Abstract
Long interspersed element-1 (LINE-1 or L1) retrotransposons account for nearly 17% of human genomic DNA and represent a major evolutionary force that has reshaped the structure and function of the human genome. However, questions remain concerning both the frequency and the developmental timing of L1 retrotransposition in vivo and whether the mobility of these retroelements commonly results in insertional and post-insertional mechanisms of genomic injury. Cells exhibiting high rates of L1 retrotransposition might be especially at risk for such injury. We assessed L1 mRNA expression and L1 retrotransposition in two biologically relevant cell types, human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), as well as in control parental human dermal fibroblasts (HDFs). Full-length L1 mRNA and the L1 open reading frame 1-encoded protein (ORF1p) were readily detected in hESCs and iPSCs, but not in HDFs. Sequencing analysis proved the expression of human-specific L1 element mRNAs in iPSCs. Bisulfite sequencing revealed that the increased L1 expression observed in iPSCs correlates with an overall decrease in CpG methylation in the L1 promoter region. Finally, retrotransposition of an engineered human L1 element was ∼10-fold more efficient in iPSCs than in parental HDFs. These findings indicate that somatic cell reprogramming is associated with marked increases in L1 expression and perhaps increases in endogenous L1 retrotransposition, which could potentially impact the genomic integrity of the resultant iPSCs.
INTRODUCTION
Human embryonic stem cells (hESCs) are pluripotent cells derived from the inner cell mass of human blastocysts (1). Recent studies have shown that the introduction of three or four defined transcription factors into lineage-restricted somatic cells (e.g. fibroblasts) leads to cellular reprogramming culminating in induced pluripotent stem cells (iPSCs). iPSCs share a similar transcriptional profile and potential for differentiation into three germ layers with hESCs (2–4). Both hESCs and iPSCs hold promise for regenerative therapies for a variety of diseases. Indeed, iPSCs may hold greater promise than hESCs as they represent a potential source of autologous cells compatible with the host immune system. However, the therapeutic utility of iPSCs and hESCs could be limited by adverse changes in genomic integrity that occur during reprogramming or subsequent expansion in vitro (5,6). For example, it is paramount to avoid introducing cells with precancerous mutations induced in the process of generating the iPSCs. Thus, it is important to understand processes that may impact genomic integrity in both iPSCs and hESCs.
Long interspersed element-1 (LINE-1 or L1) sequences are abundant retrotransposons in the human genome (7). Although most L1s have been rendered immobile by mutational processes (reviewed in 8,9), it is estimated that the average human genome harbors ∼80–100 retrotransposition-competent L1s (RC-L1s) (8–11) that can impact genome integrity by inserting into new genomic locations via the reverse transcription of an RNA intermediate (reviewed in 8,9). Human RC-L1s are ∼6 kb and contain two open reading frames (ORF1 and ORF2) whose protein products (ORF1p and ORF2p) are required for retrotransposition (12,13). The majority of these RC-L1s belong to a human-specific subfamily of L1s (L1Hs), and a small number of these elements (termed hot L1s) are responsible for the bulk of retrotransposition activity in modern day humans (10,11,14). In addition, the L1-encoded proteins also can act in trans to facilitate the retrotransposition of short interspersed elements, certain non-coding RNAs, and certain messenger RNAs to new genomic locations (15–20). Ongoing L1-mediated retrotransposition events contribute to inter-individual human genetic diversity (11,21–24) and have been implicated in a broad range of sporadic diseases, including hemophilia A, Duchenne muscular dystrophy, X-linked retinitis pigmentosa, β-thalassemia and colon cancer (25; reviewed in 8,26,27). Therefore, RC-L1 ongoing mobility have the potential to adversely impact genome integrity.
In principle, heritable L1-mediated retrotransposition events must occur in cells that give rise to gametes, during gametogenesis, or during early embryonic development. Indeed, previous studies revealed that endogenous L1s are expressed in male and female germ cells, in hESCs and in select somatic tissues (28–32,34,36,37). Consistently, genetic studies, as well as studies conducted with engineered human RC-L1s, have revealed that L1 retrotransposition can occur in the germ line, during early embryonic development, and in select somatic cells (25,32–36,38–40). Despite these findings, many questions remain about the frequency and developmental timing of L1 retrotransposition in vivo and whether L1 retrotransposition is induced as a consequence of cellular reprogramming. We now describe studies assessing L1 mRNA expression and the retrotransposition efficiency of engineered human L1 retrotransposons in hESCs, iPSCs derived from human dermal fibroblasts (HDFs) as well as parental HDFs. We demonstrate that L1 expression is reinstated upon somatic cell reprogramming and that the resultant iPSCs support levels of engineered L1 retrotransposition similar to those of hESCs.
RESULTS
Reprogramming HDFs into iPSCs induces L1 retroelement transcription
Previous studies demonstrated that mRNAs from both human-specific (L1Hs) and older L1 subfamiles are expressed in hESCs (31,37). Here, we determined the relative levels of L1 mRNA expression in hESCs, iPSCs derived from HDFs and parental HDFs. Adult HDFs from skin biopsies of two subjects were reprogrammed with retroviruses expressing four factors (Sox2, Oct3/4, Klf4 and c-Myc) (3) to generate the iPSC lines iPS-F [karyotype iPSCs and HDFs: 46X,del(x)(q24)] and iPS 8.11 (karyotype: 46,XY). Another iPSC line, iAND-4 [karyotype iPSCs: 46,XY,+der(1),t(1;17); karyotype HDFs: 46,XY], was generated by reprogramming newborn foreskin fibroblasts with a polycistronic lentiviral vector (41) (see also Supplementary Material, Table S1). A fourth previously derived iPSC line from HDFs was also used in the study (MSUH001, generated with Sox2, Oct3/4, Klf4 and Lin28) (41). The iPSC phenotype in new derived lines was verified by analyzing the transcription profiles of specific pluripotency genes, the silencing of retroviral/lentiviral expression vectors, karyotyping and the ability of the cells to differentiate into each of the three germ cell layers (W. Collins et al., manuscript in preparation and Supplementary Material, Figs S1–S3).
