Abstract
It is unclear whether mutations in fused in sarcoma (FUS) cause familial amyotrophic lateral sclerosis via a loss-of-function effect due to titrating FUS from the nucleus or a gain-of-function effect from cytoplasmic overabundance. To investigate this question, we generated a series of independent Caenorhabditis elegans lines expressing mutant or wild-type (WT) human FUS. We show that mutant FUS, but not WT-FUS, causes cytoplasmic mislocalization associated with progressive motor dysfunction and reduced lifespan. The severity of the mutant phenotype in C. elegans was directly correlated with the severity of the illness caused by the same mutation in humans, arguing that this model closely replicates key features of the human illness. Importantly, the mutant phenotype could not be rescued by overexpression of WT-FUS, even though WT-FUS had physiological intracellular localization, and was not recruited to the cytoplasmic mutant FUS aggregates. Our data suggest that FUS mutants cause neuronal dysfunction by a dominant gain-of-function effect related either to neurotoxic aggregates of mutant FUS in the cytoplasm or to dysfunction in its RNA-binding functions.
INTRODUCTION
Mutations in the fused in sarcoma (FUS; ALS6) genes (1–3) cause familial forms of amyotrophic lateral sclerosis (ALS), and FUS may contribute to sporadic cases of frontotemporal lobar degeneration (FTLD). FUS encodes an RNA-binding protein that, under physiological conditions, is almost exclusively expressed in the nucleus (4–6). However, in patients with pathogenic FUS mutations and in patients with sporadic forms of FTLD-FUS, the FUS protein accumulates in the cytoplasm of neurons in the spinal cord and brain (7–10). The majority of mutations occur in the C-terminus of the FUS protein, which is thought to contain a nuclear targeting signal (11). This has led to the conclusion that mislocalization of mutant FUS causes either a loss-of-function effect (because of titration of FUS from the nucleus) or a gain-of-function effect (arising from aberrant accumulation of FUS in the cytoplasm) (12,13). The fact that mutations in the Tar DNA-binding protein 43 (TDP-43), another RNA-binding protein, also cause familial ALS has led to the hypothesis that FUS and TDP-43 might work by the same mechanism (13). To further explore the mechanism by which mutant FUS protein causes ALS, we chose to create transgenic models bearing wild-type (WT) or mutant human FUS in Caenorhabditis elegans. We chose this model for three reasons. First, C. elegans has an orthologue of human FUS, encouraging the notion that models built in this animal would have biological validity. Secondly, the genetics of C. elegans provides a powerful tool to seek upstream and downstream partners via enhancer and suppressor screens. Thirdly, the optical transparency of C. elegans allows the application of biophysical techniques to monitor the distribution and folding state of proteins of interest (14).
RESULTS
A series of WT and mutant human FUS transgenes were generated and were then expressed in C. elegans under the control of a pan-neuronal promoter (Fig. 1). These transgenes included: (i) full-length (FL) WT human FUS (WT-FUS); (ii) four different missense mutations associated with varying clinical severity (as defined by age at onset and by disease duration) of human ALS (R514G and R521G = mild; R522G = moderate and P525L = severe) (1–3) (Supplementary Material, Table S1) and (iii) two different C-terminal-truncated FUS constructs (FUS513 and FUS501—lacking the C-terminal 13 and 25 amino acids of FUS, respectively). While the C-terminal-truncated constructs are artificial, they are very similar to several human C-terminal splicing/frame-shifting truncation mutations that are also associated with severe ALS phenotypes (15,16). Each construct was cloned into a vector containing a C. elegans pan-neuronal promoter Prgef-1and an in-frame green fluorescent protein (GFP S65T) or red fluorescent protein (TagRFP) at the N-terminus. Multiple stable integrated transgenic FUS lines were obtained for each mutant and WT-FUS construct. The levels of FUS mRNA were determined for each line by quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR), and transgenic lines with similar levels of expression (1.00–1.78-fold of WT-FUS) were chosen for subsequent experiments (Supplementary Material, Fig. S1).
Figure 1.
Transgene constructs. WT FL FUS, FUS with five different clinical mutations (R514G, R521G, R522G, R524S and P525L) and two truncated FUS (FUS513 and FUS501) were generated and injected into C. elegans. GFP, green fluorescent protein; QGSY-rich, glutamine–glycine–serine–tyrosine-rich region; G-rich, glycine-rich region; RRM, RNA recognition motif; RGG-rich, arginine–glycine–glycine-rich region.
