Abstract
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a neurodegenerative disorder associated with fragile X premutation carriers. Previous studies have shown that fragile X rCGG repeats are sufficient to cause neurodegeneration and that the rCGG-repeat-binding proteins Pur α and heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 could modulate rCGG-mediated neuronal toxicity. Mobile genetic elements or their remnants populate the genomes, and the activities of these elements are tightly controlled for the fitness of host genomes in different organisms. Here we provide both biochemical and genetic evidence to show that the activation of a specific retrotransposon, gypsy, can modulate rCGG-mediated neurodegeneration in an FXTAS Drosophila model. We find that one of the rCGG-repeat-binding proteins, hnRNP A2/B1, is involved in this process via interaction with heterochromatin protein 1. Knockdown of gypsy RNA by RNAi could suppress the neuronal toxicity caused by rCGG repeats. These data together point to a surprisingly active role for retrotransposition in neurodegeneration.
INTRODUCTION
Neurodegenerative diseases are a heterogeneous group of disorders characterized by the progressive loss of structure and/or function of neurons (1). Many neurodegenerative disorders are caused by genetic mutations within the coding regions, such as CAG repeat expansions that can directly alter the function of specific proteins; however, recent studies also suggest that toxic RNAs can directly cause several neurodegenerative disorders, among them fragile X-associated tremor/ataxia syndrome (FXTAS), which is associated with fragile X premutation carriers (2).
Fragile X syndrome (FXS) is usually caused by expansion of the CGG trinucleotide repeat in the 5′ untranslated region of the fragile X mental retardation 1 (FMR1) gene (3). Whereas normal individuals generally possess between 5 and 54 repeats, fully affected individuals have >200 CGG repeats on what are referred to as full mutation alleles (4). Premutation alleles (55–200 CGG repeats) of the FMR1 gene are known to contribute to the fragile X phenotype through genetic instability, and they can expand into the full mutation during germline transmission (5). Within the last decade, FXTAS, a late-onset neurodegenerative disorder, has been recognized among many male premutation carriers in or beyond their fifth decade of life (6), and FXTAS is distinct from the neurodevelopmental disorder, FXS. The most common clinical feature of FXTAS is a progressive action tremor with ataxia. Nearly, all autopsy studies on the brains of symptomatic premutation carriers show degeneration in the cerebellum, which includes Purkinje neuronal cell loss, Bergman gliosis, spongiosis of the deep cerebellar white matter and swollen axons (7,8). The major neuropathological hallmark and postmortem criterion for definitive FXTAS is eosinophilic, ubiquitin-positive intranuclear inclusions broadly distributed throughout the brain in neurons, astrocytes and in the spinal column (7).
One unique molecular signature of the fragile X premutation allele is that the level of FMR1 mRNA is significantly elevated, while the FMR1 protein (FMRP) remains relatively unchanged in cells from premutation carriers (9,10), so the neurodegenerative phenotypes associated with FXTAS are suspected of being caused by a gain of function in fragile X premutation rCGG-repeat RNAs (5,11). It has been hypothesized that overproduced rCGG repeats in FXTAS sequester specific RNA-binding proteins and reduce their ability to perform their normal cellular functions, thereby contributing significantly to the pathology of this disorder. The presence of FMR1 mRNA in inclusions found in the brains of FXTAS patients, as well as the formation of similar inclusions upon ectopic expression of rCGG repeats in model systems, have provided strong support for this hypothesis (11–14). Two RNA-binding proteins, Pur α and heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1, could bind rCGG repeats specifically in both mammalian and Drosophila brains (15,16). Both Pur α and hnRNP A2/B1 are found to be present in the inclusions of FXTAS brain tissues, and could modulate rCGG-mediated neuronal toxicity.
Mobile genetic elements or their remnants populate the genomes of nearly every living organism (17). Transposable elements (TEs) include members of both DNA and RNA families of transposons (retrotransposons). Retrotransposons can be further subdivided into long-terminal repeat (LTR), non-LTR (nLTR) groups, inverted repeat (IR) elements and repeat-containing elements. So far there is no evidence that the DNA elements are currently active, whereas retrotransposons are considered active (18). The potential negative effects of mobile elements on the fitness of their hosts have led to the development of strategies for transposon control in different organisms. Active retrotranspositions are reported to cause human diseases, including several types of cancer, through insertional mutagenesis of genes critical for preventing or driving malignant transformation, and active retrotranspositions contribute to inter-individual genetic variation (17,18). New retrotransposition is found to generate genomic plasticity in neurons by causing variation in genomic DNA sequences and by altering the transcriptome of individual cells (19). More recently, aberrant overexpression of satellite repeats was seen in pancreatic and other epithelial cancers (20). Furthermore, Alu RNAs are also found to directly cause age-related macular degeneration (AMD) (21). These findings point to the direct involvement of retrotransposons in the pathogenesis of human diseases.