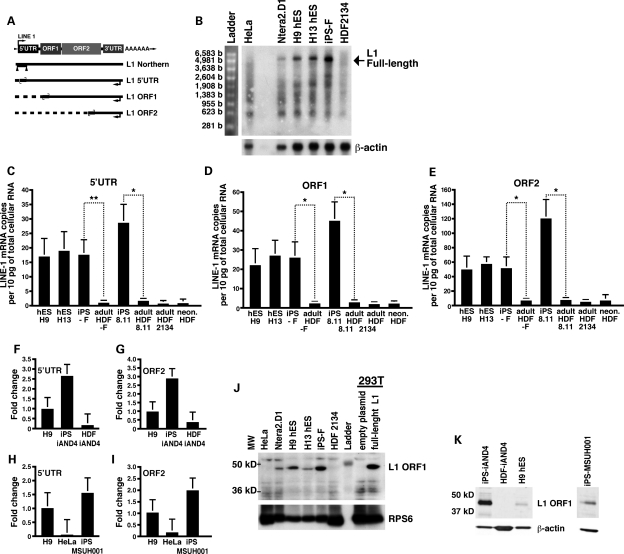
To determine whether pluripotent cells express full-length L1 mRNA, we isolated cytoplasmic poly(A)+ RNA from hESCs and iPSCs and performed northern blot analyses with an RNA probe complementary to the 5′UTR of an RC-L1 (Fig. 1A). The ∼6-kb full-length sense-strand L1 mRNA was detected in H9, H13, iPS-F and a human embryonic carcinoma cell line (Ntera2.D1) (14), but not in parental HDFs or human HeLa cells (Fig. 1B). Several smaller L1 RNA species also were evident. These shorter L1 RNAs may arise from the use of alternative polyadenylation signals and/or cryptic splice sites in the coding strand of L1 (42–44).
Figure 1.
Endogenous full-length sense-strand L1 mRNA is transcribed in pluripotent cells and is up-regulated during the reprogramming of adult HDFs. (A) A schematic depiction of an L1 retrotransposon. Depicted below the cartoon are the approximate locations of the RNA probe used in the L1 northern blot analysis in (B) (triangles) as well as the Taqman primer/probe pairs (small convergent arrows) used in RT–PCR analyses shown in (C)–(E) (large backward arrows indicate the position of the RT primer). (B) Northern blot analysis of cytoplasmic poly(A+) mRNA with a strand-specific RNA probe corresponding to the L1 5′UTR. The signal at ∼6.0 kb corresponds to full-length L1 mRNA (marked by an arrow). Endogenous expression of L1 mRNA in hESCs, iPSCs and HDFs as assessed by real-time RT–PCR using probe sets that correspond to the L1 5′UTR (C), ORF1 (D) or ORF2 (E). Standard curves were prepared using known L1 DNA concentrations and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to control for the amount of RNA input per reaction. Values are RNA copies per 10 pg of total RNA, which corresponds approximately to the total RNA per cell. Values are the mean ± SEM (n= 3). *P≤ 0.05, **P ≤ 0.01. L1 mRNA expression levels in iAND-4 (F and G) and MSUH001 iPSCs (H and I) determined by quantitative RT–PCR with a set of primers against the 5′UTR (F and H) or ORF2 (G and I) sequences as described (37). The amount of L1 RNA was determined in H9 hESCs and parental HDFs for iAND-4 and HeLa for MSUH001 as controls. The graphs show the fold change in L1 expression compared with H9 hESCs and are normalized to GAPDH RNA. (J) Western blot analysis of RNPs with an L1 ORF1p antibody (47). 293T cells transfected with a full-length L1 construct were used as a positive control. The ribosomal S6 protein (RPS6) served as a loading control. (K) Western blot analysis using a polyclonal antibody against ORF1p (kind gift of Dr G. Cristofari) in ribonucleoproteins isolated from iAND-4 and MSUH001 iPSCs. β-Actin served as a loading control. Numbers on the left side, molecular weight standards (kD).
To quantify L1 RNA produced in iPSCs and hESCs, we generated Taqman primer/probe sets that recognize specific regions in the L1 5′UTR, ORF1 and ORF2 regions of a consensus L1Hs element (10) (Fig. 1A). The probe sets were designed to discriminate between full-length L1 RNAs and truncated L1 RNAs, which may be fortuitously transcribed from other genomic sites. In addition, the reverse-transcription reaction was performed with a strand-specific primer complementary to the 3′UTR of L1Hs/L1P1, allowing the detection of only sense-strand L1 RNA transcripts (Fig. 1A). Real-time reverse transcriptase polymerase chain reaction (RT–PCR) analyses revealed similar L1 RNA levels in iPSCs and hESCs (Fig. 1C–E). However, iPSCs had on average 18.5 ± 5.4-fold higher L1 RNA levels for all three primer/probe sets when compared with parental fibroblasts. The larger amount of L1 mRNA detected with the L1 ORF2 primer/probe pair when compared with the L1 ORF1 and L1 5′UTR primer/probe pairs may be due to the presence of 5′-truncated L1s located in the introns of unspliced mRNAs or non-coding RNAs (45,46). Consistent with the above analyses, a different set of primers directed against sequences in either the L1 5′UTR or ORF2 revealed ∼15-fold higher levels of L1 mRNA in iAND-4 iPSCs when compared with the parental fibroblast cell line (Fig. 1F–G). Using the same set of primers, we also observed elevated expression of L1 mRNA (Fig. 1H–I) in an iPSC line generated with Lin28 instead of c-Myc (MSUH001, karyotype: 46,XX) (41) (Supplementary Material, Table S1 and Fig. S3).
