HeLa cell clones stably expressing different levels of CDCP1 are shown. A, anti-CDCP1 (antibody 10D7) flow cytometry analysis of four HeLa CDCP1 cell clones (numbered 1–4) that express CDCP1 at increasing levels. HeLa vector and prostate cancer PC3 cells were analyzed as negative and positive controls for CDCP1 expression (dotted lines). The mean fluorescence intensity (Mean-FI) for each cell line is shown. B, Western blot analysis of lysates from HeLa CDCP1 cell clones using anti (α)-FLAG, -p-FAK-Tyr-861 (which also recognizes p-CDCP1-Tyr-734 (20)), -p-FAK-Tyr-397, -FAK, -Src, and -GAPDH antibodies. Phosphorylated CDCP1-Tyr-734 and FAK-Tyr-861 are indicated by arrows. C, transmitted light microscopy analysis. Cells were imaged using an Olympus XX-41 microscope. Arrow, epithelial morphology; arrowhead, elongated, fibroblastic morphology. D, Western blot analysis of lysates and proteins immunoprecipitated with an anti-FLAG antibody from HeLa vector, HeLa CDCP1, and HeLa CDCP1-Y734F (clone 4) cells using antibodies against FLAG, FAK, p-tyrosine and tubulin.
As shown in Fig. 1C, Src, CDCP1, and PKCδ form a complex in adherent HeLa CDCP1 cells, and these interactions are dependent on CDCP1-Tyr-734. As our data also indicate that SFKs switch between CDCP1-Tyr-734 and FAK-Tyr-861 and this switching is dependent on CDCP1-Tyr-734, we examined whether Src, CDCP1, and FAK form a similar complex. Our Western blot analysis of immunoprecipitations performed with an anti-FLAG antibody indicated that Src, CDCP1, and FAK do not form a complex in adherent HeLa cells (Fig. 6D).
Changes in Phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 Are Inversely Related during Changes in Attachment of HeLa CDCP1 Cells
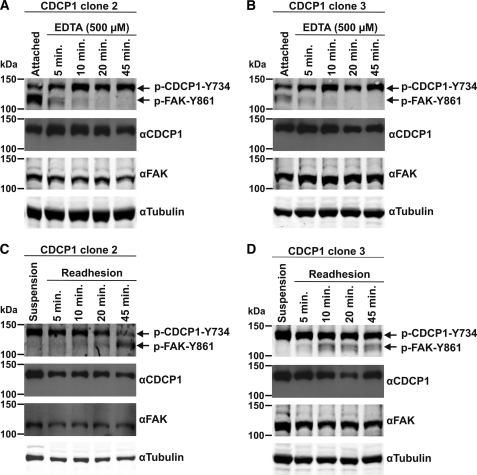
As it is known that CDCP1-Tyr-734 (20, 23, 29, 32, 42) and FAK-Tyr-861 are differentially phosphorylated during cell adhesion/de-adhesion (43), we also used changes in attachment of HeLa CDCP1 cells to initiate changes in phosphorylation of these proteins. To examine changes occurring during loss of cell adhesion, we analyzed cells that had been treated with EDTA (500 μm) to initiate de-adhesion from plastic. To examine changes occurring during cell re-adhesion, we analyzed cells that had been allowed to adhere to plastic after a period of 30 min in suspension. As shown in Fig. 7, A and B, de-adhesion of both HeLa CDCP1 clones 2 and 3 resulted in rapid reduction in phosphorylation of FAK-Tyr-861, and this was accompanied by increasing levels of p-CDCP1-Tyr-734. These changes were most obvious in clone 2 that under basal conditions has the highest levels of p-FAK-Tyr-861 and the lowest levels of p-CDCP1-Tyr-734. In cell adhesion experiments, reduction in phosphorylation of CDCP1-Tyr-734 in clone 2 was accompanied by increasing p-FAK-Tyr-861 levels (Fig. 7C). For clone 3, which has higher levels of basal p-CDCP1-Tyr-734, reduction of phosphorylation of this site during cell adhesion was not as apparent, but p-FAK-Tyr-861 levels increased during the 45-min attachment period. These data indicate that in cells stably expressing CDCP1, phosphorylation switching between FAK-Tyr-861 and CDCP1-Tyr-734 can be induced by changes in cell attachment.
FIGURE 7.
Changes in phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 are inversely related during changes in attachment of HeLa CDCP1 cells. Lysates from HeLa CDCP1 cells undergoing either de-adhesion induced by EDTA over 45 min or re-adhesion over 45 min (after suspension growth for 30 min) were analyzed by Western blot analysis using antibodies against p-FAK-Tyr-861 (which also recognizes p-CDCP1-Tyr-734 (20)), CDCP1 (#4115), FAK, and tubulin. A, HeLa CDCP1 clone 2 cells undergoing de-adhesion. B, HeLa CDCP1 clone 3 cells undergoing de-adhesion. C, HeLa CDCP1 clone 2 cells undergoing re-adhesion. D, HeLa CDCP1 clone 3 cells undergoing re-adhesion. The data in A–D are representative of two independent experiments.
