Abstract
LINE-1 repeats account for ∼17% of the human genome. Little is known about their individual methylation patterns, because their repetitive, almost identical sequences make them difficult to be individually targeted. Here, we used bisulfite conversion to study methylation at individual LINE-1 repeats. The loci studied included 39 X-linked loci and 5 autosomal loci. On the X chromosome in women, we found statistically significant less methylation at almost all L1Hs compared with men. Methylation at L1P and L1M did not correlate with the inactivation status of the host DNA, while the majority of L1Hs that were possible to be studied lie in inactivated regions. To investigate whether the male–female differences at L1Hs on the X are linked to the inactivation process itself rather than to a mere influence of gender, we analyzed six of the L1Hs loci on the X chromosome in Turners and Klinefelters which have female and male phenotype, respectively, but with reversed number of X chromosomes. We could confirm that all samples with two X chromosomes are hypomethylated at the L1Hs loci. Therefore, the inactive X is hypomethylated at L1Hs; the latter could play an exclusive role in the X chromosome inactivation process. At autosomal L1Hs, methylation levels showed a correlation tendency between methylation level and genome size, with higher methylation observed at most loci in individuals with one X chromosome and the lowest in XXY individuals. In summary, loci-specific LINE-1 methylation levels show considerable plasticity and depend on genomic position and constitution.
INTRODUCTION
Repetitive DNA sequences constitute about half of the non-coding DNA or 45% of the human genome (1). Long and short interspersed elements (LINEs and SINEs) constitute the majority of these repeats, accounting together for ∼33% of the genome (1). LINE-1 and Alu repeats are the major LINEs and SINEs; they make up ∼17 and 11% of the total DNA, respectively. About 0.5 and 1.1 million copies of LINE-1 and Alu exist in the haploid human genome, playing an important role in the overall architecture and organization of the genome. LINE-1 sequences are 2-fold enriched on the human X chromosome compared with autosomes (2). Moreover, LINE-1 sequences have been proposed to play a role in the X chromosome inactivation process by either spreading or maintaining the inactivation state (3). The complete human LINE-1 sequence consists of ∼6 kb that harbor two genes, open reading frame 1 and 2 (ORF1 and ORF2), coding for 40 and 150 kDa proteins that act as a nucleic acid chaperone with endonuclease and reverse transcriptase activities (4–6). These protein properties enable LINE-1 sequences to mobilize themselves together with other sequences (such as Alu) within the human genome, thus making it the only known active mobilizing machinery in the human genome (7).
It is believed that ∼8000–10 000 full-length LINE-1 (>6 kb) copies exist in the human genome. Most LINE-1 repeats are unable to induce retrotransposition events, either because they have been mutated in critical sequences, are truncated, or because they carry rearranged sequences. It is estimated that only ∼80–100 copies of LINE-1 are still active today in the diploid human genome (8,9). LINE-1 sequences contribute to the variability of the human genome and to inter-individual differences by variations in their insertion sites (presence or absence at a given site) and by their expression status which is proposed to influence gene expressions of neighboring genes and of host genes as has been recently suggested (10). In providing evidence for the polymorphic insertions, recently, four independent studies, using genome-wide sequencing approaches, provided data for the polymorphic occurrence of LINE-1 repeats in the human genome; their estimate of the occurrence of the polymorphic insertions ranged between 5 and 285 events per genome (11–14). On the other hand, the expression potential of a given LINE-1 element is determined in itself by two factors: the sequence and polymorphisms of the repeat (i.e. the degree of homology to the full-length expressed LINE-1) and the methylation epigenetic status of the promoter region of the repeat (10,15,16). In other words, it may not be sufficient to know the sequence and the position of a given LINE-1 repeat to predict its expression. Knowledge about its epigenetic/methylation status would also be necessary.
The majority of the LINE-1 promoter regions, whether present in full-length or truncated LINE-1, are believed to be heavily methylated at their cytosines in a CpG dinucleotide context. This phenomenon is believed to be a cellular defense system to repress the expression and, therefore, to silence the retrotransposition activities of LINE-1 (15,16). However, variability of global LINE-1 methylation is frequently observed in different tumors and by specific environmental or life style conditions (17–19). In addition, we found that gender is an important factor influencing global LINE-1 methylation, while neither natural hormone cycles nor age has a large influence on LINE-1 DNA methylation in leukocytes (20,21).
Although plenty of data exist on average genome-wide LINE-1 methylation levels in healthy and diseased human tissue, only a small number of studies has addressed methylation levels at individual LINE-1 loci in healthy individuals to date. This is mainly due to their repetitive and abundant nature in the genome, which makes measuring of methylation levels and patterns of individual LINE-1 repeats a challenge.