Here we show that fragile X rCGG repeats can induce the activation of specific retrotransposons, including gypsy, in an FXTAS Drosophila model. One of the rCGG-repeat-binding proteins, hnRNP A2/B1, could modulate the activation of gypsy via interaction with heterochromatin protein 1 (HP1). Furthermore, knockdown of gypsy RNA by RNAi could suppress the neurodegeneration caused by fragile X rCGG repeats. Our results reveal an unexpectedly active role for retrotransposition in neurodegeneration.
RESULTS
Fragile X rCGG repeats cause the activation of specific retrotransposons in the brain
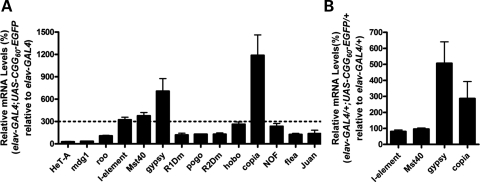
To investigate the molecular pathway(s) involved in rCGG-mediated neurodegeneration, we conducted gene expression profiling using rCGG-repeat transgenic flies that we generated previously (11). In these flies, the severity of their phenotype depends on both dosage and length of the rCGG repeat. To analyze the effect of rCGG repeats in adult brains, we used RNAs isolated from the age- and sex-matched brains of wild-type (WT) flies and flies expressing rCGG60 repeats in neurons for gene expression profiling experiments. Surprisingly, we observed a consistent upregulation of several retrotransposons. Based on this finding, we systematically examined the expression of retrotransposons in our FXTAS Drosophila model. We determined the expression levels of different retrotransposons, LTR elements (mdg1, roo, gypsy, copia and flea), nLTR elements (I-element, HeT-A, R1Dm, R2Dm and Juan), IR elements (pogo, hobo and NOF) and a repetitive locus (Mst40) in the brains of both rCGG-repeat flies (elav-GAL4; UAS-CGG60-EGFP) and control flies (elav-GAL4) by quantitative reverse transcriptase polymerase chain reaction (RT-PCR). Compared with the control flies, we saw a significant increase in three retrotransposons (I-element, gypsy and copia) and one repetitive sequence (Mst40) in rCGG-repeat transgenic flies (Fig. 1A). We further examined the expression of these transposon elements in transgenic flies expressing reduced rCGG repeats (elav-GAL4/+; UAS-CGG60-EGFP/+). We observed a reduced increase in gypsy and copia expression, but detected no elevation in the expression of I-element and Mst40 transcripts in these flies (Fig. 1B). These results together suggest that the increased expression of specific retrotransposons in rCGG-repeat transgenic flies is rCGG-repeat dosage-dependent.
Figure 1.
Activation of selective retrotransposons in rCGG-repeat transgenic flies. (A) The relative steady-state levels of retrotransposon transcripts in the brains of rCGG-repeat flies (elav-GAL4; UAS-CGG60-EGFP) versus control flies (elav-GAL4) were determined by quantitative reverse transcriptase polymerase chain reaction (RT-PCR). (B) Relative quantity of retrotransposon transcripts in rCGG-repeat heterozygous flies (elav-GAL4/+; UAS-CGG60-EGFP/+) compared with control flies (elav-GAL4/+). For all experiments, n≥ 3; error bars indicate mean ± SEM.
flamenco mutants with increased levels of gypsy RNA could modulate rCGG-mediated neurodegeneration
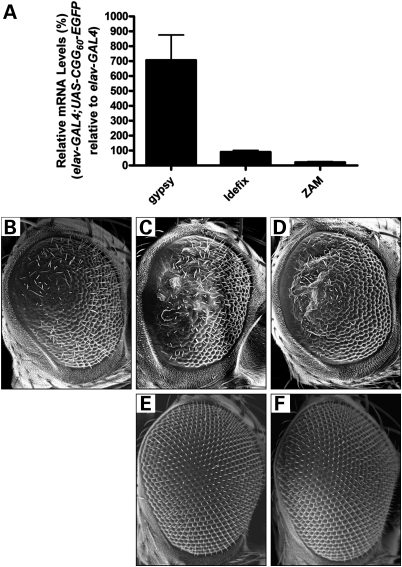
To determine the role of retrotransposon activation in rCGG-mediated neurodegeneration, we further examined the genetic interaction between specific transposons and rCGG-mediated neuronal toxicity based on the fragile X premutation rCGG-repeat-mediated neurodegenerative eye phenotype we observed previously (11). Prior genetic mapping of gypsy resistance determinants led to a discrete locus in the pericentric β-heterochromatin of the X chromosome that was named flamenco (22). Drosophila Piwi-family proteins, including Piwi, Aubergine (Aub) and Argonaute 3 (Ago3), have been implicated in transposon control in reproductive systems (23,24). Piwi-interacting RNAs (piRNAs) generated from discrete loci were found to be the master regulators of transposon activity in Drosophila (23). Recent studies have shown that flamenco is a piRNA cluster that can regulate specific retroelements, including gypsy, Idefix and ZAM (25). Indeed, the presence of one flamenco allele could further elevate the gypsy mRNA level caused by fragile X rCGG60 repeats (data not shown). We saw no significantly increased expression of either Idefix or ZAM in rCGG-repeat transgenic flies (Fig. 2A). To determine the role of gypsy in rCGG-mediated neurodegeneration, we crossed gmr-GAL4, UAS-(CGG)90-EGFP transgenic flies with two flamenco mutant fly lines that are permissive for gypsy expression (26). As shown in Figure 2B–F, derepressing gypsy expression could enhance the neurodegenerative eye phenotype of rCGG-repeat transgenic flies, suggesting that the activation of gypsy could directly modulate rCGG-mediated neurodegeneration.
