Abstract
Congenital heart disease is one of the most common human birth defects, yet many genes and pathways regulating heart development remain unknown. A recent study in humans revealed that mutations in a single Hox gene, HOXA1 (Athabascan Brainstem Dysgenesis Syndrome, Bosley-Salih-Alorainy Syndrome), can cause severe cardiovascular malformations, some of which are lethal without surgical intervention. Since the discovery of the human syndromes, there have been no reports of any Hox mouse mutants with cardiac defects, hampering studies to explore the developmental causes of the human disease. In this study, we identify severe cardiovascular malformations in a Hox mouse model, which mimic the congenital heart defects in HOXA1 syndrome patients. Hoxa1 null mice show defects such as interrupted aortic arch, aberrant subclavian artery and Tetralogy of Fallot, demonstrating that Hoxa1 is required for patterning of the great arteries and outflow tract of the heart. We show that during early embryogenesis, Hoxa1 is expressed in precursors of cardiac neural crest cells (NCCs), which populate the heart. We further demonstrate that Hoxa1 acts upstream of several genes, important for neural crest specification. Thus, our data allow us to suggest a model in which Hoxa1 regulates heart development through its influence on cardiac NCCs, providing insight into the mechanisms underlying the human disease.
INTRODUCTION
Almost 1% of all infants are born with cardiac malformations. However, not all molecular pathways regulating heart development have been identified. Genetic loss-of-function experiments in model organisms started to reveal that a variety of signaling factors and transcriptional regulators, such as Fgfs, Retinoic acid, Notch and Pbx are crucial during cardiac morphogenesis (1). One large family of transcriptional regulators, whose requirement for heart development remains obscure, is the Homeobox (Hox) gene family (2). Although Retinoic acid signaling and Hox gene expression is interdependent (3) and Pbx proteins act as Hox cofactors (4–6), no cardiac defects have been identified in any of the single or compound Hox mouse mutants. Interestingly, a recent study in humans found severe cardiovascular malformations in patients carrying a homozygous truncating mutation in HOXA1 [Bosley-Salih-Alorainy Syndrome (BSAS) (7), Athabascan Brainstem Dysgenesis Syndrome (ABDS) (8)]. Some of the malformations are lethal without surgical intervention and include interrupted aortic arch type B (IAAB), aberrant subclavian artery (ASC), ventricular septal defect (VSD), bicuspid aortic valve (BAV) and Tetralogy of Fallot (7–9). Cardiovascular malformations are present in 70% of ABDS and 31% of BSAS patients (9). In addition, some patients display cerebrovascular defects such as absent or hypoplastic internal carotid artery (ICA).
In the past two decades, several Hoxa1 mouse models were generated and analyzed (10–12). Although Hoxa1 knockout mice manifest all other symptoms present in human patients, e.g. brainstem abnormalities, hypoventilation, inner ear and craniofacial defects, cardiovascular abnormalities have not been reported in these mice (7,10,12). Due to the lack of a mouse model for the human congenital heart defects, it has not been possible to examine the developmental causes of these defects or to understand the role Hoxa1 plays during heart development.
To address the discrepancy between human patients and Hoxa1 knockout mice with respect to the cardiac phenotype, we examined a large number of Hoxa1 null embryos for cardiovascular defects. We found a variety of malformations of the cardiac outflow tract (OFT) and great arteries in Hoxa1 null embryos, many of which are identical to the defects in HOXA1 syndrome patients. We further determined that during early embryogenesis, Hoxa1 is expressed in cardiac neural crest progenitors in the neural tube and is required for the expression of several genes necessary for neural crest specification. Our study identifies the first Hox-deficient mouse with cardiac defects and suggests a model for how Hoxa1 influences cardiac development.
