Abstract
We have recently demonstrated that Notch pathway blockade by γ-secretase inhibitor (GSI) depletes cancer stem cells (CSCs) in Glioblastoma Multiforme (GBM) through reduced proliferation and induced apoptosis. However, the detailed mechanism by which the manipulation of Notch signal induces alterations on post-translational modifications such as glycosylation has not been investigated. Herein, we present a differential profiling work to detect the change of glycosylation pattern upon drug treatment in GBM CSCs. Rapid screening of differential cell surface glycan structures has been performed by lectin microarray on live cells followed by the detection of N-linked glycoproteins from cell lysates using multi-lectin chromatography and label-free quantitative mass spectrometry analysis. A total of 51 and 52 glycoproteins were identified in the CSC- and GSI-treated groups, respectively, filtered by a combination of decoy database searching and Trans-Proteomic Pipeline (TPP) processing. Although no significant changes were detected from the lectin microarray experiment, 7 differentially expressed glycoproteins with high confidence were captured after the multi-lectin column including key enzymes involved in glycan processing. Functional annotations of the altered glycoproteins suggest a phenotype transformation of CSCs toward a less tumorigenic form upon GSI treatment.
Keywords: Cancer stem cells, Gliobastoma, Glycoproteomics, Label-free, Lectin microarray, Multi-lectin chromatography
1 Introduction
The existence of cancer stem cells (CSCs) including in human brain tumors and the implications of promising new therapies that target this small subset of cells have been recently proposed [1–5]. Notch signaling has been demonstrated as one of the most important molecular mechanisms responsible for CSC properties and we and others have shown that knockdown of this pathway by γ-secretase inhibitor (GSI) results in attenuated propagation potential of CSCs in Glioblastoma Multiforme (GBM) [6–9], which is the most aggressive class of brain tumors. GSI suppresses signal transduction in Notch signaling by blocking the proteolytic step critical for releasing the functional intracellular domain of Notch receptor. However, little is known about the effect on the alteration of glycosylation upon the blockade of Notch signals. Glycoproteins play a critical role in cell–cell recognition events and glycosylation changes have been related to malignant transformation and tumor propagation. Besides the extensively studied role of phosphorylation cascade in Notch signaling, carbohydrate modification has also been shown as an essential regulation mechanism such as the modulation role of O-fucose glycans in Notch receptor function [10–12] and the tight correlation between N-glycosylation and stabilization of Nicastrin which is a component of the γ-secretase complex [13]. Therefore, it is important to study the changes of glycosylation patterns upon Notch pathway blockade by GSI in GBM in order to better understand the effects of drug treatment.
For the identification of glycoproteins, it is desirable to perform an enrichment step prior to downstream liquid chromatography (LC) coupled mass spectrometry (MS) analysis. The isolation of glycoprotein/glycopeptides is mainly implemented by hydrazide chemistry [14, 15] or affinity capture based on the recognition of different lectins to particular sugar moieties [16] or the combination of both [17]. Lectin-based affinity enrichment on the protein level facilitates potentially multiple glycosylation sites so as to strengthen the relatively weak non-covalent bindings. In addition to the use of a single lectin to capture a particular form of glycan structure, multi-lectin chromatography using lectins with broad specificities has been applied to analyze glycomes in different biological samples [18–21]. In our work, we have utilized three agarose-bound lectins: Concanavalin A (ConA), Wheat Germ Agglutinin (WGA) and Sambucus nigra Agglutinin (SNA) to produce a broad enrichment of N-linked glycoproteins simultaneously. Another technology for rapid screening of differential glycan structures lies in the development of the lectin microarray, where a large number of lectins are immobilized on a slide to profile the glycoproteins from cell lysates [22] or to obtain cell surface glycan signatures from live cells [23]. Our group has previously demonstrated the feasibility of coupling lectin microarrays for profiling live cells with LC-MS to identify cell surface glycoprotein markers [24].
In the present study, we have employed different strategies to target cell surface glycoproteins and intracellular membrane glycoproteins separately. The profiling of differential cell surface glycan structures has been performed by a fluorescent-assisted lectin microarray with a panel of 16 lectins. A larger scale profiling of N-linked glycoproteins from the soluble fraction of cell lysates has been performed by coupling multi-lectin chromatography with a label-free quantitative MS method. A selective list of differentially expressed glycoproteins following drug treatment has been found and the functional relevance of the altered glycosylation patterns has also been inferred and discussed to interpret the biological implications of our findings.
2 Materials and methods
2.1 Cell culture and treatment
GBM neurosphere cultures were obtained from the laboratory of Dr. Fan and maintained in Neurocult medium (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) supplemented with epidermal growth factor (10 ng/mL) and fibroblast growth factor (10 ng/mL) as previously described [7, 25]. The GBM neurosphere cultures possess the critical properties of neural stem cells including self-renewal and multilineage differentiation [25] and have been widely used in previous investigations [7–9]. Treatment studies were performed by growing cells in Neurocult medium overnight and replacing the next morning with medium containing GSI (Compound E, EMD Chemicals, Gibbstown) dissolved in dimethyl sulfoxide (DMSO) at 1 µM.