Figure 2.
Activation of gypsy is involved in rCGG-mediated neurodegeneration. (A) Expression of specific retrotransposons that are regulated by the flamenco locus in rCGG-repeat transgenic flies. flamenco has been shown to control three retroelements: gypsy, Idefix and ZAM. Only gypsy was significantly increased in rCGG-repeat transgenic flies. For all experiments, n ≥ 3; error bars indicate mean ± SEM. (B–F) Derepressing gypsy expression enhances the rCGG-mediated neurodegenerative eye phenotype. SEM eye images from 7-day-old flies. Flies expressing CGG90 show disorganized and fused ommatidia (B). This CGG90 eye phenotype could be enhanced by upregulating gypsy expression in flies carrying a heterozygous mutation in flamenco (C and D). flamenco itself along with gmr-GAL4 does not cause any abnormality in the eye (E and F). Genotypes are B-gmr-GAL4, UAS-CGG90-EGFP/+; C-flamBG02658/+; gmr-GAL4, UAS-CGG90-EGFP/+; D-flamKG00476/+; gmr-GAL4, UAS-CGG90-EGFP/+; E-flamBG02658/+; gmr-GAL4/+; F-flamKG00476/+; gmr-GAL/+.
HnRNP A2/B1 and its Drosophila orthologs are associated with HP1
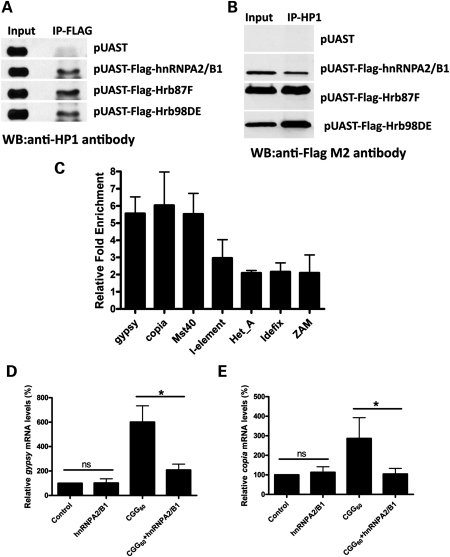
Since Piwi clade is known to be capable of controlling transposon activity in reproductive systems, we further tested the genetic interaction between Piwi proteins and rCGG-mediated neurodegeneration and found that the loss of Piwi proteins had no effect on rCGG-repeat-induced neuronal cell death (data not shown). This is consistent with the observation that Piwi proteins only function in the germline. Besides Piwi clade, transposon control also requires the presence of heterochromatin (27). Indeed, HP1, a conserved critical component of heterochromatin, is known to be required for retrotransposon silencing (28). Intriguingly, HP1 is found to interact with multiple hnRNPs to modulate heterochromatin formation (29). Given that one of the rCGG-repeat-binding proteins that we identified previously is hnRNP A2/B1, we examined the potential association between HP1 and hnRNP A2/B1 proteins. We performed immunoprecipitation using FLAG antibody from S2 cells expressing FLAG-tagged hnRNP A2/B1 and found that HP1 could be co-immunoprecipitated with hnRNP A2/B1 (Fig. 3A). Similarly, HP1 could also be co-immunoprecipitated with two Drosophila orthologs of hnRNP A2/B1, Hrb87F and Hrb98DE (Fig. 3A). These results were further confirmed by immunoprecipitating HP1 from the lysate of S2 cells expressing FLAG-tagged hnRNP A2/B1, HRB87F and HRB98DE (Fig. 3B). Our data suggest that hnRNP A2/B1 is associated with HP1, and potentially heterochromatin.
Figure 3.