SFK Switching between FAK-Tyr-861 and CDCP1-Tyr-734 Occurs Endogenously in Colorectal Cancer SW480 and HCT116 Cells and Depends on the State of Cell Adhesion
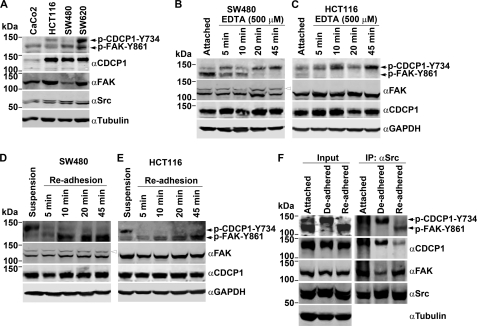
The above data indicate that in HeLa CDCP1 cells, FAK-Tyr-861 and CDCP1-Tyr-734 compete as SFK substrates. We next examined competition between these sites for SFK phosphorylation in cells endogenously expressing FAK and CDCP1. To identify appropriate cell lines for this study, we assessed the expression of 135-kDa p-CDCP1-Tyr-734 and 125-kDa p-FAK-Tyr-861 in 4 colon cancer cell lines (CaCo2, HCT116, SW480, and SW620). As shown in Fig. 8A, the highest levels of basal phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 were seen in SW620 cells, with HCT116 cells showing lower levels of basal phosphorylation of these proteins. Phosphorylated FAK-Tyr-861 was also detected in both SW480 and CaCo2 cells. In these two lines p-CDCP1-Tyr-734 was detectable only in SW480 cells and at very low levels. Two of the cell lines showing intermediate levels of p-CDCP1-Tyr-734 and p-FAK-Tyr-861, HCT116 and SW480, were selected to examine SFK switching in endogenous expressing cells. Changes in cell adhesion, as described above (Fig. 7), were used to initiate changes in phosphorylation of these proteins. As shown in Fig. 8B, de-adhesion of SW480 cells resulted in rapid reduction in phosphorylation of FAK-Tyr-861, and this was accompanied by increasing levels of p-CDCP1-Tyr-734. Loss of p-FAK-Tyr-861 in response to EDTA was even more rapid in HCT116 cells, and this was accompanied by an increase in p-CDCP1-Tyr-734 (Fig. 8C). In cell adhesion experiments, loss of p-CDCP1-Tyr-734 occurred within 5 min of adhesion to plastic of both SW480 and HCT116 cells, and this was accompanied by a gradual increase in levels of p-FAK-Tyr-861 (Fig. 8, D and E). Of note, the most pronounced SFK switching between p-FAK-Tyr-861 and p-CDCP1-Tyr-734 occurred in SW480 cells undergoing de-adhesion, which is consistent with the low levels of basal phosphorylation of p-CDCP1-Tyr-734 seen in these cells.
FIGURE 8.
Changes in phosphorylation of CDCP1-Tyr-734 and FAK-Tyr-861 are inversely related during changes in attachment of SW480 and HCT116 cells. A, anti (α)-p-FAK-Tyr-861 (which also recognizes p-CDCP1-Tyr-734 (20)), -CDCP1, -FAK, -Src, and -GAPDH Western blot analysis of lysates from colon cancer Caco2, HCT116, SW480, and SW620 cells. B and C, anti-p-FAK-Tyr-861, -CDCP1, -FAK, and -GAPDH Western blot analysis of lysates from adherent cells and cells treated for up to 45 min with EDTA. D and E, anti-p-FAK-Tyr-861, -FAK, -CDCP1, and -GAPDH Western blot analysis of lysates from cells maintained in suspension for 30 min and cells undergoing re-adhesion for up to 45 min. The data in B–E are representative of at least three independent experiments. F, Western blot analysis of lysates (input; left) and anti-Src immunoprecipitates (IP, right) from HCT116 cells either grown adhered or treated for 45 min with EDTA (De-adhered) or undergoing re-adhesion for up to 45 min (Re-adhered). Lysates and immunoprecipitated proteins were analyzed with anti-CDCP1, -FAK, -p-FAK-Tyr-861, and -Src antibodies, and lysates were also examined with an anti-tubulin antibody.