2.2 Lectin microarray
Sixteen lectins were utilized for the detection of differential cell surface glycan structures as previously described [24]. The carbohydrate specificities of these lectins are listed in Supporting Information 1. Briefly, each lectin was dissolved in PBS buffer to a concentration of 1 mg/mL and printed in three replicates on a SuperAmine slide (Arrayit, Sunnyvale, CA, USA) using a piezoelectric noncontact printer (Nano plotter; GeSiM, Germany). The slide was incubated in a humidity-controlled incubator (>45% humidity) overnight to allow lectin immobilization. After incubation, the slide was blocked with 1% BSA in PBS (pH 7.4) for 1 h. Fresh GBM CSCs and GSI-treated cells were labeled with 10 µM CFSE cell-tracing dye (Invitrogen, Carlsbad, CA, USA). The labeling procedure follows manufacture’s protocol. Generally, cells were washed with PBS thrice and reconstituted in 200 µL PBS. Nearly, 1.2 µL CFSE dye dissolved in DMSO was added and incubated at room temperature in dark for 15 min with agitation. Finally, cells were collected by centrifuging at 1500 × g for 5 min and reconstituted in 0.5mM CaCl2 and 1% BSA in PBS. Then, the labeled cells were incubated with lectin slides at room temperature for 40 min in darkness. After being washed with PBS for 5 min, the slides were air-dried and scanned with a microarray scanner (Genepix 4000A; Axon). Genepix 6.0 software was used to align microarray spots and extract the fluorescent intensity information of each spot which was further recorded in Microsoft Excel. The cell surface glycan expression was compared between non-treated CSCs and GSI-treated cells based on the average fluorescent intensity value of three spots for each cell group. Student’s t-test was used to determine the statistical significance of differences between the two cell groups.
2.3 Protein extraction
Cell pellets were resuspended in 1mL of lysis buffer (1% octyl-β-d-glucopyranoside, 150 mM NaCl and 1% protease inhibitor mixture (Sigma-Aldrich) in 20mM Tris-HCl, pH 7.4) and homogenized with 60 strokes in a Dounce glass homogenizer with a tight-fitting pestle on ice. The cell lysate was the centrifuged at 40 000 × g for 30 min at 4°C. Protein concentration from the supernatant was determined by Micro BCA™ Protein Assay Kit (Pierce/Thermo Scientific, Rockford, USA).
2.4 Multi-lectin affinity chromatography
A single Pierce disposable column was gravity-packed with 1.5mL of agarose-bound ConA, WGA and SNA at 1:1:1 v/v/v for individual samples from each biological replicate. The column was first equilibrated with 10 volume of binding buffer (20 mM Tris-HCl, Ph 7.4, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, and 1 mM MnCl2). Cell lysates containing 1 mg protein were diluted four times with binding buffer and passed through the column twice. The column was then washed with 4 volumes of binding buffer and eluted with 4 volumes of elution buffer (0.2 M methyl-α-d-mannopyronoside, 0.2 M N-acetyl-d-glucosamine, 0.2 M d-lactose and 0.5 M NaCl in 20 mM Tris pH 7.4). The eluent was buffer exchanged with 50 mM Ammonia Bicarbonate and concentrated by Microcon YM-10 centrifugal filter devices (10 k MWCO) at a final volume of 200 µL.
2.5 Online nano-RPLC and LTQ mass spectrometry
Trypsin digestion was performed by the same protocol described in our previous study [26] prior to online-RPLC (Paradigm MG4 micropump system, Michrom Biosciences, Auburn, CA, USA) connected to an LTQ mass spectrometer (Thermo Finnigan, San Jose, CA, USA). Tryptic digests were reconstituted in 100 µL of 5% ACN and 0.3% FA and introduced into an RPLC nano column (3 µm × 200Å ° , 0.1 mm × 150 mm, C18 AQ particles, Michrom) after a desalting nano trap (300 × 50 mm) (Michrom) with each injection of 20 µL. Each injection to MS analysis from the control and the GSI-treated group is referred to as a technical replicate in the following text. A 2-h linear gradient with 150 min from 5 to 40% ACN, 15 min from 5 to 80% ACN and another 15 min for equilibrium to 5% ACN was used. The remaining LTQ parameters are the same as previously described [27].
2.6 Database searching and data processing
MS/MS spectra were searched against Uniprot database using SEQUEST embedded in Proteome Discoverer (version 1.1.0.263). Searching parameters were specified as follows: (i) Fixed modification: carbamidomethylation of Cys residue with a mass shift of 57.02 Da; (ii) variable modification: oxidation of Met residue with a mass shift of 15.99 Da; (iii) two missed cleavage sites were allowed; (iv) peptide ion mass tolerance: 1.4 Da; (v) fragment ion mass tolerance: 1.0 Da; (vi) peptide charges +1, +2 and +3. Searching results were further uploaded to Scaffold (V.2.0) as msf format. Dual filtering criteria for protein identification were employed by combining FDR test from target-decoy database search with a cutoff p-value <0.05 and protein/peptide confidence above 95% probability with a minimum of two unique peptides per protein from Trans-Proteomic Pipeline (TPP) built in Scaffold. To be noted, employing either target-decoy database searching or TPP alone to filter protein identifications is commonly accepted. Thus, a combination of these two validation strategies represents a more stringent filtering criterion and further increases the identification confidence. Glycoproteins were then confirmed by searching against the post-translational modification annotation in Uniprot database. Spectral counts were parsed out and normalized by a global normalization method by setting the total sum spectral counts the same for all identified proteins in each technical run.