We next tested for the expression of the L1 ORF1p protein in iPSCs. Immunoblotting of ribonucleoprotein particles (RNPs) isolated by sucrose gradient centrifugation from the cytosolic fraction of cells with a polyclonal L1 ORF1p antibody (47) revealed detectable ORF1p protein expression in iPSCs (Fig. 1J), but not in HDFs or other differentiated cell types (i.e. HeLa cells and HEK293T cells; Fig. 1J). As described previously, L1 ORF1p also was detected in RNPs derived from H9 and H13 hESCs as well as in Ntera2.D1 cells (Fig. 1J) (31,36,48). Similarly, L1 ORF1p expression was observed in the RNP fraction isolated from the iPSC lines iPS-iAND4 and MSUH001 utilizing a different ORF1p antibody (Fig. 1K). Thus, reprogramming of fibroblasts into iPSCs is associated with a striking increase in the expression of both endogenous L1 mRNA and L1 ORF1p.
Increased expression of L1 is associated with hypomethylation of the L1 promoter region
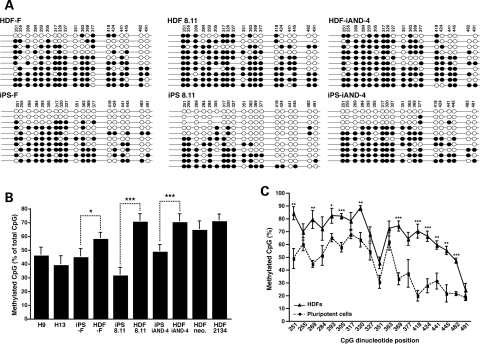
L1 expression appears to be regulated by the methylation of CpG islands in the L1 promoter region (28,49). To determine whether the L1 methylation pattern differs between iPSCs and HDFs, we performed bisulfite conversion analysis on genomic DNA corresponding to a region of the L1 5′UTR promoter containing 20 CpG dinucleotides from five pluripotent cell lines (H9 and H13 hESCs as well as iPS-F, iPS 8.11 and iPS-iAND-4 iPSCs) and five HDFs (HDF-F, HDF 8.11, HDF iAND-4, HDF 2134 and a neonatal HDF sample) (36). In general, hESCs and iPSCs exhibited similar levels of methylation that were lower than those detected in HDF cells (Fig. 2A and B). Direct comparison of three iPS/parental HDF pairs showed significantly fewer CpG methylation events in iPSCs when compared with HDFs (36.8% reduction; P= 0.029, 3.87 × 107 and 1.18 × 104; Fig. 2A and B). Analysis of the 20 CpG dinucleotides in the L1 5′UTR revealed significant differences between pluripotent cells and HDF cells, particularly in the 3′ region of the analyzed CpG island (***P ≤ 0.001; Fig. 2C and Supplementary Material, Fig. S4). Notably, fully unmethylated sequences were detected in pluripotent cells, but not HDFs (iPSCs 6 of 31 sequences versus HDFs 0 of 30 sequences; Fig. 2A). Thus, the increase in L1 expression in pluripotent cells appears to correlate with an overall decreased CpG methylation in the 3′ region of the L1 promoter. These results are in general agreement with a recently published study (36) where higher expression of L1 mRNA was demonstrated in brain compared with skin samples and correlated with a decreased CpG methylation in the L1 promoter in the brain. Although the above experiments suggest a global hypomethylation of L1 promoter sequences upon reprogramming HDFs into iPSCs, it is worth mentioning that reprogramming of other somatic cells might result in slightly different CpG methylation changes in L1 promoters, especially as variable methylation patterns between different iPSC lines were recently reported (50).
Figure 2.
Analysis of the L1 5′UTR promoter reveals significant hypomethylation in iPSCs. (A) Bisulfite sequencing results from three iPSC/parental HDF cell lines. Shown are the 10 clones with the highest sequence similarity to a hot L1 element (L1.3, accession number L19088.1) (53). Each line reflects an independent clone. For iPS 8.11, 11 clones are shown, as the two most divergent clones had the same sequence similarity. White and black circles represent unmethylated and methylated CpG dinucleotides, respectively. Mutated CpG islands are skipped (no circle). (B) Bisulfite analysis of different HDF and pluripotent cell lines show similar percentages of methylated CpG dinucleotides in hESCs and iPSCs and an overall increase in CpG methylation in HDFs. Each bar represents data from the clones shown in (A) presented as methylated CpG dinucleotides in percent of total CpG dinucleotides (mean ± SEM, *P≤ 0.05, ***P≤ 0.001, unpaired two-tailed t-test). (C) Analysis of each single CpG dinucleotide between five grouped HDF lines (HDF-F, HDF 8.11, HDF iAND-4, HDF 2134 and neonatal HDFs) and five grouped pluripotent cell lines (H9 hESCs, H13 hESCs, iPS-F, iPS 8.11 and iPS-iAND-4) reveals significant methylation differences in the 3′ region of the CpG island. Values are the mean ± SEM. *P≤ 0.05, **P≤ 0.01, ***P ± 0.001, unpaired two-tailed t-test. Values for each single cell line are shown in Supplementary Material, Fig. S4.
CpG methylation is significantly decreased in hot L1 promoter regions
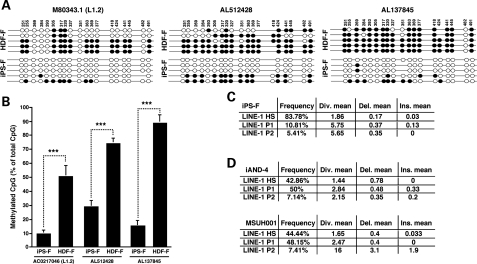
We next investigated whether alleles of known RC-L1s were hypomethylated during the establishment of iPSCs. Previous studies suggested that the bulk of L1 retrotransposition emanates from a small group of hot RC-L1s (10,11). To explore the CpG methylation state of the promoter region of alleles of known hot RC-L1s, we performed bisulfite conversion analysis on genomic DNA from the iPS-F iPSCs and parental HDFs. We chose two hot RC-L1 alleles [AL512428 (6p22.3) and AL137845 (Xq22.33)] as well as an RC-L1 allele that is the progenitor of a mutagenic insertion in man [M80343.1, L1.2A (22q11-q12)] for analysis (10,25). To study the CpG islands in these specific L1s, we performed nested PCR, first amplifying a ∼900–1200-bp region including the first 700 bp of L1 and a 200–500-bp region upstream of the specific L1, followed by the internal amplification of the L1 CpG island. In agreement with the prior results (Fig. 2), the level of CpG methylation in iPSCs was significantly lower when compared with parental HDFs [Fig. 3A and B, AL512428 (61% decrease), AL137845 (83% decrease) and M80343.1 (L1.2) (81% decrease), P≤ 0.001]. These findings suggest that the 5′UTR of these hot L1 alleles becomes hypomethylated upon somatic cell reprogramming.