To directly examine interactions between Src and FAK and between Src and CDCP1, we also performed anti-Src immunoprecipitations. For these experiments we employed HCT116 cells that express p-CDCP1-Tyr-734 and p-FAK-Tyr-861 at similar levels under adherent conditions (Fig. 8A). Immunoprecipitations were performed from adhered cells and cells that had undergone EDTA-induced de-adhesion for 45 min as well as from cells that had undergone re-adhesion for 45 min after suspension growth for 30 min. Western blot analysis of lysates and anti-Src-immunoprecipitated proteins indicated that in adhered cells both CDCP1 and FAK interact with Src and that both CDCP1 and FAK are phosphorylated (Fig. 8F). After de-adhesion, higher levels of CDCP1 and lower levels of FAK co-precipitated with Src. It was also apparent that whereas the level of phosphorylation of co-precipitating CDCP1-Tyr-734 increased, co-precipitating FAK-Tyr-861 was unphosphorylated (Fig. 8F). Also consistent with a mechanism where Src switches between phosphorylation of FAK-Tyr-861 and CDCP1-Tyr-734, re-adhesion of HCT116 cells resulted in immunoprecipitation of lower levels of CDCP1 and higher levels of FAK, with marked reduction in p-CDCP1-Tyr-734 levels and re-appearance of phosphorylated FAK-Tyr-861 (Fig. 8F). Consistent data were obtained from HeLa CDCP1 clone 3 cells (data not shown).
SFK Switching between FAK-Tyr-861 and CDCP1-Tyr-734 Does Not Involve Formation of a SFK·CDCP1·FAK Complex
It has been reported that a trimeric SFK·CDCP1·PKCδ complex forms in response to a number of stimuli including cell de-adhesion (23) and CDCP1 proteolysis (33). To examine whether an analogous complex of SFK·CDCP1·FAK forms during de-adhesion of SW480 and HCT116 cells, we performed immunoprecipitations using anti-CDCP1 and anti-FAK antibodies. To take into account that such a complex is likely to be transitory during SFK switching, immunoprecipitations were performed on lysates from both adherent cells as well as cells actively undergoing de-adhesion over a 1-h time course. As shown in supplemental Fig. 6, immunoprecipitations with both anti-CDCP1 and anti-FAK antibodies co-immunoprecipitated Src from SW480 cells. Densitometry analysis of two independent experiments indicated that Src binding to CDCP1 increased by ∼5-fold over the 1-h time course of cell de-adhesion (left panel), which was accompanied by a rapid reduction in the level of Src bound to FAK (right panel). However, both immunoprecipitations failed to provide evidence of co-immunoprecipitation of CDCP1 and FAK during de-adhesion of SW480 cells (supplemental Fig. 6A, compare left and right panel). Similar data were obtained from the same time course of de-adhesion of HCT116 cells. In these cells Src binding to CDCP1 increased by almost 5-fold over the 1-h time course of cell de-adhesion, and this was accompanied by a decrease of ∼60% in binding of Src to FAK (supplemental Fig. 6B). Once again there was no evidence indicating formation of a SFK·CDCP1·FAK complex in these cells (supplemental Fig. 6B, compare left and right panel).
Silencing of CDCP1 Induces a More Epithelial Morphology and Increases p-FAK-Tyr-861 Levels in SW480 Cells
CDCP1 was silenced in colon cancer SW480 cells to examine the relationship between CDCP1, CDCP1-Tyr-734/FAK-Tyr-861 phosphorylation and cell morphology. Quantitative shRNA-mediated knockdown of CDCP1 resulted in increased phosphorylation of p-FAK-Tyr-861 and complete loss of the already low levels of p-CDCP1-Tyr-734 apparent in control (Fig. 9A) and parental (Fig. 8A) SW480 cells. It was also accompanied by the change from an elongated morphology characteristic of control SW480 cells to a more epithelial morphology (Fig. 9B).
FIGURE 9.
Silencing of CDCP1 in SW480 cells induces a more epithelial morphology and increases p-FAK-Tyr-861 levels. A, SW480 cells were stably transfected with either a CDCP1 shRNA knockdown construct or a scramble control construct. Lysates of these cells were examined by Western blot analysis using antibodies against CDCP1 (#4115), FAK, p-FAK-Tyr-861, and tubulin. Longer exposures of the anti-p-FAK-Tyr-861 blot shows p-CDCP1-Tyr-734 in scramble control cells, but these mask the increase in p-FAK-Tyr-861 levels that accompanies knockdown of CDCP1. B, transmitted light microscopy analysis. Cells were imaged using a Nikon Eclipse TE2000-U microscope. Arrow, epithelial morphology; arrowhead, elongated, fibroblastic morphology. Bar, 100 μm,
DISCUSSION
We report evidence for a novel molecular event that involves SFK substrate switching between Tyr-861 of FAK and Tyr-734 of the transmembrane protein CDCP1. Our data suggest that this switching can be manifested by at least two mechanisms; (i) increased expression of CDCP1 and (ii) changes in cell attachment. As these events are common to a number of cancers, the identified SFK switching mechanism may be relevant to malignant transformation. Also of note, our light microscopy analyses of HeLa cells stably expressing CDCP1 and SW480 colorectal cancer cells stably silenced for this protein suggest that this SFK switching mechanism is involved in altering cell morphology. We saw that stable CDCP1 expression in HeLa cells induces a mesenchymal morphology that is dependent on CDCP1-Tyr-734. Consistently, knockdown of CDCP1 in SW480 cells causes a more epithelial morphology.