2.7 Western blot
Western Blot was performed essentially as previously described [26]. Briefly, 20µg of total proteins from each sample were separated by 4–20% SDS-PAGE and then transferred to PVDF membranes (Bio-Rad, CA, USA). After being blocked for 2 h, the membranes were incubated with antibodies including mouse monoclonal anti-BGAL, mouse polyclonal anti-P4HA1, rabbit polyclonal anti-GANAB, mouse monoclonal anti-beta Actin (Abcam, Cambridge, CA, USA), mouse monoclonal anti-CATD (BD Transduction Laboratories, Lexington, KY, USA) and, mouse monoclonal anti-THY1 (Abnova, Taibei, China) overnight. After washing thrice, the membranes were incubated with HRP-conjugated goat anti-rabbit or anti-mouse IgG (H + L) for 1 h. The blots were visualized with DAB stain (Vector Laboratory, WI, USA).
3 Results
3.1 Detection of surface glycoproteins by lectin microarray
To probe differential cell surface glycan structures, a panel of 16 lectins covering a wide range of binding specificities to different sugar moieties were printed on the slides and incubated with fluorescent-labeled live cells from the CSCs or GSI-treated group. A selection of these 16 lectins has been demonstrated to be useful in distinguishing glycosylation pattern changes using protein microarray in our previous work [28]. However, no significant intensity changes were detected as a function of the differential expression of cell surface glycans between these two groups. The detailed results can be found in Supporting Information 2. This indicates that the total amount of cell surface glycoproteins is not significantly altered after GSI treatment from a macro level point of view, although individual changes may be masked.
3.2 Profiling of intracellular glycoproteins by multi-lectin chromatography
The aforementioned negative results suggest a need for differential glycoprotein profilings on a larger scale. Thus, three widely used lectins (ConA, WGA and SNA) with major binding specificities towards α-linked mannose, N-acetyl-glucosamine and sialic acid-type structures were combined to enrich intracellular glycoproteins from the CSCs or GSI-treated group. While these lectins often recognize a range of carbohydrate structures, this multi-lectin enrichment strategy has been demonstrated to yield more complete capture of glycoproteins when compared with the use of a single lectin [19]. As shown in Fig. 1, each batch of CSCs or GSI-treated sample were processed in the same way via multi-lectin chromatography and analyzed by LC-MS/MS in triplicates. The whole cell lysates were adjusted to the same amount (1 mg) for each biological replicate of each sample prior to lectin enrichment and 10% of the eluent was introduced to the LC-MS/MS for each technical replicate. Supporting Information 3 and 4 list the information of proteins (FDR <0.05) identified from CSCs and GSI-treated cells, respectively. The dual filtering criteria by combining the threshold of FDR <0.05 from decoy database search and peptide and protein confidence probability >95% from TPP generated a total of 51 and 52 glycoprotein