RESULTS
Hoxa1 null mice exhibit severe cardiovascular malformations
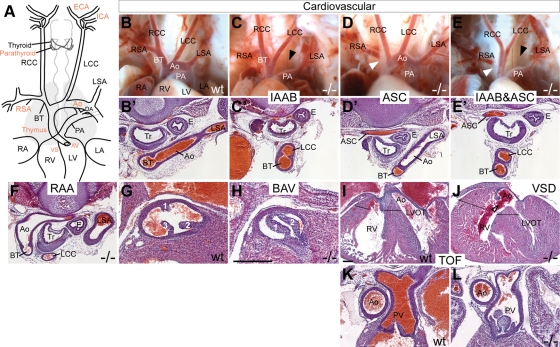
Since HOXA1 syndrome patients display congenital heart defects, which have not been reported in Hoxa1 knockout mice, we thoroughly examined Hoxa1−/− embryos for cardiac defects. For our analysis, we used a Hoxa1 null line in which GFP was inserted into the coding region of Hoxa1 (Hoxa1GFP) (11) (Fig. 1). Hoxa1GFP/GFP pups are born at normal Mendelian ratios but die at P0-P1 (n = 25), as reported previously. Upon careful examination, we found that 74% of all Hoxa1 null animals analyzed at E18.5-P1 (n = 34) showed cardiovascular defects (Fig. 2A–L; Table 1). We categorized the cardiovascular defects into aortic arch and cardiac OFT defects. Aortic arch malformations were the most severe and most penetrant, present in 68% of all mutants (Table 1). These defects include IAAB (Fig. 2C), which is a lethal heart defect present in 50% of mutants, aberrant retro-esophageal right subclavian artery (ASC) (Fig. 2D) and right aortic arch (RAA) (Fig. 2F). Cardiac OFT abnormalities were observed in 47% of mutants and include VSD (Fig. 2I and J) and BAV (Fig. 2G and H), the latter representing the most common human congenital cardiac malformation. Two animals with VSD additionally had an overriding aorta, pulmonary stenosis and hypertrophy of the right ventricle (Fig. 2I–L), which are the hallmarks of Tetralogy of Fallot. In sum, like human HOXA1 syndrome patients, Hoxa1 null mice display IAAB, ASC, VSD, BAV, Tetralogy of Fallot and carotid artery abnormalities and, therefore, represent a valuable model for the congenital heart defects in humans.
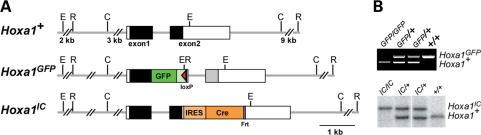
Figure 1.
Hoxa1 alleles used in this study. (A) Hoxa1GFP was generated by inserting EGFP followed by polyadenylation signals into a unique site located in the first exon of Hoxa1 resulting in a frame shift in the Hoxa1 genomic sequence 3′ of the cassette (grey box). This generates a fusion protein, lacking the homeodomain, which is encoded in exon 2. Neo was removed leaving a residual loxP site. The Hoxa1IC allele was generated by inserting an IRES-Cre-NEO cassette in the 3′UTR of Hoxa1 and subsequent removal of Neo, leaving a residual Frt site. Solid boxes represent coding regions and white boxes UTRs. (B) PCR or southern genotyping identifies the different Hoxa1 alleles.
Figure 2.
Hoxa1 null mice exhibit severe cardiovascular abnormalities that mimic the defects in HoxA1 syndrome patients. (A) Schematic of the heart, great arteries and cerebral arteries. Structures in which defects were found in Hoxa1 mutants are highlighted in red. (B–L) Cardiovascular defects in mutants include abnormalities of the great arteries and cardiac OFT. (B and B′) Anatomy and histology of great arteries in an E18.5 WT embryo. Great artery defects in mutants include IAAB (C, C′), aberrant retro-esophageal right subclavian artery (ASC, D, D′), a combination of IAAB and ASC (E, E′) and RAA (F). Defects in the OFT include BAV and VSD. The aortic valve is tricuspid in WT (G) but bicuspid in mutants (H). The ventricular septum is continuous in WT embryos at E18.5 (I), while a large VSD is present in the mutant (J). Compared with WT (I, K), this embryo also shows an overriding aorta, hypertrophy of the ventricle (J, black lines) and pulmonary stenosis (L), which are the hallmarks of Tetralogy of Fallot (TOF). Ao, aorta; BT, brachiocephalic trunk; DA, ductus arteriosus; E, esophagus; LA, left atrium; LCC, left common carotid artery; LSA, left subclavian artery; LV, left ventricle; LVOT, left ventricular outflow tract; PA, pulmonary artery; RA, right atrium; RCC, right common carotid artery; RSA, right subclavian artery; RV, right ventricle; Tr, trachea. Scale bars: anatomy panels, 0.5 mm; histology panels, 100 μm.