Evidence that FAK-Tyr-861 and CDCP1-Tyr-734 compete as SFK substrates comes from our analysis of endogenous-expressing colorectal cancer cell lines as well as HeLa cells stably expressing CDCP1. Whereas HeLa vector cells, which do not endogenously express CDCP1, exhibit phosphorylation of FAK-Tyr-861, HeLa CDCP1 cells show a complete absence of phosphorylation at this site, whereas tyrosine 734 of CDCP1 is heavily phosphorylated. The essential role for CDCP1-Tyr-734 in SFK switching between FAK and CDCP1 was indicated from analysis of HeLa CDCP1-Y734F cells, which show restored FAK-Tyr-861 phosphorylation and a complete loss of tyrosine phosphorylation of CDCP1. Also of note, silencing of CDCP1 in HeLa CDCP1 cells restored p-FAK-Tyr-861 levels as well as the epithelial morphology, reminiscent of control HeLa cells. Importantly, we also showed that two other CDCP1 tyrosines (Tyr-743 and Tyr-762), that are known to also be phosphorylated by SFKs, do not play a role in SFK switching. These changes in phosphorylation of FAK-Tyr-861 and CDCP1-Tyr-734 correlated with changes in the level of expression of CDCP1 in four different HeLa cell clones. In these clones p-FAK-Tyr-861 levels decreased as CDCP1 levels increased, whereas p-CDCP1-Tyr-734 levels increased with increasing levels of CDCP1.
Significantly, SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734 was also induced by changes in cell attachment of not only HeLa CDCP1 cells but also colorectal cancer SW480 and HCT116 cells. These analyses of cells actively undergoing changes in cell attachment indicated the dynamic nature of SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734. In these cells, de-adhesion induced by EDTA results in rapid accumulation of p-CDCP1-Tyr-734 with concomitant loss of p-FAK-Tyr-861. In re-adhering cells, the reverse biochemical changes were apparent with p-CDCP1-Tyr-734 reducing 5–10 min after commencement of re-adhesion with an attendant increase in phosphorylation of p-FAK-Tyr-861. Our proposal for an Src switching mechanism is also supported by data from anti-Src immunoprecipitations. We observed increased binding of Src to phosphorylated and total CDCP1-Tyr-734 after cell de-adhesion with concomitant loss of the Src·p-FAK-Tyr-861 complex. The converse was apparent after cell re-adhesion, with reduced levels of Src·p-CDCP1-Tyr-734 and increased levels of Src·p-FAK-Tyr-861. These data suggest that during changes in cell adhesion, binding of Src and CDCP1-Tyr-734 correlates inversely with Src binding to FAK-Tyr-861 and supports a mechanism involving Src switching between these two substrates.
Our data are largely consistent with a recent report from Spassov et al. (29) who observed that CDCP1 is tyrosine-phosphorylated on cell detachment, whereas proteins involved in stabilization of focal adhesion, FAK and paxillin, are dephosphorylated. These authors proposed that CDCP1 signaling and focal adhesion signaling function in opposition during changes in cell attachment (29, 44). This is consistent with our observations that suggest that this opposition defines an SFK-mediated switch. Our data provide novel mechanistic insight into this switch by now defining CDCP1-Tyr-734 and FAK-Tyr-861 as particular tyrosines that are differentially phosphorylated during SFK-mediated switching.