identifications for the CSC group and the GSI-treated group, respectively. We also compared the number of glycoprotein identifications by using a single FDR <0.01 alone. The number increased from 51 to 88 for the CSC group. This result is comparable to a similar study in our group where 73 glycoproteins were identified by using a different combination of lectins (ConA, WGA and PNA) for the same CSC group at FDR <0.01 [28]. Forty-seven glycoproteins were shared in common between these two studies and the remainder of the unique identifications may be due to the different glycan-binding specificity of the different lectin (SNA versus PNA) used in each study.
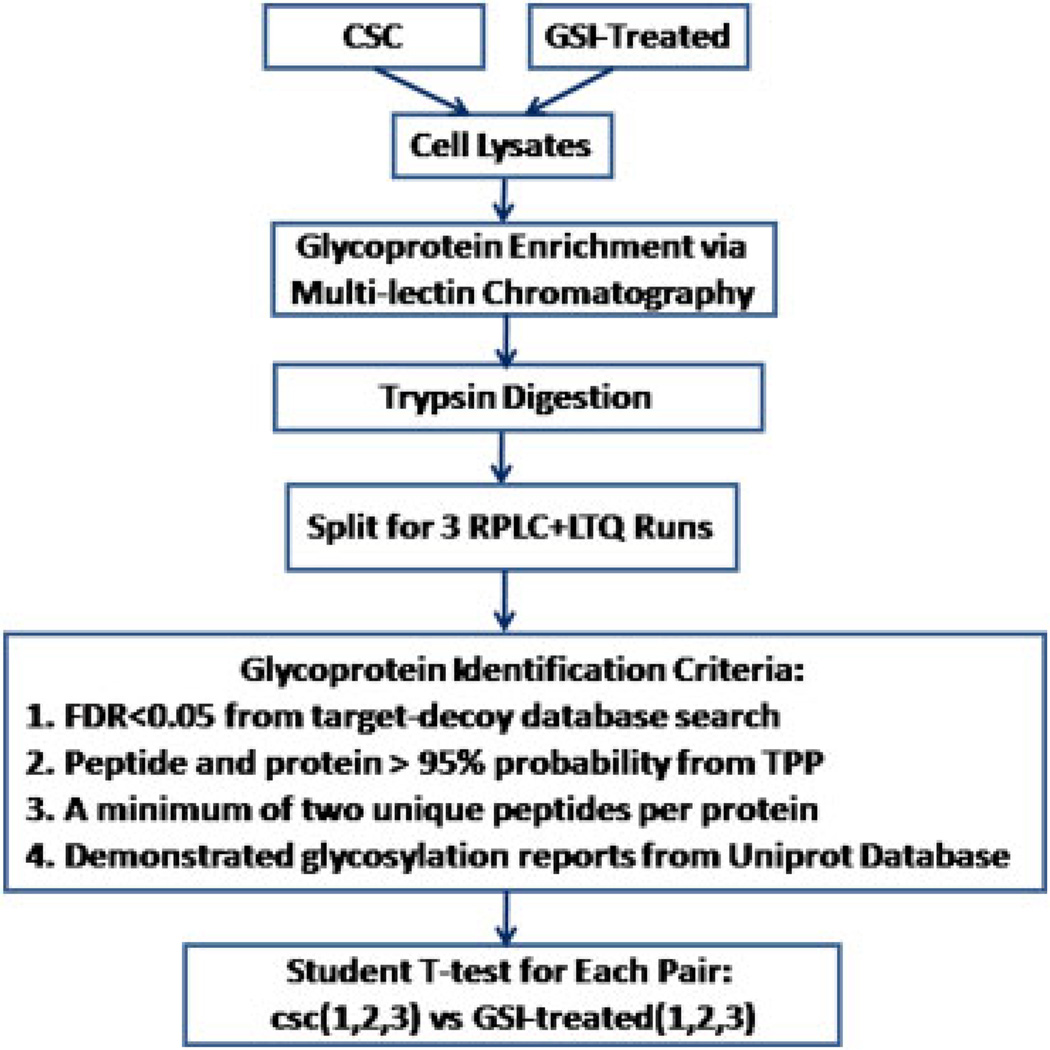
Figure 1.
Experimental workflow.
Student’s t-test was then used to capture the differentially expressed glycoproteins for each pair of biological replicates with a threshold p-value <0.05. Each treatment is essentially a split of the control group and then treated with GSI, thus, the differences resulting from the treatment effect as opposed to biological variance are more truly reflected when compared within each pair than across all pairs. Supporting Information 5 lists all the differential identifications from each pair-wise comparison. The differential expression of a protein is accepted when it has a p-value <0.05 at least two times out of the three pairs of comparisons and the direction of the changes should be consistent. This results in a total of seven differentially expressed glycoproteins after GSI treatment as listed in Table 1: β-galactosidase (BGAL), Calumenin (CALU), Deoxyribonuclease-2-α (DNS2A), Neural α-glucosidase AB (GANAB), Hypoxia up-regulated protein1 (HYOU1), Prolyl 4-hydroxylase subunit α-1 (P4HA1) and Serotransferrin (TRFE). Reproducibility of the label-free quantification data is also evaluated based on the average CV of the spectral counts across all differential expressions between technical replicates and biological replicates. The average CV across all technical replicates and biological replicates for CSC- and GSI-treated samples are 21 and 26%, respectively. Such information can also be found in Supporting Information 5.
Table 1.
Differentially expressed glycoproteins
| Protein | Entrez gene name | Location | Batch-1 | Batch-2 | Batch-3 |
|---|---|---|---|---|---|
| BGAL_HUMAN | Galactosidase, β 1 | Cytoplasm, Lysosome | 0.0022 | N/A | 0.0139 |
| CALU_HUMAN | Calumenin | ER, secreted | N/A | 0.0352 | 0.0154 |
| DNS2A_HUMAN | Deoxyribonuclease II, lysosomal | Lysosome | 0.0036 | 0.0213 | N/A |
| GANAB_HUMAN | Glucosidase, α; neutral AB | ER, Golgi | 0.0006 | N/A | 0.0010 |
| HYOU1_HUMAN | Hypoxia up-regulated 1 | ER | 0.0341 | N/A | 0.0169 |
| P4HA1_HUMAN | Prolyl 4-hydroxylase, α polypeptide 1 | ER | N/A | 0.0225 | 0.0297 |
| TRFE_HUMAN | Transferrin | Secreted | 0.0080 | N/A | 0.0441 |
Three pairs of Student’s t-test are performed on the three biological replicates with a total of 9 technical replicates for each sample.
p-Values <0.05 at least two times out of the three pairs of comparisons are accepted.