Table 1.
Incidence of cardiovascular, cerebrovascular and glandular defects in Hoxa1−/− mutants
| Cardiovascular |
Cerebrovascular | Glandular |
||||||
|---|---|---|---|---|---|---|---|---|
| Defect | IAAB | ASC | RAA | VSD | BAV | ICA/ECA | Thymic hypoplasia | Parathyroid hypoplasia or aplasia |
| n (total n analyzed) | 17 (34) | 6 (34) | 1 (34) | 4 (17) | 4 (17) | 19 (28) | 20 (28) | 20 (28) |
| % | 50 | 18 | 3 | 24 | 24 | 68 | 71 | 71 |
Seventy-four percent of mutants have at least one cardiovascular defect. Total percentage of defects exceeds 100% since some specimens have more than one defect. IAAB, interrupted aortic arch type B; ASC, aberrant retro-esophageal right subclavian artery; RAA, right aortic arch; VSD, ventricular septal defect; BAV, bicuspid aortic valve; ICA/ECA, internal and/or external carotid artery abnormalities.
Hoxa1−/− mice display cerebrovascular and glandular abnormalities
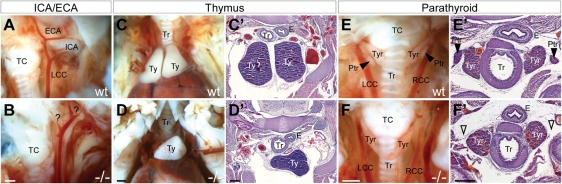
In addition to the cardiovascular defects, Hoxa1 null mice exhibit cerebrovascular and glandular defects. Cerebrovascular abnormalities include abnormal branching, hypo- or aplasia of the internal and external carotid arteries (Fig. 3A and B), which is also seen in human patients. The glandular defects include thymic hypoplasia (Fig. 3C–D′) and parathyroid hypo- or aplasia (Fig. 3E–F′). Cerebrovascular abnormalities are present in 68% of Hoxa1 mutants and glandular defects in 71% (Table 1). Notably, all Hoxa1 null animals showed at least one cardiovascular or glandular malformation (n = 34), whereas no abnormalities were detected in control littermates (n= 20).
Figure 3.
Hoxa1 null mice exhibit cerebrovascular and glandular defects. (A) In wild-type embryos, the common carotid artery branches into the external and internal carotid arteries (ICA/ECA) in a stereotyped pattern, while mutants show branching abnormalities (B). Compared with wild-type (C and C′) mutants exhibit thymic (Ty) hypoplasia (D and D′). (E and E′) The parathyroid glands (Ptr) are located adjacent to the thyroid (Tyr) in wild-type embryos but are absent in mutants (F and F′). TC, thyroid cartilage. Scale bars: anatomy panels, 0.5 mm; histology panels, 100 μm.
Hoxa1 is expressed in cardiac neural crest precursors in the hindbrain
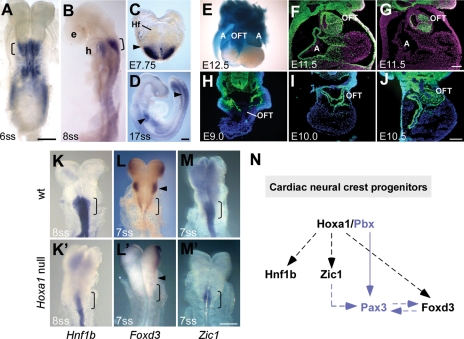
Interestingly, the cardiovascular and glandular anomalies in Hoxa1 null mice are reminiscent of the defects found in mutants in which the development of cardiac neural crest cells (NCCs) is affected (1). Cardiac NCCs originate from the posterior hindbrain (rhombomeres 6–8) and are required for remodeling of the primordial aortic arch vessels and cardiac OFT, as well as the development of the parathyroid and thymus (1,13). Hoxa1 is not expressed in the heart tube at any stage of development, but it is strongly expressed in the posterior hindbrain where cardiac NCCs arise (14,15). Hoxa1 is expressed in this region as early as E7.75, but expression recedes rapidly and is absent from the hindbrain by E9.0 (Fig. 4A–D). To examine if these early Hoxa1-expressing cells in the hindbrain contribute to the heart at later stages, we carried out lineage analysis, using a Hoxa1IC driver (15) (Fig. 1). Hoxa1 lineage contributes extensively to the cardiac OFT (Fig. 4E) (15) in a pattern equivalent to the Wnt1 lineage (Fig. 4F and G), which marks NCC (16,17), suggesting that most if not all cardiac NCCs are derived from Hoxa1-expressing cells. By examining the distribution of Hoxa1 lineage over time, we found that at E9.0 very few lineage labeled cells were present in the cardiac region (Fig. 4H). By E10.0, Hoxa1 lineage had populated the cardiac OFT (Fig. 4I) and at E10.5 a large number of labeled cells were present in this region (Fig. 4J). Our results demonstrate that Hoxa1 is expressed in cardiac neural crest precursors in the hindbrain, which contribute to the heart at later stages.