Figure 3.
Methylation analysis of the promoter region of hot RC-L1s in iPSCs. (A) Bisulfite sequencing results of the promoter region of three hot RC-L1 loci in the iPS-F/HDF-F cell lines. The specific L1 CpG islands were analyzed by nested PCR; first the hot RC-L1 region was amplified, followed by the amplification of the L1 CpG island from bisulfite converted DNAs. Shown are the five clones with the highest sequence similarity to L1.2. (M80343.1). White and black circles represent unmethylated and methylated CpG dinucleotides, respectively. Mutated CpG islands are skipped (no circle). (B) Bisulfite analysis of the iPS-F/HDF-F cell lines reveals a significant decrease in CpG methylation in iPSC lines (M80343.1/(L1.2): 81%, AL512428: 61%, AL137845: 83%). Each bar represents data from the clones shown in (A). Values are the mean ± SEM of the percentage of methylated CpGs. ***P≤ 0.001 (unpaired two-tail t-test). (C) Analysis of cloned ORF1 cDNA sequences (∼750 bp) from iPS-F cells revealed that ∼84% (31 of 37 sequences) are derived from L1Hs elements. Analysis was performed using RepeatMasker (51). Div., diversity; Del., deletion; Ins., insertion in respect to the reference sequences. (D) Analysis of cloned ORF1 cDNA sequences (236 bp) from iAND-4 and MSUH001 iPSCs revealed that ∼50% are derived from L1Hs elements. Analyses were performed using RepeatMasker (51).
iPSCs express young human-specific L1 transcripts
To determine whether L1 mRNAs expressed in iPSCs are derived from RC-L1 alleles, we isolated RNA from iPS-F cells and used it as an RT–PCR template to analyze a ∼750-bp region of ORF1. We then analyzed the sequences of 37 independent clones using RepeatMasker (http://repeatmasker.org) (51). Approximately 84% (31 of 37) of the L1 cDNAs were derived from human-specific L1s (Fig. 3C). Similar analyses conducted on a 236-bp RT–PCR amplified region of ORF1 revealed that ∼50% L1 cDNAs derived from iAND-4 (12 of 28) or MSUH001 iPSCs (12 of 27) were derived from human-specific L1s (Fig. 3D). Notably, the two different primer sets used in these studies had various specificities for the distinct L1 subtypes; the primers used to analyze the iPSC lines iAND-4 and MSUH001 could amplify L1Hs and L1P1–L1P5 subfamilies, while the primers used to analyze the iPS-F iPSCs were specific for L1Hs and L1P1. The difference in primers could explain the difference in the proportions of L1Hs cDNAs detected in the different iPSCs. Nevertheless, these data clearly reveal that L1Hs elements are expressed in iPSCs and may have the potential to retrotranspose in these cells.
Engineered human L1 retrotranspose more efficiently in iPSCs than HDFs
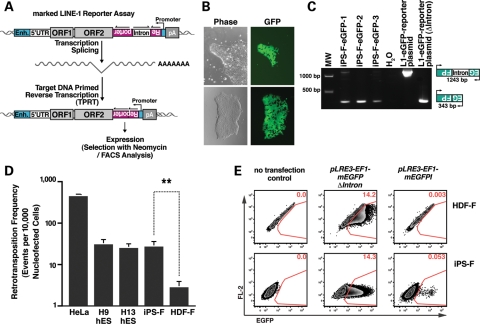
Since human-specific L1s are expressed in iPSCs, we next assessed the mobility of engineered human L1s in hESCs, iPSCs and parental HDFs. To measure L1 retrotransposition efficiency, we tagged the 3′UTR of hot RC-L1s with a retrotransposition indicator cassette that contains an antisense copy of a reporter gene interrupted by an intron in the same transcriptional orientation as the L1 (13,38,52,53). This configuration results in expression of the reporter only when the tagged L1 completes a successful cycle of retrotransposition (Fig. 4A).
Figure 4.
The retrotransposition efficiency of an engineered human L1 is higher in pluripotent stem cells than in adult HDFs. (A) An overview of the L1 retrotransposition assay. An RC-L1 is tagged with a retrotransposition indicator cassette (mneoI or mEGFPI). The indicator cassette contains an antisense copy of a selectable (NEO) or detectable (EGFP) reporter gene that is disrupted by an intron in the same transcriptional orientation as the L1 (13,52). The reporter gene contains a heterologous promoter (SV40 for the mneoI and UBC for the mEGFPI) and a poly (A) signal. An EF1a or UBC promoter (Enh, enhancer) drives L1 expression. The reporter gene can only be activated when L1 RNA is reverse transcribed, integrates into genomic DNA and is expressed from its own promoter (13). (B) An undifferentiated iPS-F colony harboring a L1 retrotransposition event from the pLRE3-EF1-mEGFPI reporter. (C) L1 retrotransposition events are stably integrated into the genome. Genomic DNA isolated from EGFP-positive colonies after nucleofection served as a PCR template. The 343-bp PCR product indicates the spliced tagged L1 (insertion); the 1243-bp product contains the intron (vector). (D) L1 retrotransposition efficiencies in different cell lines. Cells were transfected with pLRE3-EF1-mEGFPI reporter or the pLRE3-EF1-mEGFP(▵intron) plasmid as a control. Cells nucleofected with pLRE3-EF1-mEGFPI were harvested 4 days after nucleofection; cells nucleofected with pLRE3-EF1-mEGFP(▵intron) were harvested 2 days after nucleofection at the peak of EGFP expression. The frequency of new integrations (EGFP-positive cells) was analyzed by flow cytometry. For hESCs or iPSCs, ∼1–2 × 106 cells/sample were analyzed; for HDF cells, ∼0.25–0.5 × 106 cells/sample were assayed. The retrotransposition frequency was calculated as the number of EGFP-positive cells after nucleofection with the pLRE3-EF1-mEGFPI construct divided by the number of EGFP-positive cells after nucleofection with the pLRE3-EF1-mEGFP(▵intron) control plasmid. The experiments were repeated at least three times. Values are the mean ± SEM. **P≤ 0.01. (E) Flow cytometry plots of iPS-F iPSCs and HDF-F cells either untransfected or transfected with pLRE3-EF1-mEGFPI reporter (harvested 4 days post-transfection) or the pLRE3-EF1-mEGFP(▵intron) control plasmid (harvested 2 days post-transfection at the peak of EGFP expression). For iPSCs, ∼1 × 106 cells/sample were analyzed; for HDFs, ∼0.5 × 106 cells/sample were assayed. Cells were plotted against an empty channel (FL-2) to eliminate background autofluorescent cells and to increase sensitivity.