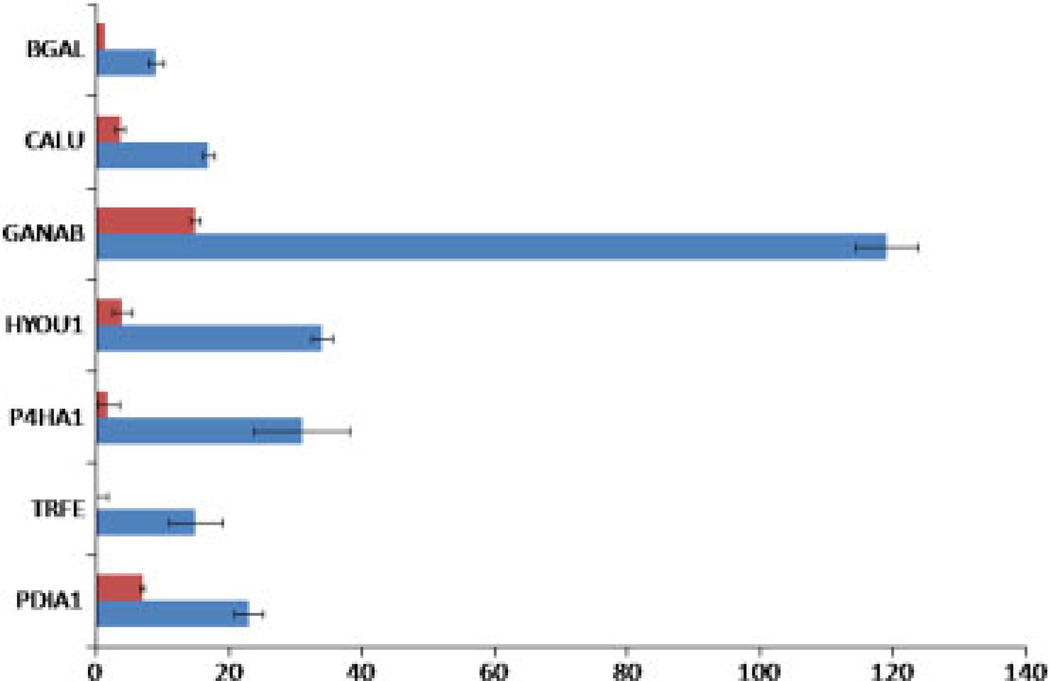
To compare the enrichment efficiency, a fraction of the whole cell lysates from the third batch was processed in the same manner excluding the step of multi-lectin chromatography and it was analyzed by LC-MS/MS in triplicate with each injection of ~1 µg. Figure 2 illustrates a comparison of the spectral counts assigned to each highly confident glycoprotein before and after multi-lectin chromatography (Note: DNS2A is not detected from batch 3.). The averaged spectral counts for each glycoprotein are significantly increased up to 17-fold after the enrichment. Protein disulfide-isomerase (PDIA) shown in the bottom of this graph is not a glycoprotein; however, binding of P4HA1 and PDIA has been previously reported [29]. Thus, the increase in PDIA after enrichment is expected as a consequence of protein–protein interaction.
Figure 2.
Comparison of protein expression level before (red bars) and after enrichment (blue bars). The horizontal axis represents spectral counts for each protein with error bars calculated in terms of standard error.
3.3 Verification of differential expression by Western blot
Five of the differentially expressed glycoproteins were selected for verification by Western Blot: GANAB, BGAL, P4HA1, Cathepsin D (CATD) and CD90 (THY1). The Western blot result of these proteins can be found in Supporting Information 6. The expression levels of the first four proteins are down-regulated after GSI treatment, whereas THY1 exhibits increased expression. The intensities of the bands are not quantitative results to compare the expression levels across different proteins. However, the lowest intensity detected for THY1 correlates well with the label-free MS results where this protein is assigned with the least spectral counts compared with others.
3.4 Protein–protein interaction network
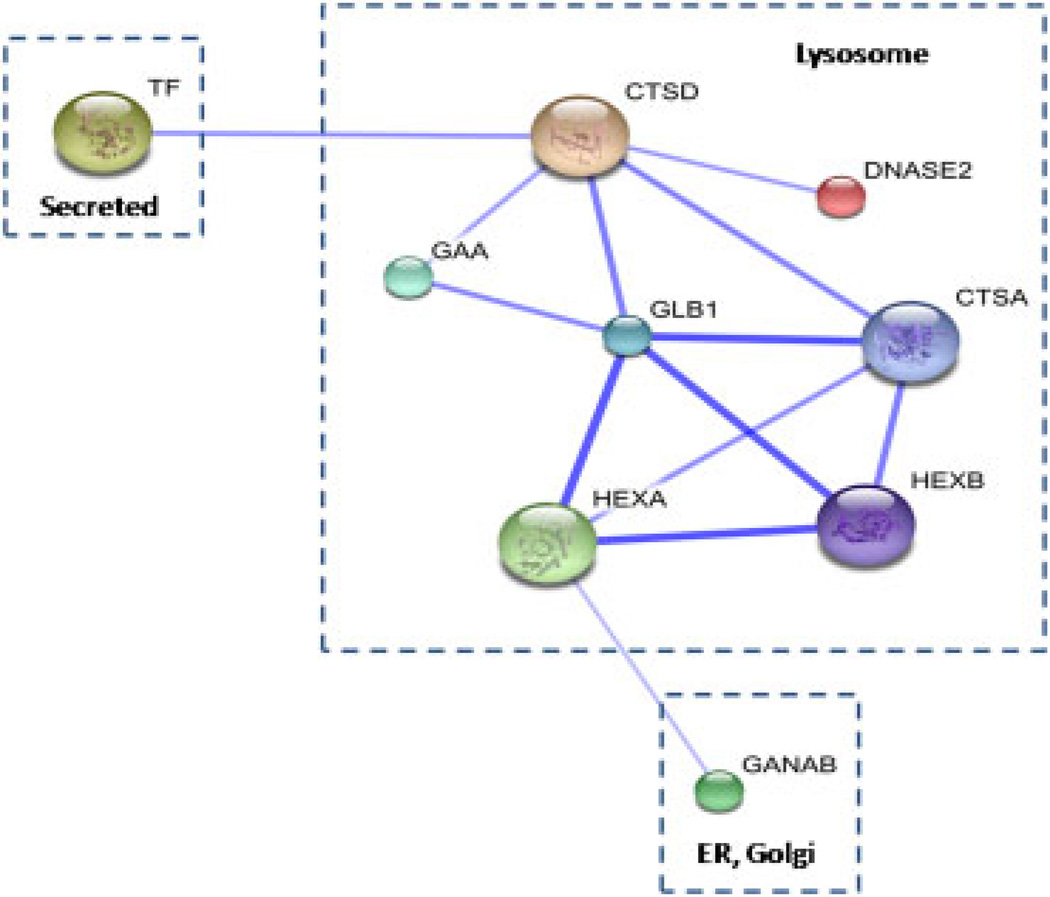
The functional relevance of the differentially expressed glycoproteins was searched against the STRING database, which enables public access to retrieve protein–protein interactions [30]. Nine glycoproteins are functionally linked to each other with medium to high confidence as displayed in Fig. 3. They are grouped according to their sub-cellular localizations and the thickness of an edge positively correlates with the level of association. The links between CTSD, GLB1, CTSA, HEXA and HEXB are of high confidence possibly due to their co-localization in the lysosome compartment. The strongest association is assigned between GLB1 and CTSA which have been demonstrated to interact to form the lysosomal multienzyme complex [31].
Figure 3.
Protein–protein interaction network generated by STRING. Each node represents a protein and each edge represents an interaction in between. A thicker line indicates stronger association.
4 Discussion
There is no significant pattern difference detected from surface glycoproteins between the drug treated and non-treated cells using the lectin microarray. In this experiment, whole cells instead of individual proteins are incubated with immobilized lectins. Thus, the signal is determined by an overall contribution from surface glycoproteins rather than individual proteins. The method itself is sensitive and specific if there is a pattern difference [22–24, 28]. However, the method cannot distinguish a difference at the individual protein level. It is not surprising that these two samples show no overall pattern difference. The treatment of GSI may not induce a significant difference on surface glycoproteins that can be detected by this method. We have therefore analyzed the intracellular glycoprotein levels using MS which will be sensitive to small differences in protein content.