Figure 4.
Hoxa1 is expressed in cardiac neural crest progenitors in the posterior hindbrain and is required for their development. (A–D) RNA in situ analysis of Hoxa1 expression in WT embryos. Strong Hoxa1 expression in the posterior hindbrain (brackets), where cardiac NCCs arise, is seen at the six-somite (A, dorsal view) and the eight-somite stage (B, lateral view). Hoxa1 is expressed in this region as early as E7.75 (headfold stage, Hf) (C) and by E9.0 it is absent from the hindbrain and is only present in the posterior neural tube and foregut (D, arrow heads). (E–J) Hoxa1 lineage contributes extensively to the OFT of the heart. (E) X-gal staining for Hoxa1 lineage (blue) in the OFT and atria (A) of a dissected E12.5 heart. (F and G) Transverse sections through the heart of WT embryos comparing Hoxa1 lineage (F) to Wnt1 lineage (G), which represents the NCC lineage, reveals that most if not all cardiac NCCs are derived from Hoxa1-expressing cells. (H–J) Temporal series showing that whereas there is almost no Hoxa1 lineage in the heart at E9.0 (H), at E10.0 the lineage has populated the cardiac region (I), giving rise to the majority of cells in the OFT, as seen at E10.5 (J). (K–M) Neural crest markers are down-regulated in the posterior hindbrain of Hoxa1 mutants. The expression of Hnf1b (K, K′), Foxd3 (L, L′) and Zic1 (M, M′) is strongly reduced in the posterior hindbrain (r6–r8; brackets) of Hoxa1 null embryos. Foxd3 expression in r4 is also absent in mutants (L, L′; arrow heads). (N) Model for the role of Hoxa1 in cardiac neural crest progenitors based on this (black) and other studies (blue) (5,21,28,29). We suggest that Hoxa1 acts upstream of Hnf1b, Zic1 and Foxd3 in neural crest development, potentially in complex with Pbx1 and upstream of Pax3 to control NCC specification. Note that dashed lines and arrows do not necessarily indicate direct gene regulation. e, eye; h, heart. Scale bars: top panels 200 μm, bottom panel 100 μm.
Hoxa1 regulates genes necessary for neural crest specification
To identify downstream targets regulated by Hoxa1 in the hindbrain, we previously carried out microarray analysis on dissected tissues from Hoxa1 null and wild-type (WT) embryos at the peak of Hoxa1 expression (18). Interestingly, our array revealed that several NC markers were down-regulated in Hoxa1 null samples. In order to determine whether any of these markers are misexpressed in cardiac NC precursors in the posterior hindbrain of Hoxa1 null embryos, we performed in situ analysis on several of these genes. We found that Hnf1b, Foxd3 and Zic1 were strongly down-regulation in the posterior hindbrain of Hoxa1 null embryos compared with wild-type controls (Fig. 4K–M′). Foxd3, Zic1 and Hnf1b are expressed in premigratory cardiac NC progenitors and have been shown to be important for NC specification (19–23). This suggests that during cardiac development, Hoxa1 acts upstream of genes required for the specification of cardiac NC cells.