We first used a modified L1 retrotransposition assay utilizing an enhanced green fluorescent protein (EGFP) retrotransposition indicator cassette (mEGFPI) (52). Briefly, cells were nucleofected with an L1 expression construct (pLRE3-EF1-mEGFPI), and the L1 retrotransposition efficiency (i.e. EGFP-positive cells) was quantified by flow cytometry 4 days post-transfection (Supplementary Material, Fig. S5A). To control for variations in transfection efficiency between different cell lines, we divided the number of EGFP-positive cells obtained after nucleofection with pLRE3-EF1-mEGFPI by the number of EGFP-positive cells obtained after nucleofection with a control construct (pLRE3-EF1-mEGFP(▵intron)). This control plasmid (pLRE3-EF1-mEGFP(▵intron)) lacks the intron in the mEGFPI gene, and thus, the expression of the EGFP protein occurs in all successfully nucleofected cells (Supplementary Material, Fig. S5B). To further increase assay sensitivity, autofluorescent cells were eliminated by plotting against an empty channel as described previously (54).
Consistent with previous findings, control experiments conducted in H9 and H13 hESCs revealed ∼30 ± 5 EGFP-positive cells per 10 000 nucleofected cells (Fig. 4D). The retrotransposition efficiency in H9 and H13 cells was highly reproducible and was ∼10% of the level of retrotransposition detected in HeLa cells (Fig. 4D). PCR analyses on genomic DNA derived from clonal H9 hESC lines revealed precise splicing of the intron from the mEGFPI indicator cassette gene, which is indicative of retrotransposition (Supplementary Material, Fig. S6C) (31,36,55). Further, treatment with lamivudine (3TC, 100 µm), a reverse transcriptase inhibitor that potently suppresses L1 retrotransposition (56), reduced retrotransposition by ∼95% (Supplementary Material, Fig. S5A and D), but did not affect the expression of EGFP in cells transfected with control plasmid pLRE3-EF1-mEGFP(▵intron) (Supplementary Material, Fig. S5D).
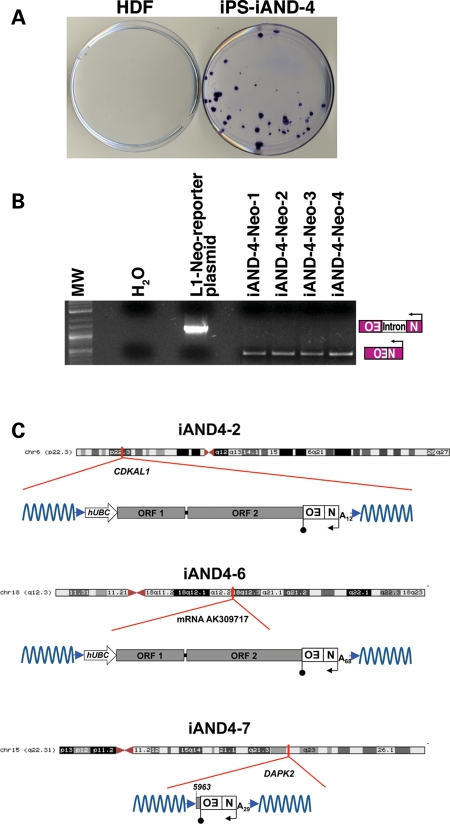
We next used the same strategy to examine the level of engineered L1 retrotransposition in iPSCs and parental fibroblasts. The level of L1 retrotransposition in the iPS-F cells was similar to that detected in H9 and H13 hESCs. Strikingly, L1 retrotransposition was ∼10-fold higher in iPSCs than in the parental HDFs (Fig. 4D and E). Additional controls revealed robust EGFP expression in iPS-F clones (Fig. 4B) and precise splicing of the engineered intron as demonstrated by PCR using genomic DNA isolated from iPS-F clones harboring a retrotransposition event (Fig. 4C). In addition, experiments conducted with an L1 containing an mneoI retrotransposition indicator cassette (pKUB102/L1.3-sv+, Supplementary Material, Fig. S6A) (13,57) indicated that iAND-4 cells support ∼20–30-fold more L1 retrotransposition than parental fibroblasts (an average of 32 foci in iAND-4 versus 0–2 foci in the parental HDFs; Fig. 5A). Notably, we did not detect any L1 retrotransposition events from an RT-defective L1 allele (data not shown). In addition, PCR using genomic DNA derived from four independent G418-resistant iAND-4 clonal lines revealed precise splicing of the intron from the mneoI cassette (Fig. 5B). Together, these findings demonstrate that reprogramming fibroblasts to iPSCs is associated with an approximate one log10 increase in the retrotransposition efficiency of an engineered human L1 element, at least when the L1 mRNA is produced by a strong exogenous promoter. Further, comparable levels of retrotransposition were detected in hESCs and iPSCs, indicating that reprogramming does not lead to elevated levels of retroelement mobility beyond that found in hESCs. In addition, these data confirm that primary human fibroblasts only accommodate very low levels of retrotransposition (54,58).
Figure 5.