HnRNP A2/B1, an rCGG-repeat-binding protein, could modulate retrotransposon activation via interaction with HP1. (A) HP1 could be co-immunoprecipitated with hnRNP A2/B1 and its Drosophila orthologs. Immunoprecipitation (IP) was performed in S2 cell lysates transfected with Flag-hnRNPA2/B1, Flag-Hrb87F and Flag-Hrb98DE, respectively, using anti-Flag M2 antibody. The immunoprecipitates were analyzed by western blot with anti-HP1 antibody. S2 cells transfected with pUAST vector were used as a negative control. (B) HnRNP A2/B1 and its Drosophila orthologs could be co-immunoprecipitated with HP1. Immunoprecipitation (IP) was performed in S2 cell lysates transfected with Flag-hnRNPA2/B1, Flag-Hrb87F and Flag-Hrb98DE, respectively, using anti-HP1 antibody. The immunoprecipitates were analyzed by western blot with anti-FLAG M2 antibody. S2 cells transfected with the pUAST vector were used as a negative control. (C) HnRNP A2/B1 associates with selective retrotransposons in vivo. HnRNP A2/B1 ChIP assay indicates that hnRNP A2/B1 could bind to the genomic regions containing selective retrotransposons, such as gypsy, copia, Mst40 and I-element, but not Het_A, Idefix or ZAM. Relative enrichment is calculated relative to IgG-only non-specific control and normalized to the empty vector (n = 3, error bars indicate mean ± SEM). (D) Overexpression of hnRNPA2/B1 only (elav-GAL4/+; UAS-hnRNPA2/B1/+) shows no effect on gypsy transcript expression compared with control (elav-Gal4/+). However, overexpression of hnRNPA2/B1 (elav-GAL4/+; UAS-CGG60-EGFP/+; UAS-hnRNPA2/B1/+) could suppress the upregulated gypsy transcript expression caused by rCGG repeats (elav-GAL4/+; UAS-CGG60-EGFP/+) (n = 3 for all experiments, *P < 0.05; ns, P > 0.05). (E) Overexpression of hnRNPA2/B1 in normal fly brain (elav-GAL4/+; UAS-hnRNPA2/B1/+) has no effect on copia activity, whereas overexpression of hnRNPA2/B1 in rCGG-repeat transgenic flies (elav-GAL4/+; UAS-CGG60-EGFP/+; UAS-hnRNPA2/B1/+) could rescue the elevated expression of copia (n = 3 for all experiments, *P < 0.05; ns, P > 0.05).
HnRNP A2/B1 binds to selective retrotransposon DNAs and modulates their expression
HP1 is required for retrotransposon silencing (28), and previous chromatin immunoprecipitation (ChIP) assays indicated that HP1 could associate with the majority of the flamenco locus to control retroelement activity (27). Since hnRNP A2/B1 and its Drosophila orthologs are associated with HP1, we examined whether hnRNP A2/B1 could directly bind to genomic DNA. Given that fragile X rCGG repeats could cause the activation of selective retrotransposons and hnRNP A2/B1 is one of the rCGG-binding proteins, we determined the interaction between hnRNP A2/B1 and retrotransposons induced by fragile X rCGG repeats using a ChIP assay. As shown in Figure 3C, hnRNP A2/B1 is significantly associated with the genomic regions containing retrotransposons with increased expression in the presence of fragile X rCGG repeats, including gypsy, copia and Mst40. The retrotransposons without altered expression in the presence of fragile X rCGG repeats are not associated with hnRNP A2/B1 (Fig. 3C).
To further examine the role of hnRNP A2/B1 in regulating retrotransposons, we chose to test the possible interaction between hnRNP A2/B1 and selective retrotransposons. As shown in Figure 3D, overexpression of hnRNPA2/B1 in control fly brain has no effect on gypsy expression, whereas overexpression of hnRNPA2/B1 in rCGG-repeat transgenic flies could suppress upregulated gypsy expression; similar suppression was also seen with copia expression (Fig. 3E). Our results from co-immunoprecipitation experiments, ChIP assays and genetic studies suggest that hnRNP A2/B1 could bind to selective retrotransposons and recruit HP1 for transposon silencing. In the presence of fragile X rCGG repeats, hnRNP A2/B1 will be sequestered by excess rCGG repeats. The depletion of hnRNP A2/B1 potentially leads to less HP1 being recruited to the genomic regions containing those retrotransposons, and subsequent activation of retrotransposons.
Reduction of gypsy expression by RNAi suppresses rCGG-mediated neurodegeneration
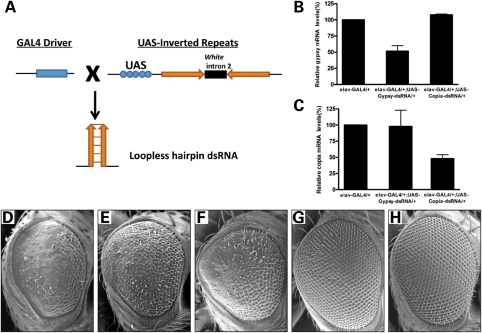
To further investigate the physiological relevance of retrotransposon activation in fragile X premutation rCGG-mediated neurodegeneration, we generated fly UAS lines that could express dsRNAs against either gypsy or copia in the presence of a GAL4 driver (Fig. 4A). Once synthesized, those dsRNAs could produce siRNAs against corresponding retrotransposons and knockdown their expressions (Fig. 4B and C). We then crossed these transgenic lines with the rCGG-repeat transgenic lines that exhibit eye neurodegeneration. To our surprise, we found that expression of dsRNAs against gypsy, but not copia, could suppress rCGG-mediated neurodegeneration (Fig. 4D–F). Expression of these dsRNAs has no effect on control fly eyes (Fig. 4G and H). This observation strongly indicates that the dysregulation of retrotransposon activation plays a significant role in the molecular pathogenesis of rCGG-mediated neuronal toxicity.