DISCUSSION
Although Hox genes are essential for the development of many different tissues and organs in the embryo, almost nothing is known about their role in cardiac development. Even after cardiac malformations were identified in human patients with homozygous truncating mutations in HOXA1 in 2005 (7), there were no reports of cardiac defects in any Hox mouse mutants. Our study is the first to identify a Hox mouse model with severe cardiovascular defects, which are identical to the malformations in HoxA1 syndrome patients. Our analyses demonstrate that Hoxa1 is required for patterning of the great arteries and OFT of the heart. We further show that Hoxa1 is expressed in cardiac NC precursors in the hindbrain. These cells are required for remodeling of the cardiac OFT and aortic arch arteries III, IV and VI, which will contribute to specific segments of the carotid arteries, subclavian arteries and aortic arch (1,24), all of which are affected in Hoxa1 mutants.
We identified three genes, Foxd3, Zic1 and Hnf1b, which are regulated by Hoxa1 and are necessary for NC specification. Foxd3 is expressed in premigratory NCCs in the hindbrain (19) and has been shown to promote the development of neural crest from neural tube progenitors (22). Deletion of Foxd3 in NCC leads to subtle defects in cardiovascular development (25), but when combined with one null allele of Pax3, severe abnormalities such as persistent truncus arteriosus are observed (21). While one null allele of Pax3 does not lead to cardiac defects, Pax3 homozygous null mice have profound defects in the OFT, great vessels and glands (26,27). Similarly, mutations in members of the Zic gene family have been shown to lead to congenital heart defects in humans and mice (1), and it was demonstrated that Zic1 acts upstream of Pax3 in NC induction (28). A recent study further demonstrated that Pbx1 is required upstream of Pax3 in premigratory cardiac NCCs and suggested a model in which a Pbx1–Hox complex carries out this function (29). In fact, it has previously been shown that Pbx1 acts as a cofactor for Hoxa1 (5) and like Hoxa1 null embryos, Pbx1 mutants display cardiac OFT defects including VSD and great artery anomalies such as ASC and RAA (29,30). Based on our findings, as well as those from previous studies (21,28,29), we propose a model, in which Hoxa1 acts upstream of Foxd3, Zic1 and Hnf1b in cardiac NC development, potentially in a complex with Pbx1 upstream of Pax3 (Fig. 4N).
Our study suggests that the cardiovascular defects in mice and humans could, at least in part, be due to a role of Hoxa1 in cardiac neural crest development. Future studies will be necessary to determine whether Hoxa1 plays a cell autonomous or non-cell autonomous function during cardiac neural crest development and which specific step of neural crest maturation is regulated by Hoxa1. It will also be interesting to determine how Hoxa1 and possibly other Hox genes intersect with some of the known regulators of cardiac development, such as Fgfs, Retinoids and Pbx. Our Hoxa1 mouse mutant provides a valuable model to study the molecular mechanisms through which Hox genes regulate cardiac morphogenesis, which will help uncover the developmental causes for the cardiac malformations in human patients.
MATERIALS AND METHODS
Mouse lines and genotyping
The Hoxa1GFP allele (null allele) (11) was generated by inserting an EGFP-PolyA-loxP-MC1-Neo-loxP cassette into a unique AatII site in exon 1 of Hoxa1 [previously referred to as exon 2 (11)], which results in a fusion protein lacking the homeodomain (Fig. 1A). Neo was removed using a Cre ‘deleter’ mouse (31). The Hoxa1-IRES-Cre (Hoxa1IC) allele was generated by inserting an IRES-Cre-frt-MC1-Neo-frt cassette downstream of the stop codon and subsequently removing the Neo allele by crossing to an FLPe deleter (15) (Fig. 1A). Genotyping was performed using multiplex polymerase chain reaction (PCR) (Fig. 1B) with the following primers for Hoxa1GFP: WT forward NM5 5′-ACT CCT TAT CCC CTC TCC AC-3′, WT reverse NM6 5′-CCT CCT TCT CAC GCT TCT TC-3′, GFP reverse NM7 5′-TTG TAC TCC AGC TTG TGC C-3′, generating an 821 bp WT and 488 bp engineered band; and the following primers for Hoxa1IC: WT forward NM1 5′-AGC GAT GAG AAA ACG GAA G-3′, WT reverse NM4 5′-GGG ACG AGA AAG GAA GAG AG-3′, Cre NM3 5′-CAA TAC CGG AGA TCA TGC AAG-3′, generating a 220 bp WT and 382 bp engineered band. Both mouse lines were maintained on a C57BL/6 background. Lineage analysis was carried out using the previously described R26R-EYFP and R26R-lacZ lines (32,33). All mouse use complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee.