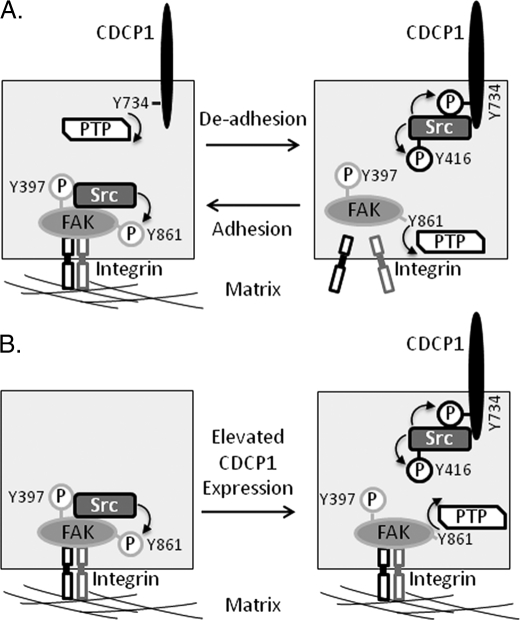
A model of molecular events potentially associated with this SFK switching is shown in Fig. 10. In adhered cells it is known that Src binds to p-FAK-Tyr-397 and phosphorylates several other FAK tyrosines including Tyr-861 (45). Consistent with previous reports (20, 23, 29), we have shown here that CDCP1-Tyr-734 is rapidly dephosphorylated during cell adhesion. However, the protein-tyrosine phosphatase that mediates this dephosphorylation is not yet known (Fig 10A). Our data indicate that during cell de-adhesion, Src switches from FAK-Tyr-861 to phosphorylation of CDCP1-Tyr-734. The rapid loss of phosphorylation of FAK-Tyr-861 during this event also indicates the action of an as yet unidentified protein-tyrosine phosphatase.
FIGURE 10.
SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734. A, in adherent cells Src binds to p-FAK-Tyr-397 and phosphorylates several other FAK tyrosines including Tyr-861 (45). We and others (20, 23, 29) have shown that CDCP1-Tyr-734 is rapidly dephosphorylated during cell adhesion, but the protein-tyrosine phosphatase (PTP) that mediates this dephosphorylation is not yet known. Src phosphorylation of CDCP1-Tyr-734 also results in increased phosphorylation of Src-Tyr-416. In de-adhered cells our data indicate that Src switches to phosphorylation of CDCP1-Tyr-734, and we have shown that FAK-Tyr-861 is rapidly dephosphorylated in this setting. The PTP that mediates FAK-Tyr-861 dephosphorylation is also not known. B, in cells with elevated expression of CDCP1, Src phosphorylates CDCP1-Tyr-734, which is accompanied by dephosphorylation of FAK-Tyr-861. In HeLa cells stably expressing CDCP1, phosphorylation of FAK-Tyr-397 is maintained, indicating that the kinase activity of FAK, and its role in focal adhesion is retained.
In our experiments in which we increased expression of CDCP1, we also observed robust SFK-mediated phosphorylation of CDCP1-Tyr-734 that was accompanied by dephosphorylation of FAK-Tyr-861 (Fig 10B). Interestingly, whereas we showed that HeLa cells stably expressing CDCP1 retain phosphorylation of FAK-Tyr-397, Spassov et al. (29) noted that induction of expression of CDCP1 in MDA-MB-468 breast cancer cells results in loss of phosphorylation of this site. As p-FAK-Tyr-397 is required for dynamic focal adhesion assembly and disassembly (46), this difference in phosphorylation of FAK-Tyr-397 may explain at least in part the different changes in morphology induced in HeLa and MDA-MB-468 cells by CDCP1 expression. Although HeLa CDCP1 cells acquire a fibroblast-like appearance contrasting with the epithelial morphology of parental and vector-transfected cells, MDA-MB-468-CDCP1 cells lose adhesion to the underlying matrix. The difference in phosphorylation of FAK-Tyr-397 in these cells likely also indicates that the protein-tyrosine phosphatase mediating p-FAK-Tyr-861 dephosphorylation in MDA-MB-468 cells is absent in HeLa cells.
We also note that whereas we observed increased p-FAK-Tyr-861 levels in colon cancer SW480 cells silenced for CDCP1, Razorenova et al. (27) recently reported that CDCP1 silencing in renal cell carcinoma 786-O cells did not alter phosphorylation of FAK-Tyr-861 or four other FAK tyrosines. Interestingly, although we saw a more epithelial morphology in SW480-shCDCP1 cells compared with control cells, Razorenova et al. (27) did not report a change in morphology of 786-O cells after CDCP1 knockdown. We have not explored the reason for these differences. However, we speculate that they may indicate that induction of changes in cell morphology, caused by reduced CDCP1 expression, is finely tuned to Src switching between FAK-Tyr-861 and CDCP1-Tyr-734. If this is correct, an altered cell morphology would only be expected where down-regulation of CDCP1 induces increased levels of p-FAK-Tyr-861.
Consistent with other reports showing that substrate binding accentuates phosphorylation of Src-Tyr-416 (39–41), our data indicate that Src binding to CDCP1-Tyr-734 induces increased p-Src-Tyr-416 levels. Consistently, an activating anti-CDCP1 antibody that induces increased phosphorylation of CDCP1-Tyr-734 also increases p-Src-Tyr-416 levels (47). These observations from cell lines are consistent with in vivo data showing that p-CDCP1-Tyr-734 and p-Src-Tyr-416 levels are elevated in tumor nodules of gastric cancer 44As3 cells during peritoneal invasion in mice (30). Also, Liu et al. (25) have shown recently that the ability of Src to enhance metastasis of melanoma cells in mice is dependent on a “feed-forward” loop involving CDCP1-Tyr-734-mediated enhancement of Src activation.