Engineered L1 retrotransposition events in pluripotent stem cells exhibit hallmarks of retrotransposition. (A) An mneoI-based L1 retrotransposition assay shows higher levels of retrotransposition in the iPSC line iAND-4 than in parental HDFs. Cell were transfected with the pKUB102/L1.3-sv+ engineered L1 plasmid and treated with G418 for 2 weeks. G418-resistant foci were stained with crystal violet for visualization. In parental HDFs, typically 0–2 foci/experiment were observed. (B) G418-resistant clones harbor a stable integrated event derived from pKUB102/L1.3-sv+. Genomic DNA isolated from four G418-resistant clones served as a PCR template. The 468-bp PCR product represents the spliced product (insertion); the 1361-bp product contains the intron (vector). (C) The cartoons show schematic representations of the L1 retrotransposition events in iAND-4 iPSCs. Names of the insertions are indicated at the tops of the schematics. Red lines indicate the genomic site of insertion and whether the insertion occurred within a gene. Blue arrowheads indicate target site duplications. The approximate length of the L1 poly(A) tail and the position of the L1 where 5′ truncation occurred also are indicated and are based on the L1.3 reference sequence (L19088.1) (53). Additional details can be found in Supplementary Material, Fig. S6.
Retrotransposed L1s in iPSCs exhibit hallmarks of retrotransposition
To determine whether L1 retrotransposition events occurring in iPSCs exhibited bona fide structural hallmarks, we used inverse PCR to characterize four retrotransposition events from iAND-4 iPSCs. Three insertions (iAND4-2, iAND4-6 and iAND4-7) were completely characterized by DNA sequencing. A fourth insertion (iAND4-E) could not be completely analyzed because it was located in α-satellite DNA and was truncated at the restriction enzyme site used in the inverse PCR (data not shown). The iAND4-2 and iAND4-6 insertions were full length and contained part of the exogenous UBC promoter used to drive L1 expression (Fig. 5C), while the iAND4-7 insertion was 5′-truncated. Each insertion exhibited typical L1 structural hallmarks. They ended in poly(A) tails, were flanked by target-site duplications that ranged in size from 8 to 14 bp and appeared to integrate into a sequence resembling a consensus L1 endonuclease cleavage site (12,19,59) (Fig. 5C and Supplementary Material, Fig. S6B–D). Notably, the iAND4-2 and iAND4-7 insertions occurred within the introns of known genes (CDKAL1 and DAPK2), which is consistent with previous studies (31,32,36,55).
DISCUSSION
While iPSCs might one day be of great value in the treatment of currently incurable human diseases (2,3), several challenges posed by these cells must be overcome. Chief among these is ensuring that the genomic integrity of iPSCs is not compromised during their generation or ex vivo culture.
In this study, we demonstrate for the first time that cytoplasmic full-length L1 mRNAs are expressed in hESCs and iPSCs, but are not detectable in HDFs and HeLa cells under our experimental conditions. In addition, we detected endogenous L1 ORF1 protein in ribonucleoprotein fractions from iPSCs and hESCs. Both cytoplasmic full-length L1 RNA and RNPs containing the L1 ORF proteins are required for L1 retrotransposition (60–62).
It is thought that L1 transcription is suppressed by the CpG methylation of its 5′UTR in most somatic cells (49). We have detected decreased DNA methylation in CpG islands located within the L1 5′UTR promoter region in hESCs and iPSCs. Notably, sequencing revealed that L1 cDNAs in iPSCs are derived from human-specific L1s. These findings suggest that L1 expression is a feature of pluripotent cells and indicate that hot L1s might actively retrotranspose in iPSCs. Genomic quantitative PCR (36), second-generation sequencing (21,22,63) and fosmid-based, paired-end DNA sequencing (11) approaches have recently been used to detect and/or quantify new L1 integration sites in various cell types. Future adaptations of these approaches could be used to examine whether the retrotransposition of endogenous L1s is increased in iPSCs compared with HDFs.
We also demonstrated that an engineered human L1 can retrotranspose with ∼10-fold higher efficiency in iPSCs than in parental HDFs, and the characterization of three retrotransposition events in iPSCs revealed canonical L1 structural hallmarks. Thus, reprogramming of somatic fibroblasts not only reinstates L1 transcription, but also appears to create an intracellular milieu that supports at least a 10-fold higher level of engineered L1 retrotransposition than observed in parental fibroblasts. Interestingly, a recent study suggested engineered L1 retrotransposition events could be epigenetically silenced in certain human embryonic carcinoma cells and that this silencing was attenuated in differentiating cells (55). Thus, our studies may actually underestimate the efficiency of engineered human L1 retrotransposons in iPSCs. In addition, high levels of L1 expression and higher engineered L1 retrotransposition efficiencies in pluripotent cells may explain why some L1 insertions in humans seem to be of early embryonic origin (31,38–40).
Two recent reports demonstrated that somatic coding mutations (64) and copy number variations (5) during iPSC reprogramming could lead to genetic mosaicism. Genomic instability can also occur during the culturing of hESCs (6). Thus, it is tempting to speculate that L1-mediated processes may occasionally lead to the generation of genomic variability in cultured pluripotent cells by a variety of mechanisms (19,65–72). Clearly, a better understanding of how L1-mediated retrotransposition events affect genomic stability in hESCs and iPSCs is warranted before these cells, or more likely their progeny, are utilized as cellular therapies.
MATERIAL AND METHODS
Cell lines and culture conditions
iPSC lines were derived as described (3,41). HDFs, H9 and H13B (hereafter referred as H13) hESCs and iPSCs were cultured and expanded under standard conditions (Supplementary Material and Methods).