Histology, β-galactosidase staining and RNA in situ hybridization
Embryos were harvested at E18.5, fixed in formalin overnight, washed in phosphate buffered saline (PBS) and stored in 70% ethanol for thoracic dissections. For histology, embryos were further dehydrated, embedded in wax, sectioned at 18 μm and stained with hematoxylin and eosin. For β-gal staining, hearts were dissected in PBS, pH 7.4 with 2 mm MgCl2, fixed in 1% formaldehyde, 0.2% glutaraldehyde, 25 mm EGTA, 2 mm MgCl2, 0.02% NP40 in PBS, washed in PBS with 2 mm MgCl2 and stained in X-gal solution (0.8 mg/ml X-gal, 25 mm K3Fe(CN)6, 25 mm K4Fe(CN)6–3H2O, 2 mm MgCl2, 0.01% Na deoxycholate, 0.02% NP40 in PBS) over night at room temperature. For RNA in situ hybridization, digoxigenin-labeled antisense RNA probes were generated from plasmids carrying cDNA fragments. The following cloned mouse cDNAs were obtained, sequenced and used to prepare riboprobes: Foxd3 (19), Hnf1b (34), Zic1 (35) and a 216 bp Hoxa1 exon1 fragment. Whole-mount in situ hybridization was performed on embryos isolated from timed pregnancies essentially as described (36).
Immunostaining and analysis
Tissues were fixed at 4°C for 1–2h in 4% formaldehyde, rinsed in PBS, equilibrated to 30% sucrose and embedded in OCT. Cryosections were cut at 10 µm, washed in PBS and preincubated in blocking solution (2% BSA, 10% NGS, 0.1% Triton in PBS, pH 7.2). Rabbit anti-GFP (Abcam; 1:4000 or Molecular probes; 1:2000) primary antibody was applied overnight at 4°C in a humid chamber, followed by secondary detection using Alexa Fluor conjugated secondary antibodies (Molecular Probes). Immunodetection was carried out using an SP5 confocal system (Leica) or an inverted microscope (Axiovert 200M, Zeiss) equipped with a SensiCam camera (The Cooke Cooperation). Data were acquired using the LAS AF or SlideBookTM software and processed using Adobe Photoshop.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the NIH (NIH5R01GM021168-34) and Howard Hughes Medical Institute to M.R.C. and the Boehringer Ingelheim Fonds PhD fellowship and the University of Utah Graduate Research Fellowship to N.M.
ACKNOWLEDGEMENTS
We thank Deborah Frank for her helpful insight on phenotypic analysis; members of our tissue culture and mouse facility for ES cell culture, injection and mouse care; Ruth Arkell, Patricia Labosky, Xiaojing Ma for plasmids used to generate riboprobes. This manuscript was improved by helpful comments from Anne Boulet and Daniel Kopinke.