Figure 4.
Knockdown of gypsy expression by RNAi suppresses rCGG-mediated neurodegeneration. (A) Shown is the diagram depicting the strategy for generation of transgenic dsRNA lines. A DNA fragment corresponding to the gene of interest is inserted twice into the pWIZ vector, with inserts in opposite orientations on each side of the white intron. IRs that are head–head or tail–tail are placed downstream of the UAS promoter. When these UAS lines are crossed to GAL4 driver lines, the F1 progeny generate tissue- and cell-specific expression of loopless hairpin RNA to induce RNAi in Drosophila. (B) Knockdown efficiency of gypsy and copia expression in transgenic dsRNA fly lines. Real-time PCR data indicate that both gypsy and copia could be efficiently downregulated in transgenic dsRNA fly lines. Data are plotted as mean ± SEM. (D–H) Reduction in gypsy RNA suppresses rCGG-mediated neuronal toxicity. SEM (D–H) eye images from 14-day-old flies. Flies expressing CGG90 show disorganized, fused ommatidia (D). Knockdown of gypsy could rescue the neurodegenerative eye phenotype (E). However, knockdown of copia has no such effect on the eye phenotype (F). Knockdown of gypsy or copia alone does not cause an abnormal eye phenotype (G–H). Genotypes are D-gmr-GAL4, UAS-CGG90-EGFP/+; E-gmr-GAL4, UAS-CGG90-EGFP/UAS-Gypsy-dsRNA; F-gmr-GAL4, UAS-CGG90-EGFP/UAS-Copia-dsRNA; G-gmr-GAL4/UAS-Gypsy-dsRNA; H-gmr-GAL4 /UAS-Copia-dsRNA.
DISCUSSION
FXTAS is a neurodegenerative disorder associated with fragile X premutation carriers. Previous studies have shown that fragile X rCGG repeats are sufficient to cause neurodegeneration and that the rCGG-repeat-binding proteins Pur α and hnRNP A2/B1 could modulate rCGG-mediated neuronal toxicity. Mobile genetic elements or their remnants populate the genomes, and the activities of these elements are tightly controlled for the fitness of host genomes in different organisms. Here we demonstrate that activation of specific retrotransposons could contribute to fragile X rCGG-repeat-mediated neurodegeneration using a FXTAS fly model. We show that fragile X premutation rCGG repeats could induce the activation of specific retrotransposons. HnRNP A2/B1, one of the rCGG-binding proteins that we identified previously, could regulate the activity of these retrotransposons by interacting with HP1. More importantly, reduction in a specific retrotransposon, gypsy, could suppress rCGG-repeat-mediated neuronal toxicity. Our biochemical and genetic studies demonstrate a surprisingly active role for retrotransposition in fragile X premutation rCGG-repeat-mediated neurodegeneration.
TEs make up a significant portion of most eukaryotic genomes: for example, 80% of the maize (30), 45% of the human (31) and 5.3% of the fruit fly (32,33) genomes are known to consist of TEs. These elements can impact genome diversity and human disease either through insertional mutation or by contributing recombination substrates, both during and long after their integration (17). Barbara McClintock has proposed that, in addition to causing chromosome breakage and acting as insertional mutagens, these transposons might also act as ‘controlling elements’ to play an important role in gene regulation (34). Recent work suggests TEs may upregulate the expression of host genes (35). They may function as part of genome-wide regulatory networks (36). LINE-1 (L1) elements, the major group of nLTR retrotransposons, are known to play important roles in mammalian genome evolution (18,37,38). Early studies of L1 expression put a strong emphasis on primarily germline expression of these elements (39,40). Recent evidence suggests a somatic function for L1 transcripts, including cell proliferation (41), differentiation (42) and early embryo development (42). Moreover, somatic retrotransposition events have been found in the human brain, and the retrotransposition may have the potential to contribute to neurogenesis and/or affect neuronal function (43). More recently, aberrant overexpression of satellite repeats has been seen in pancreatic and other epithelial cancers (20). Furthermore, Alu RNAs are also known to directly cause AMD, and knockdown of Alu could mitigate AMD (21). These findings point to the direct involvement of retrotransposons in the pathogenesis of human diseases. In this study, for the first time, we show an active role for retrotransposition in neurodegeneration, as well. We saw a significant increase in several retrotransposon transcripts in fly brains from a FXTAS Drosophila model. More importantly, we show that the activation of these retrotransposons is directly related to neuronal cell death, since the reduction in a specific retrotransposon RNA, gypsy RNA, could suppress rCGG-mediated neurodegeneration.