Mutations in FUS cause its accumulation in cytoplasmic inclusions
Transgenic animals expressing only GFP under the control of the same pan-neuronal Prgef-1 promoter showed diffuse nuclear and cytoplasmic signals in neurons (data not shown). In contrast, the GFP-tagged FL WT-FUS was detected only in the nuclei of neurons (nuclear FUS/total FUS ratio = 93 ± 3%, n = 20 animals per transgenic strain) (Figs 2A and B and 3A). The nuclear localization of WT-FUS is consistent with the physiological subcellular localization of FUS in healthy human neurons (1,2). Immunostaining with anti-FUS antibody revealed that the FUS immunoreactivity fully replicated the subcellular distribution of the GFP signal in the same animals, indicating that the GFP tag was a reliable indicator of FUS localization in vivo (Supplementary Material, Fig. S2).
Figure 2.
Representative images of transgenic FUS worms. (A) Live fluorescent images of neurons in the head and body and (B) confocal microscopic images of immunostained animals. Non-transgenic N2 animals show no fluorescent signals with either wide-field (A) or confocal (B) microscope. GFP signals are observed almost exclusively in the nuclei in WT-FUS (WT), R514G and R521G (A, arrow). GFP signals in WT and R514G and DAPI perfectly overlap, suggesting that FUS in these two strains is exclusively expressed in the nucleus (B, WT, arrow). R522G animals show both nuclear (A and B, R522G, arrow) and cytoplasmic FUS (A and B, arrowhead), but nuclear signal is still stronger (A and B). P525L, FUS513 and FUS501 animals show marked cytoplasmic GFP signals (A and B, arrowheads), which are rather stronger than those in nuclei (A and B, arrows). Staining patterns with anti-GFP and anti-FUS antibodies are almost the same (B; Supplementary Material, Fig. S2). There is no difference in the distribution of GFP-positive neurons in each line. Scale bars: 20 µm (A) and 2.0 µm (B).
Figure 3.
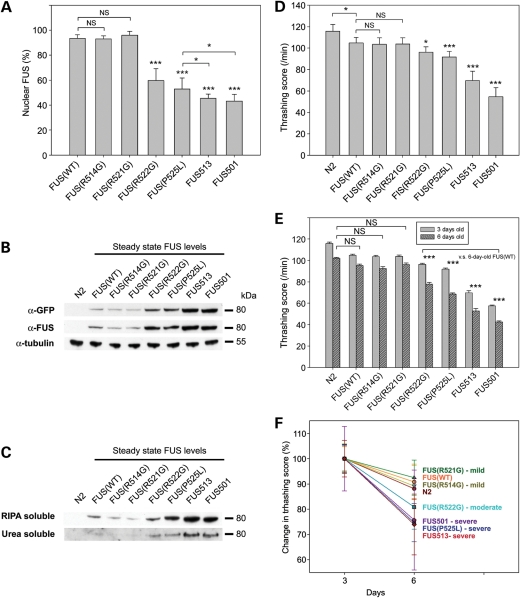
Features of transgenic worms. (A) Ratio of nuclear FUS in each transgenic strain. In WT-FUS animals (WT), 93 ± 3% of FUS is in the nucleus. Nuclear FUS in R514G and R521G do not differ from that in WT (92 ± 3 and 96 ± 3%, respectively). However, R522G and P525L show significantly lower ratio than WT (59 ± 10 and 52 ± 9%, respectively). FUS513 and FUS501 show even lower scores (44 ± 4 and 43 ± 5% of WT, respectively). NS, not significant. *P< 0.05 and ***P< 0.001. (B and C) Immunoblot analyses of FUS aggregation. (B) Whole lysates (80 μg) probed with anti-FUS or anti-GFP antibodies. α-Tubulin was used as a loading control. (C) RIPA-soluble and RIPA-insoluble/urea soluble fractions from the whole lysates were separated by sequential extraction and probed with anti-FUS antibody. (D–F) Thrashing assays. (D) Motor activity as measured by the thrashing assay at 3 days of age. WT-FUS animals (WT) show slightly lower score than non-transgenic N2 animals (P< 0.05). Compared with WT, there is no decrease in R514G and R521G, but R522G and P525L animals show significantly lower score by 8.2 ± 4.6 and 12.5 ± 5.0%, respectively (P< 0.05 for R521G and P< 0.001 for P525L). All truncated strains (FUS513 and FUS501) show marked decrease by ∼33.5 ± 8.5 and 45.2 ± 4.0% of WT (P< 0.001). (E) Motor activity, as measured by thrashing scores for 3-day-old and 6-day-old animals, shows age-related reductions in motor activity in all strains (11.9 ± 3.7% decrease in movement activity) in N2 non-transgenic animals. The extents of decline in movement activity in WT-FUS, R514G and R521G animals were not different from that of N2 animals. However, all of the other FUS mutants, including those associated with moderate and severe ALS phenotypes in humans (R522G, P525L, FUS513 and FUS501), show significantly larger decreases at 6 days of age (P< 0.001). NS, not significant. *P< 0.05 and ***P< 0.001. (F) The rate of progression of motor impairment normalized to the activity at day 3 shows that the R522L (moderate), R525L, FUS 513 and FUS501 (severe) animals have greater rates of progression in proportion to the severity of the human ALS phenotypes associated with those mutants.