Several recent reports on FAK-Tyr-861 and growing literature on CDCP1, particularly CDCP1-Tyr-734, indicate that SFK switching between FAK-Tyr-861 and CDCP1-Tyr-734 will be relevant in physiological and pathological settings including those requiring changes in cell attachment. For example, FAK-Tyr-861 is phosphorylated under disparate conditions that are most clearly linked with changes in cell attachment. This site is phosphorylated in mouse mammary NMuMG cells during both migration and TGF β1-induced EMT (48). Also, phosphorylation of FAK-Tyr-861 is induced in HeLa cells grown on collagen (49) and in endothelial cells by vascular endothelial growth factor (50). In prostate cancer LNCaP cells, this site is dephosphorylated in cells grown in suspension, whereas in other prostate cancer lines, PC3 and DU145, changes in adhesion do not alter p-FAK-Tyr-861 levels (51). In addition, SFK-mediated phosphorylation of FAK-Tyr-861 and subsequent re-organization of filamentous actin can initiate an anti-apoptotic cascade that protects a range of epithelial cells from hyperosmotic stress (52). Furthermore, although phosphorylation of other FAK tyrosines was largely unaltered, p-FAK-Tyr-861 levels increased markedly on formation of rat bladder tumor NBT-II cell-cell contacts (43).
In contrast with FAK-Tyr-861, which does not have a clearly defined pathological role, dysregulated CDCP1 expression is associated with cancer progression, and CDCP1-Tyr-734 is known to mediate cancer promoting phenotypes in vitro and in vivo. For example, increased CDCP1 levels correlate with poor prognosis in lung cancer (53), renal cell carcinoma (54), and pancreatic cancer (26), and studies of small numbers of patients indicate that elevated CDCP1 expression may also occur in colorectal (19, 55) and breast (56) cancer. Consistent with a functional role in human malignancies for SFK-mediated switching between phosphorylation of FAK-Tyr-861 and CDCP1-Tyr-734, levels of this second phosphotyrosine are elevated in human gastric cancers in tumor cells invading the gastric wall (30), and in lung cancer patient samples p-CDCP1-Tyr-734 is largely present in invading tumor cells (53). CDCP1-Tyr-734 has also been shown to regulate anoikis of lung cancer cells in vitro via a mechanism involving recruitment of PKCδ to CDCP1-Tyr-762 and the formation of a SFK·CDCP1·PKCδ complex (23). In addition, this site regulates in vitro pancreatic cancer cell migration, invasion, and extracellular matrix degradation in a tyrosine phosphorylation- and PKCδ-dependent manner (26), and CDCP1-Tyr-734 is required for melanoma cell dispersive growth in vitro and metastasis in mice (25). Our data and previous reports indicate that formation of the SFK·CDCP1·PKCδ complex can be driven by at least three mechanisms that are often dysregulated in cancer; that is, cell de-adhesion (23), proteolysis mediated by serine proteases (33), and as shown in this report, overexpression of CDCP1.
In summary, we have demonstrated a novel SFK switching mechanism between the substrates FAK-Tyr-861 and CDCP1-Tyr-734. In settings of increased expression of CDCP1 and loss of cell attachment, SFK almost exclusively phosphorylates CDCP1-Tyr-734 in preference to FAK-Tyr-861. Therapeutic targeting of this SFK switch may be a rational approach to treating diseases such as cancer.
Supplementary Material
Acknowledgments
We thank Dr. Leonore de Boer for expert technical assistance with confocal microscopy experiments, Dr. Brett Hollier and Abhishek Kashyap for assistance with virus-mediated silencing, and Dr. Ying Dong for provision of antibodies.
This work was supported by a National Health and Medical Research Council of Australia Fellowship 339732 (to J. D. H.), a Cancer Council Queensland grant (to J. D. H.), Wesley Research Institute Grant 2008/06 (to J. D. H.), a Ph.D. scholarship from the Cancer and Bowel Research Trust (to A. W.), and an Australian Research Council Futures Fellowship FT0991446 (to N. J. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- SFK
- Src family kinase
- CDCP1
- CUB-domain-containing protein 1
- EMT
- epithelial-mesenchymal transition
- FAK
- focal adhesion kinase.