Plasmids
Cloning strategies are available upon request. L1 retrotransposition assays using the neomycin indicator cassette (mneoI) were performed with the pKUB102/L1.3-sv+ plasmid, which contains an L1.3 element (53) without its 5′UTR, the mneoI indicator cassette (57) and the SV40 late polyadenylation signal 3′ of the engineered L1. The engineered L1 was expressed from a modified version of pBSKS-II (Stratagene) that contains a human UBC promoter upstream of the engineered L1 (36). pLRE3-EF1-mEGFPI contains a full-length LRE3 element (38) under the control of a heterologous EF-1α promoter and the internal 5′UTR promoter, and the EGFP retrotransposition indicator cassette is under the control of an ubiquitin promoter and the SV40 late polyadenylation signal; the construct was cloned into pBSKS-II (Stratagene). The positive control pLRE3-EF1-mEGFP(▵intron) is identical to pLRE3-EF1-mEGFPI but lacks the intron in the mEGFPI indicator cassette.
Nucleofection, retrotransposition assay and flow cytometry analysis
L1 retrotransposition assays using the L1 mneoI indicator cassette were performed as follows. Cells (4 × 106) were nucleofected with an Amaxa nucleofector, using 4 µg of plasmid DNA, hESC solution 2 (Amaxa) and the A-23 program. The transfected cells were cultured in iPSC medium supplemented with 10 µm Rho-associated coiled kinase (ROCK) inhibitor (73). Cells were selected with 50 µg/ml G418 for 7 days and 100 µg/ml G418 for an additional 7 days, fixed in 0.2% glutaraldehyde and 2% formaldehyde and stained with crystal violet to visualize foci. For mEGFPI-based retrotransposition assays, cells were nucleofected with an Amaxa nucleofector, using the V-Kit solution (Amaxa) and the A-23 program. Transfected cells were cultured in conditioned hESC medium supplemented with 10 µm ROCK inhibitor (73). Two [pLRE3-EF1-mEGFP(▵intron)] or 4 days (pLRE3-EF1-mEGFPI) after nucleofection, cells were analyzed by flow cytometry (hES or iPS, ∼1–2 × 106 cells/sample; HDF, ∼0.25–0.5 × 106 cells/sample). Data were analyzed with FlowJo software (Tree Star).
L1 promoter methylation studies
Bisulfite analysis was performed as described (36). Briefly, genomic DNA was extracted with a DNeasy kit (Qiagen). Bisulfite conversion was performed with the Epitect kit (Qiagen), according to the manufacturer's instructions. For whole-genomic L1 CpG analysis, PCR was performed to amplify a 363-bp region in the 5′UTR harboring 20 CpG dinucleotides (2 min at 95°C, 35 cycles of 30 s at 94°C followed by 30 s at 54°C and 60 s at 72°C, and a final extension of 5 min at 72°C; primers are listed in Supplementary Material, Table S2). To analyze hot L1 promoter regions, we first amplified a ∼900–1200-bp region including the first 700 bp of L1 and a 200–500-bp region, upstream (3 min at 95°C, 35 cycles of 30 s at 94°C followed by 30 s at 50°C and 2 min at 72°C, and a final extension of 7 min at 72°C; primers are listed in Supplementary Material, Table S2). Two microliters of the PCR product were used for the second round of PCR, using the same primer and condition as for the whole-genomic bisulfite analysis. The PCR products were subcloned and analyzed with online software (QUantification tool for Methylation Analysis, QUMA; quma.cdb.riken.jp) (74). Only sequences with a CpG conversion rate >95% were accepted for analysis; the final analysis included the clones with the highest sequence similarity with a hot L1 element (L1.3, accession number L19088.1) (53).
Real-time RT–PCR
Total RNA was extracted with Trizol (Invitrogen), treated with a Turbo DNA-free kit (Ambion), enriched with an RNeasy kit (Qiagen) and reverse transcribed with the Superscript III first-strand synthesis system, using sense-strand L1 primers. PCR and real-time RT–PCR were performed according to standard procedures (Supplementary Experimental Procedures). Sequences and specifications of primers are shown in Supplementary Material, Table S2.
Northern blot analysis
Northern blot analysis was performed by standard procedures (Supplementary Experimental Procedures). Briefly, 2.5 µg of cytoplasmic poly(A)-selected RNA were fractionated on a 1% agarose-formaldehyde gel. The RNA was transferred onto a Hybond-N+-Nylon membrane (Amersham), hybridized with alpha 32P-UTP-labeled RNA probes overnight at 68°C and subjected to two 10-min low-stringency washes at 20°C and two 20-min high-stringency washes at 68°C. β-Actin mRNA served as the loading control.
Isolation of genomic DNA for integration analysis
Fully spliced L1 reporter constructs in the genomic DNA were detected as described (13,52).
Inverse PCR analysis
Inverse PCR on clonal iPSC lines was conducted as described (17,31).
Analysis of expressed L1 mRNAs
Briefly, total RNA was used in RT–PCRs using ORF1p primers as described (31,37,55). Amplified products were cloned and at least 25 independent clones sequenced. Sequences were analyzed with RepeatMasker (http://repeatmasker.org.).
Statistical analysis
Statistical analysis was performed with the unpaired two-tailed t-test.
Ethical approvals
This study was approved by the Human Gamete, Embryo and Stem Cell Research Committee UCSF (GESCR numbers 7396-29609 and H51338-32135-03) and the Andalusian and Spanish Embryo and Cellular Reprogramming Ethical Institutional Review Board.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by funding from the California Institute for Regenerative Medicine (CIRM) to W.C.G. (RS1-00210-1 and TRI-01227) and a CIRM scholarship (TG2/01160) and by a fellowship from the German Academy of Sciences Leopoldina (BMBF-LPD 9901/8-144) to S.W. J.L.G.-P.'s laboratory is supported by ISCIII-CSJA-FEDER (EMER07/056), a Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG), CICE (P09-CTS-4980), Proyectos en Salud PI-002 from Junta de Andalucia (Spain) and the Spanish Ministry of Health (FIS-FEDER PI08171). J.V.M. is supported by NIH (GM082970 and GM060518) and is an Investigator of the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs J.L. Goodier and G. Cristofari for polyclonal L1 ORF1 antibodies, Dr H.H. Kazazian for the LRE3 construct, Drs Jose Cibelli and Steve Suhr for the lentiviral construct ThOKSIM and the Gladstone Stem Cell Core for technical assistance. We also thank Purificacion Catalina, Ivan Gutierrez-Aranda, Rene Rodriguez, Paola Leone and Pablo Menendez (Andalusian Stem Cell Bank) for assistance with experiments. We thank S. Ordway, G. Howard and S. Richardson for editorial assistance, and S. Cammack, R. Givens and J. Carroll for assistance in preparation of the manuscript and graphics.