In animals expressing R514G and R521G mutations, which have a mild ALS phenotype in human ALS cases (1), the pattern of GFP-FUS was identical to WT-FUS at all ages (nuclear FUS/total FUS ratio = 92 ± 3 and 96 ± 3%, respectively) (Figs 2A and B and 3A). In animals expressing the R522G mutation, which has a moderate human ALS phenotype (1), FUS was also present in the nuclei of neurons, but there was some FUS diffusely present in the cytoplasm (nuclear FUS/total FUS ratio = 59 ± 10%, P< 0.001 compared with WT-FUS) (Figs 2A and B and 3A). In animals expressing the clinically severe P525L mutation, nearly half of the total neuronal FUS was located in the cytoplasm in an uneven, clumpy distribution (nuclear FUS/total FUS ratio = 52 ± 9%, P< 0.001 compared with WT-FUS) (Figs 2A and B and 3A). Both truncated FUS constructs also showed strong, clumpy, cytoplasmic FUS with less nuclear FUS (nuclear FUS/total FUS ratio = 44 ± 4 and 43 ± 5% for FUS513 and FUS501, respectively) (Figs 2A and B and 3A).
Mutant FUS accumulates and aggregates in neuronal cytoplasm in vivo
To determine whether the cytoplasmic inclusions contained insoluble aggregates of FUS, we undertook sequential extraction of animals with RIPA and then with urea buffer. This analysis revealed that there were increased steady-state levels of FUS in animals that had cytoplasmic FUS accumulation (Fig. 3B) and that some of this accumulated protein was in the form of sodium dodecyl sulphate-insoluble, urea-soluble aggregates (Fig. 3C). All animals had similar levels of transgene mRNA expression, and there was no correlation between the level of transgene expression and the abundance of FUS (Supplementary Material, Fig. S1). The differences in steady-state levels of FUS protein, therefore, likely reflect differences in the amount of cytoplasmic aggregated FUS, which in turn could reflect differences in the intrinsic aggregation propensity of the various mutant FUS proteins.
Animals with FUS aggregates show progressive motor defects and die prematurely
To determine whether the cytoplasmic aggregation of FUS was accompanied by functional deficits, we assessed the motor activity of the transgenic animals by employing a standard thrashing assay (http://www.wormbook.org/chapters/www_behavior/behavior.html#sec7). This assay measured motor activity by counting the frequency of body bends when animals were placed in liquid M9 buffer. At 3 days of age (i.e. young adults), animals expressing only a GFP transgene had a slight motor disturbance (100.7 ± 5.6/min) compared with non-transgenic N2 animals of the same age (n= 20, 115.7 ± 6.3/min, P< 0.05, Fig. 3D). Animals expressing WT-FUS had a similar minor impairment of motor function (104.8 ± 5.0/min), indicating that expression of WT-FUS per se has little or no toxicity. The motor function of animals expressing the clinically mild R514G and R521G mutations, which had predominantly nuclear FUS, was indistinguishable from that of WT-FUS and GFP-only animals (R514G 103.5 ± 6.0/min and R521G = 103.8 ± 5.9/min). In contrast, animals expressing either the moderate R522G (96.3 ± 4.9/min) or the severe P525L mutation (91.8 ± 5.2/min) showed significantly impaired motor function compared with WT-FUS (P< 0.05 and P< 0.001, respectively). Animals expressing C-terminal-truncated FUS513 (69.7 ± 8.9/min) or FUS501 (54.6 ± 8.7/min) also had significantly impaired motor function compared with WT-FUS (P< 0.001). Representative locomotor behaviours of the animals are shown in Supplementary Material, Movies SM1–SM5.