LTR retroelements are found to amplify to high levels, resulting in major modifications of the host genome in several species (44,45). In Drosophila, gypsy is the most abundant among the four main groups of LTR retroelements (gypsy, copia, DEL and DIRS) (32,46). In our study, we observed much higher gypsy and copia RNA expression in the rCGG-repeat transgenic fly brain than in normal fly brain. Moreover, the increased expression of these retrotransposons is rCGG-repeat dosage-dependent. Intriguingly, the knockdown of gypsy RNA only, but not copia RNA, could suppress rCGG-mediated neuronal toxicity. This may imply that different active retrotransposons could influence the fate of neuronal cells differently, which requires further investigation.
One unique molecular signature of the fragile X premutation allele is that the level of FMR1 mRNA is significantly elevated, while the FMRP remains relatively unchanged in cells from premutation carriers (9,10). Given that the neurodegenerative phenotype of FXTAS is associated specifically with premutation carriers, but not with the full mutation, FMRP deficiency per se is not likely to be the culprit behind FXTAS (6). Instead, the neurodegenerative phenotypes associated with FXTAS are suspected of being caused by a gain of function in fragile X premutation rCGG-repeat RNAs (5,11). It has been hypothesized that overproduced rCGG repeats in FXTAS sequester specific RNA-binding proteins and reduce their ability to perform their normal cellular functions, thereby contributing significantly to the pathology of this disorder. The presence of FMR1 mRNA in inclusions found in the brains of FXTAS patients, as well as the formation of similar inclusions upon ectopic expression of rCGG repeats in model systems, provide strong support for this hypothesis (11–14). Two RNA-binding proteins, Pur α and hnRNP A2/B1, have been shown to bind rCGG repeats specifically in both mammalian and Drosophila brains (15,16). Both Pur α and hnRNP A2/B1 are present in the inclusions of FXTAS brain tissues. Furthermore, overexpression of either Pur α or hnRNP A2/B1 can alleviate neurodegeneration in the fly model of FXTAS (15,16). In this study, we show that hnRNP A2/B1 could also directly bind to the genomic regions containing specific retrotransposons and interact with HP1, a conserved critical component of heterochromatin that is known to be required for retrotransposon silencing (28). Moreover, our study found that overexpression of hnRNP A2/B1 could suppress upregulated gypsy and copia expression in rCGG-repeat flies. These data together suggest that hnRNP A2/B1 is also required in retrotransposon modulation. In FXTAS, our results imply that the sequestration of the rCGG-repeat-binding proteins, particularly hnRNP A2/B1, could limit the amount of hnRNP A2/B1 protein that can bind to retrotransposons and recruit HP1 for efficient retrotransposon silencing.
In summary, we provide both biochemical and genetic data to support an active role for retrotransposon activation in rCGG-mediated neurodegeneration. The consequences of such retrotransposon activation in post-mitotic neurons could be genomic instability and neuronal apoptosis. It would be interesting to explore further whether the activation of TEs could be a common mechanism underlying neurodegeneration in general.
MATERIALS AND METHODS
Drosophila genetics
All flies were maintained under standard culture conditions. The rCGG-repeat transgenic flies (UAS-CGG60-EGFP and UAS-CGG90-EGFP) and UAS-hnRNPA2/B1 were generated in the lab as described previously. The flamenco mutant fly lines (flamBG02658 and flamKG00476) were obtained from Bloomington Stock Center. The UAS-Gypsy-dsRNA transgenic flies were generated as described previously (47). In brief, a 595 bp DNA fragment corresponding to gypsy was amplified by PCR (forward primer: GCTCTAGAAAGTGGTATCGGTGCAGTCC; reverse primer: GCTCTAGAGCAGTGAATAGCGTTCACGA). The PCR products were inserted twice by two ligation steps into the pWIZ vector. After the second ligation step, inserts were in an opposite orientation. IRs that are head–head or tail–tail repeats were confirmed by DNA sequencing. The constructs were then injected in the w1118 strain by standard methods.