Among the five differentially expressed glycoproteins validated by Western blot, GANAB is the α-subunit of an important ER-resident enzyme glucosidase II which sequentially cleaves inner α-1,3-linked glucose residues [32] and it has been previously reported to be purified by ConA [33]. The decreased expression after GSI treatment may suggest altered glycan processing effects on the maturation of glycoproteins. BGAL is an essential enzyme that catalyzes the hydrolysis of β-galactosides into monosaccharides. A previous report shows it can be purified from either a ConA or WGA lectin column [34]. The potential role in GBM CSCs remains unknown.
P4HA1 is another key enzyme in collagen synthesis which catalyzes the formation of 4-hydroxyproline and it exhibits binding to ConA. Decreased P4HA1 expression has been observed after knocking down its transcriptional regulator hypoxia-induced-factor 1-alpha(H1F-1α) in GBM CSCs [35]. In addition, the translocation of H1F-1α to bind Notch ICD and the convergence required to maintain stem cell properties have been proposed [36]. Therefore, the similar down-regulated pattern of P4HA1 after GSI treatment may be due to the blockade of Notch and the reduced level of Notch ICD.
CATD which is an essential lysosomal aspartyl endopeptidase is also detected to be down-regulated after treatment. It has been reported that the N-linked oligosaccharide chains consist of a cluster of mannose 6-sulfate residues [37] enabling the purification of this protein by ConA [38]. Overexpression of CATD stimulates breast cancer tumorigenicity and metastasis [39]. It has been reported to play an essential role at multiple breast cancer progression steps by promoting cell proliferation and angiogenesis via inhibition of tumor apoptosis [40]. Therefore, the reduced level of CATD after treatment may suggest a transformation of the phenotype towards a less tumorigenic form. This hypothesis is also in line with the detection of increased THY1 (also known as CD90) after treatment which points to the same inference. THY1 is a heavily N-glycosylated cell surface antigen with ConA-binding sites [41]. Previous studies proposed that THY1 is a putative tumor suppressor gene in ovarian cancer [42, 43] and nasopharyngeal carcinoma [44]. The protein expression level was found to be exclusively in the non-tumorigenic models as compared to their tumorigenic counterparts [42–44].
Three of the five proteins mentioned above are selected based on the criteria we described. The other two proteins (CATD and THY1) are selected because they represent interesting biological relevance although they do not appear to be statistically different according to our selection criteria. Also, there is no simple robust statistical test currently available for our case which represents a hierarchical data structure. One of the few would be the Generalized Linear Mixed Effects Model (GLMM) [45] which can be used to handle such a hierarchical data structure. Future efforts are needed to fit the data with GLMM to distinguish between the true group difference and the noise from technical/biological variance.