Because human FUS mutations differ both in their age of onset and rates of progression, we next assessed rates of progression by comparing motor performance of 3-day-old and 6-day-old adult animals (n = 20 for each strain). The 6-day-old non-transgenic WT (N2), WT-FUS, R514G and R521G animals showed minor degrees of age-related decline compared with the 3-day-old N2 animals (−11.0 ± 0.8, −9.2 ± 1.3, −10.8 ± 1.7 and −7.5 ± 1.4%, respectively; P= NS) (Fig. 3E and F). However, the decline in R522G, P525L, FUS513 and FUS501 animals (−19.1 ± 1.6, −25.3 ± 1.3, −27.7 ± 1.8 and −22.3 ± 2.1%, respectively) was significantly greater than that in N2 and WT-FUS animals (P< 0.001). These differences in rates of progression in mutant FUS animals closely parallel the differences in rates of progression of ALS in humans with the same FUS mutations.
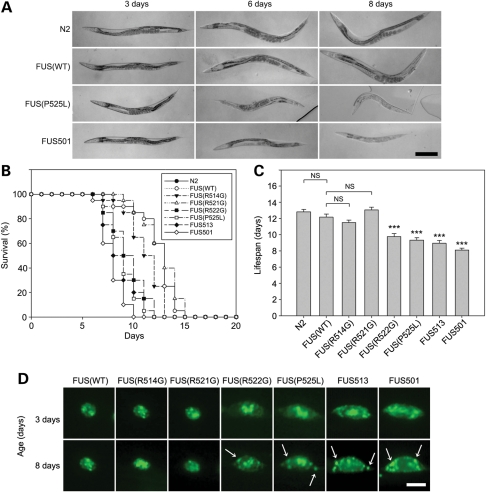
As is the case with human ALS, this progressive motor paralysis was also accompanied by accelerated mortality. At 3 days of adult age, all transgenic animals appeared morphologically similar to non-transgenic N2 animals and were not distinguishable in size (Fig. 4A). However, at about 6 days of age, P525L, FUS513 and FUS501 animals became smaller compared with WT-FUS animals of the same age. By 8 days of age, most of the P525L, FUS513 and FUS501 animals were partially or completely paralysed, severely shrunken and many were dead (Fig. 4A). Kaplan–Meier analyses (n = 20 animals/transgenic strain) revealed that under our culture conditions, the mean lifespans of animals expressing WT-FUS or the mild ALS mutations (R514G and R521G) were not different from those of N2 animals (Fig. 4B and C). In contrast, the mean lifespans of animals expressing the R522G, P525L, FUS513 and FUS501 mutants were reduced to 9.7 ± 0.4, 9.3 ± 0.3, 8.9 ± 0.3 and 8.1 ± 0.2 days, respectively, and were significantly shorter than those of non-transgenic N2 (12.8 ± 0.3 days, P< 0.001) and WT-FUS animals (12.1 ± 0.4 days, P< 0.001). Neurons from 6–8-day-old R522G, P525L, FUS513 and FUS501 animals also had abundant FUS-positive granules in the cytoplasm that were not present at younger ages (Fig. 4D).
Figure 4.
Age-dependent changes of FUS-expressing C. elegans. (A) Representative microscopic images of transgenic FUS animals. At 3 days of age, all transgenic lines look similar in size, whereas at 6 days of age, P525L and FUS501 animals are slightly shrunk compared with WT-FUS animals. At 8 days of age, P525L and FUS501 animals are obviously smaller and pale. Scale bar: 0.2 mm for all images. (B) Kaplan–Meier survival curve and (C) lifespans. (B) WT-FUS (WT), R514G and R521G animals live up to around 14 days, but most R522G, P525L, FUS513 and FUS501 animals died by 11 days. (C) The lifespans of WT, R514G and R521G animals are not different for N2 animals, but lifespans of R522G, P525L, FUS513 and FUS501 are significantly shorter than those of both N2 and WT-FUS animals. NS, not significant; ***P< 0.001. (D) Fluorescent microscopic images of FUS aggregates in transgenic FUS animals. At 8 days of age (9 days for R522G), R522G, P525L, FUS513 and FUS501 animals show abundant FUS granules in the cytoplasm (arrows), which are not obvious in younger animals (3-day-old). Scale bar: 2.0 µm.