REFERENCES
- 1.Srivastava D. Genetic assembly of the heart: implications for congenital heart disease. Annu. Rev. Physiol. 2001;63:451–469. doi: 10.1146/annurev.physiol.63.1.451. [DOI] [PubMed] [Google Scholar]
- 2.Alexander T., Nolte C., Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 3.Vitobello A., Ferretti E., Lampe X., Vilain N., Ducret S., Ori M., Spetz J.F., Selleri L., Rijli F.M. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C.P., Brocchieri L., Shen W.F., Largman C., Cleary M.L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phelan M.L., Rambaldi I., Featherstone M.S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol. Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann R.S., Chan S.K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 7.Tischfield M.A., Bosley T.M., Salih M.A., Alorainy I.A., Sener E.C., Nester M.J., Oystreck D.T., Chan W.M., Andrews C., Erickson R.P., et al. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat. Genet. 2005;37:1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- 8.Holve S., Friedman B., Hoyme H.E., Tarby T.J., Johnstone S.J., Erickson R.P., Clericuzio C.L., Cunniff C. Athabascan brainstem dysgenesis syndrome. Am. J. Med. Genet. A. 2003;120:169–173. doi: 10.1002/ajmg.a.20087. [DOI] [PubMed] [Google Scholar]
- 9.Bosley T.M., Alorainy I.A., Salih M.A., Aldhalaan H.M., Abu-Amero K.K., Oystreck D.T., Tischfield M.A., Engle E.C., Erickson R.P. The clinical spectrum of homozygous HOXA1 mutations. Am. J. Med. Genet. A. 2008;146A:1235–1240. doi: 10.1002/ajmg.a.32262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chisaka O., Musci T.S., Capecchi M.R. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–520. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- 11.Godwin A.R., Stadler H.S., Nakamura K., Capecchi M.R. Detection of targeted GFP-Hox gene fusions during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1998;95:13042–13047. doi: 10.1073/pnas.95.22.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lufkin T., Dierich A., LeMeur M., Mark M., Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 13.Gebbia M., Ferrero G.B., Pilia G., Bassi M.T., Aylsworth A., Penman-Splitt M., Bird L.M., Bamforth J.S., Burn J., Schlessinger D., et al. X-linked situs abnormalities result from mutations in ZIC3. Nat. Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 14.Bertrand N., Roux M., Ryckebusch L., Niederreither K., Dolle P., Moon A., Capecchi M., Zaffran S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev. Biol. 2011;353:266–274. doi: 10.1016/j.ydbio.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makki N., Capecchi M.R. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev. Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danielian P.S., Muccino D., Rowitch D.H., Michael S.K., McMahon A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X., Rowitch D.H., Soriano P., McMahon A.P., Sucov H.M. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 18.Makki N., Capecchi M.R. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol. 2011;357:295–304. doi: 10.1016/j.ydbio.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labosky P.A., Kaestner K.H. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech. Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 20.Barbacci E., Reber M., Ott M.O., Breillat C., Huetz F., Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- 21.Nelms B.L., Pfaltzgraff E.R., Labosky P.A. Functional interaction between Foxd3 and Pax3 in cardiac neural crest development. Genesis. 2010;49:10–23. doi: 10.1002/dvg.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dottori M., Gross M.K., Labosky P., Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- 23.Aruga J. The role of Zic genes in neural development. Mol. Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Waldo K., Miyagawa-Tomita S., Kumiski D., Kirby M.L. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev. Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- 25.Teng L., Mundell N.A., Frist A.Y., Wang Q., Labosky P.A. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway S.J., Henderson D.J., Kirby M.L., Anderson R.H., Copp A.J. Development of a lethal congenital heart defect in the splotch (Pax3) mutant mouse. Cardiovasc. Res. 1997;36:163–173. doi: 10.1016/s0008-6363(97)00172-7. [DOI] [PubMed] [Google Scholar]
- 27.Epstein J.A. Pax3, neural crest and cardiovascular development. Trends Cardiovasc. Med. 1996;6:255–260. doi: 10.1016/S1050-1738(96)00110-7. [DOI] [PubMed] [Google Scholar]
- 28.Li B., Kuriyama S., Moreno M., Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–3278. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C.P., Stankunas K., Shang C., Kao S.C., Twu K.Y., Cleary M.L. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development. 2008;135:3577–3586. doi: 10.1242/dev.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stankunas K., Shang C., Twu K.Y., Kao S.C., Jenkins N.A., Copeland N.G., Sanyal M., Selleri L., Cleary M.L., Chang C.P. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwenk F., Baron U., Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 34.Gray P.A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D.I., Tsung E.F., Cai Z., et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 35.Elms P., Scurry A., Davies J., Willoughby C., Hacker T., Bogani D., Arkell R. Overlapping and distinct expression domains of Zic2 and Zic3 during mouse gastrulation. Gene Expr. Patterns. 2004;4:505–511. doi: 10.1016/j.modgep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Henrique D., Adam J., Myat A., Chitnis A., Lewis J., Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]