In addition to N-linked glycosylated proteins, non-glycoproteins were also detected by MS analysis after glycoprotein enrichment. The identifications of non-glycosylated proteins are mainly due to protein–protein interactions. Totally, 1% octyl-β-d-glucopyranoside used as the detergent in the lysis is considered to be a mild MS compatible surfactant [46] which may not disrupt strong bindings between proteins. For example, Ribophorin 1 is a glycoprotein detected with reduced expression level after treatment. A non-glycosylated protein Ubiqilin 4 is also detected with reduced level possibly due to its demonstrated binding to Ribophorin 1 [47]. The elution of a larger protein complex resulting from co-precipitation occurs in the case where the glycoprotein P4HA1 is the primary target. P4HA1 binds to PDIA exhibiting direct interaction with 1433G [48] which further binds to three other proteins. Future improvements on optimizing the lysis buffer or washing conditions will be beneficial to reduce the number of off-target identifications. Also, the incorporation of isotope labeling by inducing a larger mass shift that can be recognized by the mass spectrometer will be helpful to discover novel glycosylation sites.
In conclusion, in this work we investigate the alteration of glycosylation pattern upon treatment of GSI in GBM CSCs. We have utilized a combination of lectin microarray and muti-lectin chromatography coupled RPLC-MS analysis to target cell surface glycoproteins and glycoproteins from cell lysates, respectively. While no significant changes have been detected from microarray screening, several differentially expressed intracellular membrane proteins and plasma membrane proteins were captured by the multi-lectin enrichment approach. The finding of down-regulation of GANAB and BGAL may suggest an altered glycan processing while reduced level of CATD and increased expression of THY1 may imply attenuated proliferation and elevated apoptosis upon GSI treatment. Future improvements may involve the optimization of detergent condition and incorporation of isotope labeling to increase the percentage of glycoprotein identifications. Overall, our study provides information regarding the influence of GSI treatment on glycosylation in GBM CSCs which may lead to an improved understanding of drug mechanism.
Supplementary Material
Acknowledgments
This work was funded under the National Institute of Health Grants R01 49500 (D.M.L.) and R21 CA134623 (D.M.L.). The authors acknowledge grant support to Dr. Fan from Accelerate Brain Cancer Cure Project Award, American Brain Tumor Association Translational Grant, and Voices Against Brain Cancer Research Grant.
Abbreviations
- BGAL
β-galactosidase
- CATD
cathepsin D
- ConA
concanavalin A
- CSC
cancer stem cell
- GANAB
neural α-glucosidase AB
- GBM
Glioblastoma Multiforme
- GSI
γ-secretase inhibitor
- PDIA
protein disulfide-isomerase
- P4HAI
prolyl 4-hydroxylase subunit α-1
- SNA
Sambucus nigra Agglutinin
- TPP
Trans-Proteomic Pipeline
- WGA
Wheat Germ Agglutinin
Footnotes
The authors have declared no conflict of interest.
References
- 1.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat. Rev. Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 2.Read TA, Fogarty M, Markant SL, McLendon RE, et al. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward RJ, Lee L, Graham K, Satkunendran T, et al. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009;69:4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- 4.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert CA, Ross AH. Cancer stem cells: cell culture, markers, and targets for new therapies. J. Cell Biochem. 2009;108:1031–1038. doi: 10.1002/jcb.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockhausen MT, Kristoffersen K, Poulsen HS. The functional role of Notch signaling in human gliomas. Neuro Oncol. 2010;12:199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X, Khaki L, Zhu TS, Soules ME, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan X, Matsui W, Khaki L, Stearns D, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 9.Fan X, Eberhart CG. Medulloblastoma stem cells. J. Clin. Oncol. 2008;26:2821–2827. doi: 10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanley P. Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jafar-Nejad H, Leonardi J, Fernandez-Valdivia R. Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology. 2010;20:931–949. doi: 10.1093/glycob/cwq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi HHR. Role of glycosylation of Notch in development. Semin. Cell Dev. Biol. 2010;21:638–645. doi: 10.1016/j.semcdb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomita T, Katayama R, Takikawa R, Iwatsubo T. Complex N-glycosylated form of nicastrin is stabilized and selectively bound to presenilin fragments. FEBS Lett. 2002;520:117–121. doi: 10.1016/s0014-5793(02)02802-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 15.Chen R, Jiang X, Sun D, Han G, et al. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 16.Xiong L, Andrews D, Regnier F. Comparative proteomics of glycoproteins based on lectin selection and isotope coding. J. Proteome Res. 2003;8:651–661. doi: 10.1021/pr0340274. [DOI] [PubMed] [Google Scholar]