Cellular stress exacerbates both cytoplasmic mislocalization of FUS and motor dysfunction
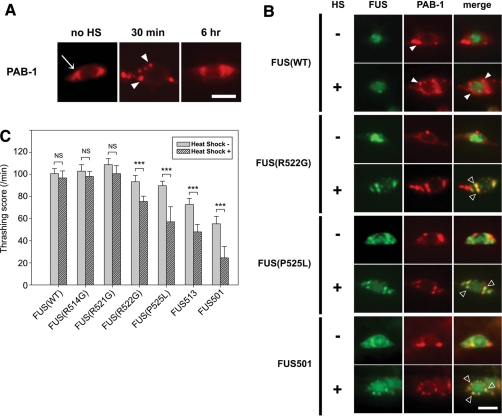
Because heat stress has been previously shown to cause mislocalization of FUS, we applied brief periods of thermal stress (35°C for 30 min) and then monitored both the subcellular distribution of FUS and the severity of motor dysfunction. Because FUS is an RNA-binding protein, we were particularly interested to determine whether it might co-localize with cytoplasmic stress granules, which can be monitored by poly (A)-binding protein (PAB-1) (17). At all ages, animals over-expressing PAB-1 tagged at the N-terminus with mCherry showed only diffuse cytoplasmic fluorescent signals with no apparent stress granules under normal conditions (Fig. 5A, no HS). However, 30 min after application of 35°C heat shock, PAB-1 coalesced into numerous cytoplasmic granules (Fig. 5A). These PAB-1-positive stress granules slowly disappeared, and PAB-1 staining reverted to the basal diffuse cytoplasmic distribution by 6 h after heat shock (Fig. 5A).
Figure 5.
Effects of heat shock on transgenic FUS animals. (A) PAB-1 expression and (B) effect of heat shock. Under basal conditions, PAB-1 is diffusely expressed in cytoplasm (A, no HS, arrow). Thirty minutes after heat shock, neurons developed some stress granules in the cytoplasm (A, 30 min, arrowheads), but most of them disappear 6 h after heat shock (A, 6 h). (B) Transgenic animals after single heat shock. There are more stress granules in WT-FUS (WT) animals after heat shock, but still FUS remains in the nucleolus [(B), WT/HS(+), arrowheads]. In contrast, R522G, P525L and FUS501 animals show many granules that are both FUS- and PAB-1-positive [R522G/HS(+), P525L/HS(+) and FUS501/HS(+), open arrowheads]. Scale bar: 2.0 μm. (C) Thrashing assay of transgenic animals with and without heat shock. WT-FUS (WT), R514G and R521G animals show no significant decrease after recovery from the heat shock, but all other strains (R522G, P525L, FUS513 and FUS501) show significantly decreased motor activity after heat shock. NS, not significant; ***P< 0.001.
When PAB-1 was co-expressed with either WT-FUS or mutant FUS, a small amount of PAB-1-positive stress granules was observed under basal conditions (Fig. 5B). In the double transgenic animals expressing WT or mutant FUS and PAB-1, under the same basal conditions, the distribution and morphology of FUS were identical to those in single transgenic FUS animals, indicating that PAB-1 does not affect localization of either WT-FUS or mutant FUS animals per se. Following the heat shock, WT-FUS animals exhibited a small increment in the number of stress granules in the cytoplasm, but there was no corresponding cytoplasmic accumulation of FUS (Fig. 5B). In sharp contrast, when the same heat shock stimulus was applied to the R522G, P525L or FUS501 animals, all three mutant FUS lines showed more stress granules and obvious recruitment of the mutant FUS to the heat-stress-induced granules.
Concomitantly, we assessed motor function in these transgenic animals. All animals showed impaired motility immediately after the heat shock. However, animals expressing WT-FUS and animals expressing the very mild ALS mutants (R514G and R521G) all rapidly recovered to baseline motor function within 30 min. In sharp contrast, all of the transgenic animals expressing the moderate and severe ALS mutants that cause cytoplasmic aggregates of FUS showed persistent motor deficits at 30 min after the heat shock (Fig. 5C, P< 0.001, compared with WT-FUS animals). Other assays of neuronal activity, such as escaping behaviour following body touch, also showed a persistent post-heat-shock motor defect in the animals with persistent cytoplasmic FUS (Supplementary Material, Fig. S3). These results support the notion that cytoplasmic accumulation of FUS is neurotoxic, but does not exclude the possibility that mutant FUS binds WT-FUS and titrates it from the nucleus.