REFERENCES
- 1. Parsons S. J., Parsons J. T. (2004) Oncogene 23, 7906–7909 [DOI] [PubMed] [Google Scholar]
- 2. Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 3. Irby R. B., Mao W., Coppola D., Kang J., Loubeau J. M., Trudeau W., Karl R., Fujita D. J., Jove R., Yeatman T. J. (1999) Nat. Genet. 21, 187–190 [DOI] [PubMed] [Google Scholar]
- 4. Irby R. B., Yeatman T. J. (2000) Oncogene 19, 5636–5642 [DOI] [PubMed] [Google Scholar]
- 5. Boyer B., Bourgeois Y., Poupon M. F. (2002) Oncogene 21, 2347–2356 [DOI] [PubMed] [Google Scholar]
- 6. Myoui A., Nishimura R., Williams P. J., Hiraga T., Tamura D., Michigami T., Mundy G. R., Yoneda T. (2003) Cancer Res. 63, 5028–5033 [PubMed] [Google Scholar]
- 7. Wheeler D. L., Iida M., Dunn E. F. (2009) Oncologist 14, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frame M. C. (2002) Biochim. Biophys. Acta 1602, 114–130 [DOI] [PubMed] [Google Scholar]
- 9. Thomas J. W., Ellis B., Boerner R. J., Knight W. B., White G. C., 2nd, Schaller M. D. (1998) J. Biol. Chem. 273, 577–583 [DOI] [PubMed] [Google Scholar]
- 10. Frisch S. M., Vuori K., Ruoslahti E., Chan-Hui P. Y. (1996) J. Cell Biol. 134, 793–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanks S. K., Polte T. R. (1997) Bioessays 19, 137–145 [DOI] [PubMed] [Google Scholar]
- 12. Gabarra-Niecko V., Schaller M. D., Dunty J. M. (2003) Cancer Metastasis Rev. 22, 359–374 [DOI] [PubMed] [Google Scholar]
- 13. Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol.. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 14. Schaller M. D., Hildebrand J. D., Shannon J. D., Fox J. W., Vines R. R., Parsons J. T. (1994) Mol. Cell. Biol. 14, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlaepfer D. D., Hanks S. K., Hunter T., van der Geer P. (1994) Nature 372, 786–791 [DOI] [PubMed] [Google Scholar]
- 16. Schlaepfer D. D., Hunter T. (1996) Mol. Cell. Biol. 16, 5623–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calalb M. B., Polte T. R., Hanks S. K. (1995) Mol. Cell. Biol. 15, 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calalb M. B., Zhang X., Polte T. R., Hanks S. K. (1996) Biochem. Biophys. Res. Commun. 228, 662–668 [DOI] [PubMed] [Google Scholar]
- 19. Hooper J. D., Zijlstra A., Aimes R. T., Liang H., Claassen G. F., Tarin D., Testa J. E., Quigley J. P. (2003) Oncogene 22, 1783–1794 [DOI] [PubMed] [Google Scholar]
- 20. Brown T. A., Yang T. M., Zaitsevskaia T., Xia Y., Dunn C. A., Sigle R. O., Knudsen B., Carter W. G. (2004) J. Biol. Chem. 279, 14772–14783 [DOI] [PubMed] [Google Scholar]
- 21. Benes C. H., Wu N., Elia A. E., Dharia T., Cantley L. C., Soltoff S. P. (2005) Cell 121, 271–280 [DOI] [PubMed] [Google Scholar]
- 22. Bhatt A. S., Erdjument-Bromage H., Tempst P., Craik C. S., Moasser M. M. (2005) Oncogene 24, 5333–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uekita T., Jia L., Narisawa-Saito M., Yokota J., Kiyono T., Sakai R. (2007) Mol. Cell. Biol. 27, 7649–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deryugina E. I., Conn E. M., Wortmann A., Partridge J. J., Kupriyanova T. A., Ardi V. C., Hooper J. D., Quigley J. P. (2009) Mol. Cancer Res. 7, 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H., Ong S. E., Badu-Nkansah K., Schindler J., White F. M., Hynes R. O. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyazawa Y., Uekita T., Hiraoka N., Fujii S., Kosuge T., Kanai Y., Nojima Y., Sakai R. (2010) Cancer Res. 70, 5136–5146 [DOI] [PubMed] [Google Scholar]
- 27. Razorenova O. V., Finger E. C., Colavitti R., Chernikova S. B., Boiko A. D., Chan C. K., Krieg A., Bedogni B., LaGory E., Weissman I. L., Broome-Powell M., Giaccia A. J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siva A. C., Wild M. A., Kirkland R. E., Nolan M. J., Lin B., Maruyama T., Yantiri-Wernimont F., Frederickson S., Bowdish K. S., Xin H. (2008) Cancer Res. 68, 3759–3766 [DOI] [PubMed] [Google Scholar]