- 17.Kaji H, Saito H, Yamauchi Y, Shinkawa T, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J. Chromatogr. A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 19.Yang Z, Hancock WS. Monitoring glycosylation pattern changes of glycoproteins using multi-lectin affinity chromatography. J. Chromatogr. A. 2005;1070:57–64. doi: 10.1016/j.chroma.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Orazine CI, Hincapie M, Hancock WS, Hattersley M, Hanke JH. A proteomic analysis of the plasma glycoproteins of a MCF-7 mouse xenograft: amodel system for the detection of tumor markers. J. Proteome Res. 2008;7:1542–1554. doi: 10.1021/pr7008516. [DOI] [PubMed] [Google Scholar]
- 21.Lee HJ, Na K, Choi EY, Kim KS, et al. Simple method for quantitative analysis of N-linked glycoproteins in hepatocellular carcinoma specimens. J. Proteome Res. 2010;9:308–318. doi: 10.1021/pr900649b. [DOI] [PubMed] [Google Scholar]
- 22.Pilobello KT, Slawek D, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl. Acad. Sci. USA. 2007;104:11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao SC, Li Y, Zhou J, Qian J, et al. Lectin microarrays identify cell-specific and functionally significant cell surface glycan workers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Liui Y, Xie X, Zhu T, et al. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC-MS/MS approach. J. Proteome Res. 2010;9:2565–2572. doi: 10.1021/pr100012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galli R, Binda E, Orfanelli U, Cipelletti B, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 26.Dai L, Li C, Shedden KA, Misek DE, Lubman DM. Comparative proteomic study of two closely related ovarian endometrioid adenocarcinoma cell lines using cIEF fractionation and pathway analysis. Electrophoresis. 2009;30:1119–1131. doi: 10.1002/elps.200800505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai L, Li C, Shedden KA, Lee CJ, et al. Quantitative proteomic profiling studies of pancreatic cancer stem cells. J. Proteome Res. 2010;9:3394–3402. doi: 10.1021/pr100231m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Liu Y, Zhu TS, Xie X, et al. Glycoproteomic analysis of glioblastoma stem cell differentiation. J. Proteome Res. 2011;10:330–338. doi: 10.1021/pr101158p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J. Biol. Chem. 2003;278:47685–47693. doi: 10.1074/jbc.M306806200. [DOI] [PubMed] [Google Scholar]
- 30.Jensen LJ, Kuhn M, Stark M, Chaffron S, et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santamaria R, Chabás A, Callahan JW, Grinberg D, Vilageliu L. Expression and characterization of 14 GLB1 mutant alleles found in GM1-gangliosidosis and Morquio B patients. J. Lipid Res. 2007;48:2275–2282. doi: 10.1194/jlr.M700308-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier MF, Marsil A, Sevigny G, Jakob CA, et al. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology. 2000;10:815–827. doi: 10.1093/glycob/10.8.815. [DOI] [PubMed] [Google Scholar]
- 33.Martiniuk F, Ellenbogen A, Hirschhorn R. Identity of neutral alpha-glucosidase AB and the glycoprotein processing enzyme glucosidase II. Biochemical and genetic studies. J. Biol. Chem. 1985;260:1238–1242. [PubMed] [Google Scholar]
- 34.Heyworth CM, Wynn C. The binding of human liver acid beta-galactosidase to wheat-germ lectin is influenced by aggregation state of the enzyme. Biochem. J. 1982;201:615–619. doi: 10.1042/bj2010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Méndez O, Zavadil J, Esencay M, Lukyanov Y, et al. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol. Cancer. 2010;9:133. doi: 10.1186/1476-4598-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustafsson MV, Zheng X, Pereira T, Gradin K, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Journet A, C.hapel A, Jehan S, Adessi C, et al. Characterization of Dictyostelium discoideum cathepsin D. J. Cell Sci. 1999;112:3833–3843. doi: 10.1242/jcs.112.21.3833. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava PN, Ninjoor V. Isolation of rabbit testicular cathepsin D and its role in the activation of proacrosin. Biochem. Biophys. Res. Commun. 1982;109:63–69. doi: 10.1016/0006-291x(82)91566-2. [DOI] [PubMed] [Google Scholar]
- 39.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, et al. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Berchem G, Glondu M, Gleizes M, Brouillet JP, et al. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002;21:5951–5955. doi: 10.1038/sj.onc.1205745. [DOI] [PubMed] [Google Scholar]
- 41.Tamaki K. Heterogeneity of epidermal Thy-1-positive cells defined by lectin-binding sites. J. Invest. Dermatol. 1986;86:222–225. doi: 10.1111/1523-1747.ep12285165. [DOI] [PubMed] [Google Scholar]
- 42.Abeysinghe HR, Cao Q, Xu J, Pollock S, et al. THY1 expression is associated with tumor suppression of human ovarian cancer. Cancer Genet. Cytogenet. 2003;143:125–132. doi: 10.1016/s0165-4608(02)00855-5. [DOI] [PubMed] [Google Scholar]
- 43.Abeysinghe HR, Pollock S, Guckert NL, Veyberman Y, et al. The role of the THY1 gene in human ovarian cancer suppression based on transfection studies. Cancer Genet. Cytogenet. 2004;149:1–10. doi: 10.1016/S0165-4608(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 44.Lung HL, Bangarusamy D, Xie D, Cheung AK, et al. THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene. 2005;24:6525–6532. doi: 10.1038/sj.onc.1208812. [DOI] [PubMed] [Google Scholar]
- 45.Zeger SL, Karim MR. Generalized linear models with random effects; a Gibbs sampling approach. J. Am. Stat. Assoc. 1991;86:79–86. [Google Scholar]
- 46.McDonald CA, Yang J, Marathe V, Yen TY, Macher BA. Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol. Cell. Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim J, Hao T, Shaw C, Patel AJ, et al. Protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 48.Jin J, Smith F, Stark C, Wells CD, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.