WT-FUS does not rescue mutant FUS and is not recruited to mutant FUS aggregates
To test the hypothesis that mutant FUS might remove WT-FUS from the nucleus, and thereby cause a loss-of-function effect, we generated transgenic animals expressing WT-FUS labelled with TagRFP (TagRFP-WT-FUS) under the same pan-neuronal promoter. TagRFP-WT-FUS, like the previously described GFP-WT-FUS, was expressed in the nucleus, but was not recruited onto stress granules following heat shock and was not associated with significant neurological deficits (Fig. 6A and B, n = 10 each). In sharp contrast, the double transgenic animals (TagRFP-WT-FUS and GFP-FUS-P525L) showed the presence of TagRFP-WT-FUS in the nucleus and GFP-FUS-P525L in the nucleus and cytoplasm under basal conditions (Fig. 6A). Following heat shock, significant quantities of GFP-FUS-P525L were recruited to stress granules, but the TagRFP-WT-FUS remained in the nucleus (Fig. 6A). Importantly, these double transgenic animals and the single transgenic GFP-FUS-P525L animals had equivalent degrees of impaired motor activity and equivalent degrees of sensitivity to heat stress (Fig. 6B). Mutant FUS is therefore likely to cause ALS through a dominant gain-of-function effect, instead of a loss-of-function effect through titrating and reducing the WT-FUS function.
Figure 6.

Localization of WT-FUS in animals expressing mutant FUS. (A) Representative fluorescence microscopic images of TagRFP-WT-FUS and GFP-FUS P525L double transgenic animals with or without heat shock. The distribution of TagRFP-WT-FUS and GFP-FUS P525L proteins was not changed in double transgenic animals. After heat shock, GFP-FUS P525L protein formed numerous cytoplasmic granules (arrows), whereas TagRFP-WT-FUS remained in the nucleus. (B) Thrashing assay of the single and double transgenic animals. Co-expression of WT-FUS did not rescue motor activity impairment of double transgenic animals under basal conditions. After heat shock, WT-FUS animals recovered within 30 min, whereas both the single GFP-FUS P525L and double transgenic animals showed persistent motor impairment. NS, not significant; ***P< 0.001.
DISCUSSION
The experiments reported here demonstrate that pathogenic missense mutations in FUS that cause ALS in humans also cause neuronal dysfunction and death of C. elegans expressing these same mutant proteins. Importantly, there are strong parallels in the genotype-to-phenotype correlations between the human disease and the phenotypes in C. elegans described here. These similarities strongly argue that the mutant human proteins, when expressed in C. elegans, are closely modelling a fundamental property of these mutations in humans.
Our data indicate that ALS-associated mutations in FUS likely cause neurodegeneration via a toxic gain-of-function effect that is likely to be a consequence of the accumulation of FUS in the cytoplasm, rather than titration of FUS from its physiological location in the nucleus. This toxic gain-of-function effect could be mediated by either of two mechanisms. First, as is widely held in the field, it is conceivable that this toxic effect could be attributable to changes in RNA metabolism induced by the mutant FUS. Secondly, it is also possible that the aggregated FUS itself is the neurotoxic moiety, as is the case for several other neurotoxic intracellular and extracellular protein aggregates (e.g. tau, alpha-synuclein and huntingtin). Under such circumstances, the mutant, aggregation-prone forms may have a novel function unrelated or only partly related to the native function. Such is the case with ALS-inducing mutations in SOD1, which do not appear to work via alterations in superoxide dismutase activity (18). The animal models described here will be of use in the necessary work that will be required to discriminate these two possibilities. Additional work will also be required to understand the functional significance of binding of mutant FUS to stress granules, which have a role in cell viability and recovery after stress (11,19,20).
MATERIALS AND METHODS
Transgenic FUS worms
Animals were maintained on nematode growth medium (NGM) plates (1.8% agar, 0.3% NaCl, 0.25% peptone, 25 mm KPO4, 1 mm MgSO4, 1 mm CaCl2 and 5 µg/ml cholesterol) at 22°C. FL human FUS/TLS cDNA (NM_004960) (WT-FUS) was mutated to contain either one of the five clinical mutations (R514G, R521G, R522G, R524S and P525L). Two C-terminal-truncated FUS constructs were generated (FUS513 and FUS501), lacking the C-terminal 13 and 25 amino acids of FUS, respectively. Each construct was injected together with lin-15 rescuing plasmid into a host worm with temperature-sensitive lin-15 (n765) allele, and non-Muv (multivulva) animals were selected for further analysis (21). These transgenes were integrated into the genome to generate stable transgenic lines by UV irradiation, and the lines were outcrossed more than six times with N2 WT strain. Constructs used in this study are summarized in Figure 1.