Quantitative RT–PCR
The fly heads from genotypes indicated were collected. Trizol (Invitrogen) was used to isolate total RNA from each genotype. RNA samples were reverse-transcribed into cDNA with oligo(dT)20 and SuperScript III (Invitrogen). Real-time PCR was performed with gene-specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems) using the 7500 Standard Real-Time PCR System (Applied Biosystems). RpL32 (Qiagen) was used as an endogenous control for all samples. Primers for retrotransposon transcripts were designed using Primer Express 3.0 software (Applied Biosystems) and were as follows:
- Primer name
Sequence (forward primer, reverse primer)
- Gypsy
CGTCTACCGTTCGGCTTGAG, CACATCGTCTAGGGCTCTTTGA
- HeT-A
CGCGCGGAACCCATCTTCAGA, CGCCGCAGTCGTTTGGTGAGT
- Mdg1
CTTCAGTACCCAGATTTCAGCAAA, CCGCTCCACATGCTTGTTTA
- I-element
TCGACTTCGGACTGTCTACTCTATGT, GTGACGAGGGAGGTGTGCAT
- Mst40
CCTAAGTCCCTCGCAATCAAGT, ACGCTTAAAGTGCGATCATCAG
- Roo
GTCTGAGGCATCCGTTTGGT, CGTGTGGTGAGGTTTACGACAA
- R1Dm
TGCGCCTACGGTGAGGTT, CCGACAGCTACGCACATACG
- Pogo
CCAAAAATTTAACGACGCCTTT, GCGCCAGCGCCAAA
- R2Dm
CCGACCCTCCGTGGATATC, GTAGCCGCTGCGTTTGGTT
- Hobo
CAAGTGCGACCGTCGACAT, AAGTGATGCCCAAAAAGTTTCTTT
- Copia
GAGGTTGTGCCTCCACTTAAGATT, CAATACCACGCTTAGTGGCATAAA
- NOF
TGCATCGAAGCTGTTTGCA, GAGTTTTAACGGTCTTGCGTTTC
- Flea
TTGTAGCTGGCCGCAAGAT, TTGACCCCTGTGACTTCGTATG
- Juan
GACCGTTTTAATATACCCGAATACACTA, CGCCGCCTCCCCTACTT
All real-time PCR reactions were performed in triplicate, and RQs were calculated using the ΔΔCt method, with calibration to control samples.
Scanning electron microscopy
For scanning electron microscopy (SEM) images, whole flies were dehydrated in gradient concentration ethanol (25, 50, 75 and 100%), dried with hexamethyldisilazane (Sigma), and analyzed with an ISI DS-130 LaB6 SEM/STEM microscope.
Immunoprecipitation
S2 cells were co-transfected with pMT-GAL4 and pUAST plasmid coding for Hrb87F, Hrb98DE and hnRNPA2/B1 protein, all FLAG-tagged using Effectene transfection reagent (Qiagen) following the manufacturer's procedure. Cells were induced 24 h with 500 μm copper sulfate (Sigma) beginning 12 h after transfection. Cells were then harvested by centrifugation and incubated in lysis buffer [10 mm Tris (pH 7.4), 150 mm NaCl, 30 mm EDTA, 0.5% Triton X-100] with 2× complete protease inhibitors on ice for 30 min, followed by centrifugation at 15 000g for 20 min. The nuclear lysate was precleared with 100 μl recombinant protein A agarose (Invitrogen) for 1 h. Ten-microgram precleared nuclear lysate was used as an input in western blot analysis. The remaining nuclear lysate was immunoprecipitated with anti-Flag M2 beads (Sigma) or anti-HP1 antibody (Covance) with protein A agarose (Invitrogen) at 4°C overnight. The precipitated complexes were used for western blot. Anti-HP1 antibody (Covance) at a dilution of 1:500 or anti-Flag M2 antibody at a dilution of 1:1000 was used for western blot. Detection of horseradish peroxidase was performed using ECL Western Blotting Detection reagents (GE Healthcare).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using a ChIP Assay Kit (Millipore). S2 cells were crosslinked with 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. Chromatin was fragmented to an average size of 500 bp by sonication (Sonicator 3000; Misonix) and immunoprecipitated with an anti-Flag M2 antibody (sigma). Immunoprecipitated and purified DNA fragments were diluted to 1 ng/µl in nuclease-free water. We used 8 ng of DNA in 20 µl SYBR Green real-time PCR reactions consisting of 1× Power SYBR Green Master Mix and 0.5 µm forward and reverse primers. Reactions were run on an SDS 7500 Fast Instrument (Applied Biosystems). Primers were designed using Primer Express 3.0 software (Applied Biosystems) and were as above. DNA relative enrichment was determined by taking the absolute quantity ratios of specific IPs to non-specific IPs (normal mouse IgG only), IP/IgG and normalizing to control (pUAST only). Independent chromatins were prepared for all ChIP experiments, and real-time PCR reactions were performed in triplicate for each sample on each amplicon.
FUNDING
H.T. is supported by the China Scholarship Council. A.Q. was supported by and is a recipient of National Ataxia Foundation Postdoctoral Award. H.L. is supported by the National Natural Science Foundation of China 30430260 and 30225024. P.J. is supported by NIH grants (R01 NS051630 and R21 NS067461). P.J. is a recipient of the Beckman Young Investigator Award and the Basil O'Connor Scholar Research Award, as well as an Alfred P Sloan Research Fellow in Neuroscience.