- 29. Spassov D. S., Wong C. H., Sergina N., Ahuja D., Fried M., Sheppard D., Moasser M. M. (2011) Mol. Cell. Biol. 31, 766–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uekita T., Tanaka M., Takigahira M., Miyazawa Y., Nakanishi Y., Kanai Y., Yanagihara K., Sakai R. (2008) Am. J. Pathol. 172, 1729–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spassov D. S., Baehner F. L., Wong C. H., McDonough S., Moasser M. M. (2009) Am. J. Pathol. 174, 1756–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong C. H., Baehner F. L., Spassov D. S., Ahuja D., Wang D., Hann B., Blair J., Shokat K., Welm A. L., Moasser M. M. (2009) Clin. Cancer Res. 15, 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He Y., Wortmann A., Burke L. J., Reid J. C., Adams M. N., Abdul-Jabbar I., Quigley J. P., Leduc R., Kirchhofer D., Hooper J. D. (2010) J. Biol. Chem. 285, 26162–26173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brooks P. C., Lin J. M., French D. L., Quigley J. P. (1993) J. Cell Biol. 122, 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. (2000) Mol. Cell. Biol. 20, 9018–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hakkinen K. M., Harunaga J. S., Doyle A. D., Yamada K. M. (2011) Tissue Eng. Part A 17, 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sedelies K. A., Ciccone A., Clarke C. J., Oliaro J., Sutton V. R., Scott F. L., Silke J., Susanto O., Green D. R., Johnstone R. W., Bird P. I., Trapani J. A., Waterhouse N. J. (2008) Cell Death Differ. 15, 708–717 [DOI] [PubMed] [Google Scholar]
- 38. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xing L., Ge C., Zeltser R., Maskevitch G., Mayer B. J., Alexandropoulos K. (2000) Mol. Cell. Biol. 20, 7363–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arias-Salgado E. G., Lizano S., Sarkar S., Brugge J. S., Ginsberg M. H., Shattil S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13298–13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yadav S. S., Miller W. T. (2007) Cancer Lett. 257, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wortmann A., He Y., Deryugina E. I., Quigley J. P., Hooper J. D. (2009) IUBMB Life 61, 723–730 [DOI] [PubMed] [Google Scholar]
- 43. Playford M. P., Vadali K., Cai X., Burridge K., Schaller M. D. (2008) Exp. Cell Res. 314, 3187–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spassov D. S., Wong C. H., Moasser M. M. (2011) Cell Cycle 10, 1225–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McLean G. W., Carragher N. O., Avizienyte E., Evans J., Brunton V. G., Frame M. C. (2005) Nat. Rev. Cancer 5, 505–515 [DOI] [PubMed] [Google Scholar]
- 46. Hamadi A., Bouali M., Dontenwill M., Stoeckel H., Takeda K., Rondé P. (2005) J. Cell Sci. 118, 4415–4425 [DOI] [PubMed] [Google Scholar]
- 47. Alvares S. M., Dunn C. A., Brown T. A., Wayner E. E., Carter W. G. (2008) Biochim. Biophys. Acta 1780, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamura K., Yano H., Schaefer E., Sabe H. A. (2001) Oncogene 20, 2626–2635 [DOI] [PubMed] [Google Scholar]
- 49. Yano H., Mazaki Y., Kurokawa K., Hanks S. K., Matsuda M., Sabe H. (2004) J. Cell Biol. 166, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abu-Ghazaleh R., Kabir J., Jia H., Lobo M., Zachary I. (2001) Biochem. J. 360, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Slack J. K., Adams R. B., Rovin J. D., Bissonette E. A., Stoker C. E., Parsons J. T. (2001) Oncogene 20, 1152–1163 [DOI] [PubMed] [Google Scholar]
- 52. Lunn J. A., Jacamo R., Rozengurt E. (2007) J. Biol. Chem. 282, 10370–10379 [DOI] [PubMed] [Google Scholar]
- 53. Ikeda J., Oda T., Inoue M., Uekita T., Sakai R., Okumura M., Aozasa K., Morii E. (2009) Cancer Sci. 100, 429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Awakura Y., Nakamura E., Takahashi T., Kotani H., Mikami Y., Kadowaki T., Myoumoto A., Akiyama H., Ito N., Kamoto T., Manabe T., Nobumasa H., Tsujimoto G., Ogawa O. (2008) J. Cancer Res. Clin. Oncol. 134, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scherl-Mostageer M., Sommergruber W., Abseher R., Hauptmann R., Ambros P., Schweifer N. (2001) Oncogene 20, 4402–4408 [DOI] [PubMed] [Google Scholar]
- 56. Ikeda J. I., Morii E., Kimura H., Tomita Y., Takakuwa T., Hasegawa J. I., Kim Y. K., Miyoshi Y., Noguchi S., Nishida T., Aozasa K. (2006) J. Pathol. 210, 75–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.