REFERENCES
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Wernig M., Lengner C.J., Hanna J., Lodato M.A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Narva E., Ng S., Sourour M., Hamalainen R., Olsson C., et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 6.Narva E., Autio R., Rahkonen N., Kong L., Harrison N., Kitsberg D., Borghese L., Itskovitz-Eldor J., Rasool O., Dvorak P., et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat. Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Babushok D.V., Kazazian H.H., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 9.Goodier J.L., Kazazian H.H., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 13.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 14.Skowronski J., Fanning T.G., Singer M.F. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 16.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 17.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzdin A., Gogvadze E., Kovalskaya E., Volchkov P., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Perez J.L., Doucet A.J., Bucheton A., Moran J.V., Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C.R., Schneider A.M., Lu Y., Niranjan T., Shen P., Robinson M.A., Steranka J.P., Valle D., Civin C.I., Wang T., et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witherspoon D.J., Xing J., Zhang Y., Watkins W.S., Batzer M.A., Jorde L.B. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hormozdiari F., Alkan C., Ventura M., Hajirasouliha I., Malig M., Hach F., Yorukoglu D., Dao P., Bakhshi M., Sahinalp S.C., et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazazian H.H., Jr, Wong C., Youssoufian H., Scott A.F., Phillips D.G., Antonarakis S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 26.Belancio V.P., Hedges D.J., Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 27.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourc'his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 29.Trelogan S.A., Martin S.L. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin S.L., Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell Biol. 1993;13:5383–5392. doi: 10.1128/mcb.13.9.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Perez J.L., Marchetto M.C., Muotri A.R., Coufal N.G., Gage F.H., O'Shea K.S., Moran J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 32.Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 33.Ostertag E.M., DeBerardinis R.J., Goodier J.L., Zhang Y., Yang N., Gerton G.L., Kazazian H.H., Jr A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 34.Georgiou I., Noutsopoulos D., Dimitriadou E., Markopoulos G., Apergi A., Lazaros L., Vaxevanoglou T., Pantos K., Syrrou M., Tzavaras T. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum. Mol. Genet. 2009;18:1221–1228. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 35.Prak E.T., Dodson A.W., Farkash E.A., Kazazian H.H., Jr. Tracking an embryonic L1 retrotransposition event. Proc. Natl Acad. Sci. USA. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O'Shea K.S., Moran J.V., Gage F.H. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macia A., Munoz-Lopez M., Cortes J.L., Hastings R.K., Morell S., Lucena-Aguilar G., Marchal J.A., Badge R.M., Garcia-Perez J.L. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol. Cell Biol. 2011;31:300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouha B., Meischl C., Ostertag E., de Boer M., Zhang Y., Neijens H., Roos D., Kazazian H.H., Jr Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Hurk J.A., Meij I.C., Seleme M.C., Kano H., Nikopoulos K., Hoefsloot L.H., Sistermans E.A., de Wijs I.J., Mukhopadhyay A., Plomp A.S., et al. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 40.Kano H., Godoy I., Courtney C., Vetter M.R., Gerton G.L., Ostertag E.M., Kazazian H.H., Jr L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross P.J., Suhr S., Rodriguez R.M., Chang E.A., Wang K., Siripattarapravat K., Ko T., Cibelli J.B. Human induced pluripotent stem cells produced under xeno-free conditions. Stem Cells Dev. 2009;19:1221–1229. doi: 10.1089/scd.2009.0459. [DOI] [PubMed] [Google Scholar]
- 42.Belancio V.P., Roy-Engel A.M., Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perepelitsa-Belancio V., Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 44.Belancio V.P., Roy-Engel A.M., Pochampally R.R., Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 46.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 47.Goodier J.L., Ostertag E.M., Engleka K.A., Seleme M.C., Kazazian H.H., Jr A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 48.Leibold D.M., Swergold G.D., Singer M.F., Thayer R.E., Dombroski B.A., Fanning T.G. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl Acad. Sci. USA. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 50.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smit A.F.A., Hubley R., Green P. RepeatMasker. http://repeatmasker.org .
- 52.Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 54.Shi X., Seluanov A., Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Perez J.L., Morell M., Scheys J.O., Kulpa D.A., Morell S., Carter C.C., Hammer G.D., Collins K.L., O'Shea K.S., Menendez P., et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones R.B., Garrison K.E., Wong J.C., Duan E.H., Nixon D.F., Ostrowski M.A. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One. 2008;3:e1547. doi: 10.1371/journal.pone.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freeman J.D., Goodchild N.L., Mager D.L. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques. 1994;17:48–49. , 52. [PubMed] [Google Scholar]
- 58.Kubo S., Seleme M.C., Soifer H.S., Perez J.L., Moran J.V., Kazazian H.H., Jr, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulpa D.A., Moran J.V. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 61.Kulpa D.A., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 62.Doucet A.J., Hulme A.E., Sahinovic E., Kulpa D.A., Moldovan J.B., Kopera H.C., Athanikar J.N., Hasnaoui M., Bucheton A., Moran J.V., et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belancio V.P., Deininger P.L., Roy-Engel A.M. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E., et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 66.Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roman-Gomez J., Jimenez-Velasco A., Agirre X., Cervantes F., Sanchez J., Garate L., Barrios M., Castillejo J.A., Navarro G., Colomer D., et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 68.Daskalos A., Nikolaidis G., Xinarianos G., Savvari P., Cassidy A., Zakopoulou R., Kotsinas A., Gorgoulis V., Field J.K., Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int. J. Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 69.Kazazian H.H., Jr., Goodier J.L. LINE drive. retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 70.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 71.Belgnaoui S.M., Gosden R.G., Semmes O.J., Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wallace N.A., Belancio V.P., Deininger P.L. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 74.Kumaki Y., Oda M., Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.