ACKNOWLEDGEMENTS
The authors would like to thank J. Taylor of The Integrated Microscopy and Microanalytical Facility for her help with SEM, the members of the Jin lab for their assistance and C. Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Zoghbi H.Y., Orr H.T. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranum L.P., Day J.W. Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet. 2004;20:506–512. doi: 10.1016/j.tig.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Warren S.T., Sherman S.L. The Metabolic & Molecular Bases of Inherited Disease. In: Scriver C.R., Beaudet A.L., Valle D., Childs B., Kinzler K.W., Vogelstein B., editors. Vol. 1. New York: McGraw-Hill Companies; 2001. pp. 1257–1290. [Google Scholar]
- 4.Sherman S. Fragile X Syndrome: Diagnosis, Treatment and Research. In: Hagerman R.J., editor. Baltimore, MD: The Johns Hopkins University Press; 2002. pp. 136–168. [Google Scholar]
- 5.Hagerman R.J., Hagerman P.J. The fragile X premutation: into the phenotypic fold. Curr. Opin. Genet. Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- 6.Hagerman P.J., Hagerman R.J. The fragile-X premutation: a maturing perspective. Am. J. Hum. Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greco C.M., Berman R.F., Martin R.M., Tassone F., Schwartz P.H., Chang A., Trapp B.D., Iwahashi C., Brunberg J., Grigsby J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 8.Greco C.M., Hagerman R.J., Tassone F., Chudley A.E., Del Bigio M.R., Jacquemont S., Leehey M., Hagerman P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 9.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenneson A., Zhang F., Hagedorn C.H., Warren S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 11.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 12.Willemsen R., Hoogeveen-Westerveld M., Reis S., Holstege J., Severijnen L.A., Nieuwenhuizen I.M., Schrier M., van Unen L., Tassone F., Hoogeveen A.T., et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 13.Tassone F., Iwahashi C., Hagerman P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 14.Arocena D.G., Iwahashi C.K., Won N., Beilina A., Ludwig A.L., Tassone F., Schwartz P.H., Hagerman P.J. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum. Mol. Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- 15.Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 20.Ting D.T., Lipson D., Paul S., Brannigan B.W., Akhavanfard S., Coffman E.J., Contino G., Deshpande V., Iafrate A.J., Letovsky S., et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelisson A., Song S.U., Prud'homme N., Smith P.A., Bucheton A., Corces V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravin A.A., Hannon G.J. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- 25.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 26.Mevel-Ninio M., Pelisson A., Kinder J., Campos A.R., Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moshkovich N., Lei E.P. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000880. e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klattenhoff C., Xi H., Li C., Lee S., Xu J., Khurana J.S., Zhang F., Schultz N., Koppetsch B.S., Nowosielska A., et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piacentini L., Fanti L., Negri R., Del Vescovo V., Fatica A., Altieri F., Pimpinelli S. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000670. e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.SanMiguel P., Tikhonov A., Jin Y.K., Motchoulskaia N., Zakharov D., Melake-Berhan A., Springer P.S., Edwards K.J., Lee M., Avramova Z., et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 31.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 32.Kaminker J.S., Bergman C.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0084. RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesneville H., Bergman C.M., Andrieu O., Autard D., Nouaud D., Ashburner M., Anxolabehere D. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput. Biol. 2005;1:166–175. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 35.Naito K., Zhang F., Tsukiyama T., Saito H., Hancock C.N., Richardson A.O., Okumoto Y., Tanisaka T., Wessler S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 36.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotea V., Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Xing J., Wang H., Belancio V.P., Cordaux R., Deininger P.L., Batzer M.A. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl Acad. Sci. USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branciforte D., Martin S.L. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostertag E.M., DeBerardinis R.J., Goodier J.L., Zhang Y., Yang N., Gerton G.L., Kazazian H.H., Jr. A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 41.Kuo K.W., Sheu H.M., Huang Y.S., Leung W.C. Expression of transposon LINE-1 is relatively human-specific and function of the transcripts may be proliferation-essential. Biochem. Biophys. Res. Commun. 1998;253:566–570. doi: 10.1006/bbrc.1998.9811. [DOI] [PubMed] [Google Scholar]
- 42.Mangiacasale R., Pittoggi C., Sciamanna I., Careddu A., Mattei E., Lorenzini R., Travaglini L., Landriscina M., Barone C., Nervi C., et al. Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene. 2003;22:2750–2761. doi: 10.1038/sj.onc.1206354. [DOI] [PubMed] [Google Scholar]
- 43.Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O'Shea K.S., Moran J.V., Gage F.H. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baucom R.S., Estill J.C., Leebens-Mack J., Bennetzen J.L. Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res. 2009;19:243–254. doi: 10.1101/gr.083360.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piegu B., Guyot R., Picault N., Roulin A., Saniyal A., Kim H., Collura K., Brar D.S., Jackson S., Wing R.A., et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin-Shan X., Qing-You X., Jun L., Guo-Qing P., Ze-Yang Z. Survey of long terminal repeat retrotransposons of domesticated silkworm (Bombyx mori) Insect Biochem. Mol. Biol. 2005;35:921–929. doi: 10.1016/j.ibmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y.S., Carthew R.W. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]