Poly (A)-binding protein (PAB-1) was used as a marker for stress granules (17). PAB-1 was cloned into the pJH897 vector with the tag replaced with mCherry at the N-terminus and was injected, solely or together with FUS constructs, along with the lin-15 rescuing plasmid into lin-15 worms. The N2 strain and transgenic animals expressing only GFP (GFP-only animals) were used as negative controls.
Quantitative RT–PCR
Transgene expression in each integrated strain was determined using quantitative real-time PCR. Total RNA was prepared from L4 stage animals with TRIzol reagent (Invitrogen) and RNeasy kit (Qiagen, USA), according to the manufacturer's’ instructions. cDNA was synthesized with AffinityScript reverse transcriptase (Stratagene, USA) and was amplified with primers: forward, 5′-TAAATTTGGTGGCCCTCGGG-3′ and reverse, 5′-TCACCCAGGCCTTGCACAAA-3′. The amplification was monitored using SYBR Green Premix Ex Taq reagent (Takara Bio, Japan) in ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, USA) with gpd-2 as an internal control.
Immunohistochemistry
FUS localization in neurons was assessed by fixation of the animals with 4% paraformaldehyde in MRWB buffer (80 mm KCl, 20 mm NaCl, 10 mm EGTA, 5 mm spermidine HCl, 15 mm Na PIPES and 25% methanol, pH 7.4), reduction with 1% 2-mercaptoethanol in BO3 buffer (25 mm H2BO3 and 125 mm NaOH), then oxidation in 0.3% H2O2/BO3 buffer and incubation with primary antibody [monoclonal anti-GFP antibody (Invitrogen, 1:200 dilution) or rabbit polyclonal anti-FUS antibody (Abcam, 1:200 dilution)] overnight at room temperature. After several washes, the animals were incubated with secondary antibody (1:400 dilution), washed again and then mounted in DAPI to stain nuclei (Molecular Probes, USA).
Nuclear FUS quantification
To quantify the ratio of nuclear to cytoplasmic FUS, fluorescent microscopic images of neurons in the body were used, so that signals from other nearby neurons were not counted. Using Image J, the nucleus and cytoplasm of the neuron were cropped and the pixels were counted (3-day-old animals, n = 20 animals, 10 neurons from each). Pixels that overlapped with DAPI were considered to be nuclear, and pixels that did not were considered to be cytoplasmic.
Thrashing assay
Animals from each transgenic line were picked at L4 stage (n = 20). Sixteen hours later, the 3-day-old young adult animals were transferred into 5 μl of M9 buffer (0.6% Na2PO4, 0.3% KH2PO4, 0.5% NaCl and 0.01% NH4Cl) on a glass slide. Thirty seconds after the transfer, the frequency of the body bending was counted for 1min (22). The same protocol was used to assess the motor function in 6-day-old adults.
Heat shock treatment
Animals were picked at L4 stage (n = 16) and kept at 22°C. Sixteen hours later, the animals were subjected to heat shock by incubation at 35°C for 30 min. After heat shock, the animals were returned to the 22°C incubator for recovery and were then assessed for functional deficits at 30 min after heat shock.
Fluorescence microscopy
Transgenic FUS animals with or without heat shock were anaesthetized in 20 mm sodium azide in M9 buffer, mounted on an agarose pad with cover slip and examined with a standard fluorescent microscope or confocal microscope (Nikon H600L, Japan).
Lifespan analysis
Animals were placed on NGM plates (n = 20) and replated daily, and the number of live animals was counted. Immobile, shrunken animals were considered to have died.
Western blot analysis
Animals were lysed in RIPA buffer with sonication. After removing undissolved debris by centrifugation, protein concentrations of the lysates were determined by bicinchoninic acid assay. Equal amounts of protein were subjected to NuPAGE Bis–Tris (Invitrogen); transferred onto nitrocellulose membrane (Whatman, Germany) and detected by western blotting with anti-GFP (Abcam, USA), anti-FUS (Santa Cruz Biotechnology, USA) or anti-tubulin (Abcam) antibodies. For analysis of the solubility of FUS proteins, RIPA lysates were centrifuged at 100 000g for 30 min at 4°C. The pellets were resuspended in urea buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris–HCl, pH 8.5), sonicated and centrifuged at 100 000g for 30 min at room temperature (10).
Statistical analysis
For statistical analysis, the unpaired Student's t-tests and analysis of variance were performed with SigmaPlot version 11.0.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We thank Drs Seydoux (Johns Hopkins University) and Schisa (Central Michigan University, USA) for PAB-1 construct.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the Wellcome Trust, Canadian Institutes of Health Research, the Medical Research Council, the Biotechnology and Biological Sciences Research Council, the Howard Hughes Medical Institute, Safra Foundation, Canada Research Chairs and the Alzheimer Society of Ontario. S.-P.Y. received a post-doctoral fellowship from the Korean National Research Foundation. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust, and Canadian Institutes of Health Research.
REFERENCES
- 1.Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. doi:10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 2.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. doi:10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broustal O., Camuzat A., Guillot-Noel L., Guy N., Millecamps S., Deffond D., Lacomblez L., Golfier V., Hannequin D., Salachas F., et al. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J. Alzheimers Dis. 2010;22:765–769. [PubMed] [Google Scholar]
- 4.Andersson M.K., Stahlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., Nilsson O., Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. doi:10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackenzie I.R., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. doi:10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 6.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. doi:10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackenzie I.R., Munoz D.G., Kusaka H., Yokota O., Ishihara K., Roeber S., Kretzschmar H.A., Cairns N.J., Neumann M. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol. 2011;121:207–218. doi: 10.1007/s00401-010-0764-0. doi:10.1007/s00401-010-0764-0. [DOI] [PubMed] [Google Scholar]
- 8.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 9.Deng H.X., Zhai H., Bigio E.H., Yan J., Fecto F., Ajroud K., Mishra M., Ajroud-Driss S., Heller S., Sufit R., et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. doi:10.1001/archneurol.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winton M.J., Igaz L.M., Wong M.M., Kwong L.K., Trojanowski J.Q., Lee V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. doi:10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. doi:10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sendtner M. TDP-43: multiple targets, multiple disease mechanisms? Nat. Neurosci. 2011;14:403–405. doi: 10.1038/nn.2784. doi:10.1038/nn.2784. [DOI] [PubMed] [Google Scholar]
- 13.Dormann D., Haass C. TDP-43 and FUS: a nuclear affair. Trends Neurosci. 2011;34:339–348. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Kaminski Schierle G.S., Bertoncini C.W., Chan F.T., van der Goot A.T., Schwedler S., Skepper J., Schlachter S., van Ham T., Esposito A., Kumita J.R., et al. A FRET sensor for non-invasive imaging of amyloid formation in vivo. Chemphyschem. 2011;12:673–680. doi: 10.1002/cphc.201000996. doi:10.1002/cphc.201000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belzil V.V., St-Onge J., Daoud H., Desjarlais A., Bouchard J.P., Dupre N., Camu W., Dion P.A., Rouleau G.A. Identification of a FUS splicing mutation in a large family with amyotrophic lateral sclerosis. J. Hum. Genet. 2010;56:247–249. doi: 10.1038/jhg.2010.162. [DOI] [PubMed] [Google Scholar]
- 16.DeJesus-Hernandez M., Kocerha J., Finch N., Crook R., Baker M., Desaro P., Johnston A., Rutherford N., Wojtas A., Kennelly K., et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 2010;31:E1377–E1389. doi: 10.1002/humu.21241. doi:10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jud M.C., Czerwinski M.J., Wood M.P., Young R.A., Gallo C.M., Bickel J.S., Petty E.L., Mason J.M., Little B.A., Padilla P.A., et al. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 2008;318:38–51. doi: 10.1016/j.ydbio.2008.02.059. doi:10.1016/j.ydbio.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. doi:10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito D., Seki M., Tsunoda Y., Uchiyama H., Suzuki N. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann. Neurol. 2011;69:152–162. doi: 10.1002/ana.22246. doi:10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- 20.Kino Y., Washizu C., Aquilanti E., Okuno M., Kurosawa M., Yamada M., Doi H., Nukina N. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 2010;39:2781–2798. doi: 10.1093/nar/gkq1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh E., Kawano T., Ng S., Fetter R., Hung W., Wang Y., Zhen M. Caenorhabditis elegans innexins regulate active zone differentiation. J. Neurosci. 2009;29:5207–5217. doi: 10.1523/JNEUROSCI.0637-09.2009. doi:10.1523/JNEUROSCI.0637-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao S., Zhen M. Action potentials drive body wall muscle contractions in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2011;108:2557–2562. doi: 10.1073/pnas.1012346108. doi:10.1073/pnas.1012346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.