To date, Phokaew et al. (22) have provided the only detailed study directly analyzing methylation at locus-specific LINE-1 repeats in a quantitative manner. They studied 17 full-length autosomal loci in four different cell types. The authors found cell type as well as locus-specific variations. Only a limited number of studies on genome-wide methylation analysis using bisulfite deep sequencing could map a number of locus-specific LINE-1 repeats (based on existing polymorphisms in specific individual repeats) (23,24). However, only specific cell types were analyzed, such as ES cell lines, primordial germ cells or fibroblast cell lines, and their methylation results are far from being quantitative at specific loci. In addition, none of these studies compared X-linked with autosomal LINE-1 loci.
Therefore, a study on both autosomal and X-linked locus-specific LINE-1 loci using a highly quantitative method from healthy blood tissue of a large number of individuals is still lacking. To this end, we used bisulfite analysis combined with semi-nested polymerase chain reaction (PCR) followed by highly quantitative methods to measure methylation at individual LINE-1 sequences. We addressed the influence of inter-individual variations and the effect of genomic position and chromosomal constitution on the level of methylation in a large number of individuals, including Turner syndrome patients (45,X), healthy male (46,XY), healthy female (46,XX) and Klinefelter syndrome patients (47,XXY). Our analysis revealed three novel observations. First, a considerable variability in the level of methylation at different loci and between different individuals at some loci was observed. Secondly, L1Hs subtype in X chromosomal regions that are subject to inactivation are significantly less methylated on the inactive allele, while L1P are similarly methylated at both active and non-active X and in both inactivated and non-inactivated regions. Thirdly, a tendency towards higher methylation at autosomal LINE-1s in smaller genomes (Turner syndrome versus Klinefelter syndrome) was observed, suggesting limited methylation capability within cells that could be depleted faster in bigger genomes and/or in rapidly dividing cells such as the studied blood-derived leukocytes.
RESULTS
Individual LINE-1s are heavily methylated, but show loci-specific variation as well as inter-individual variations but only at some loci
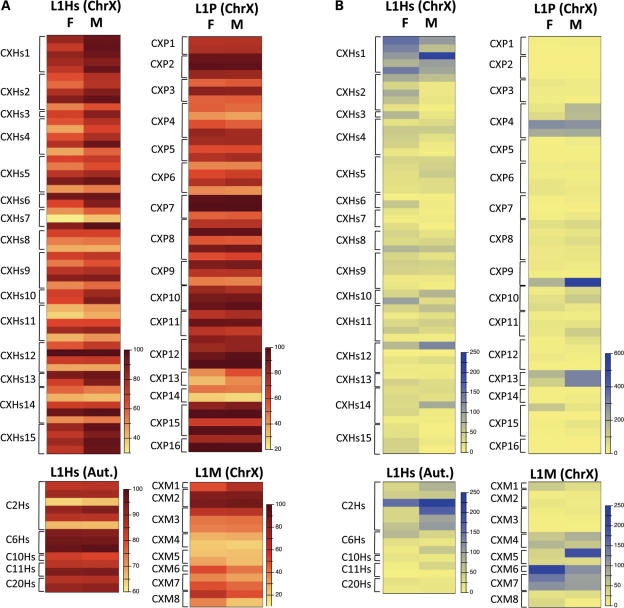
We studied 44 specific LINE-1 loci with 39 (15 L1Hs, 8 L1M, 16 L1P) on the X chromosomes and 5 (L1Hs) on different autosomes. The first noticeable characteristic of all studied LINE-1 CpGs was high methylation regardless of whether autosomal or X-linked (Fig. 1A). However, considerable inter-loci and inter-individual variability were seen, specifically at some and not all loci (Fig. 1B and Supplementary Material, Table S1).
Figure 1.
Heat map presentations of (A) the average of methylation and (B) the variance of methylation at the studied LINE-1 loci. Linear scale is used to represent the data.
For example, the average methylation level at the autosomal loci in the male and female groups showed a range of 63% (C2Hs-CpG3) to 99% (C6Hs-CpG3) (Supplementary Material, Table S1). At the X-linked studied regions, L1Hs showed a wider range of methylation between different loci, with a range of as low as 30% (CXHs7-CpG2) to 100% (CXHs12-CpG2); L1P loci ranged between 26% (CXP14-CpG2) and 100% (several loci); and L1M loci were considerably less methylated with a range of 21% (CXM4-CpG2) to 90% (CXM2-CpG2).
Most importantly, variability in methylation was not only observed between different CpG sites but also between different individuals at a given CpG site. This inter-individual difference could be reflected by the variance parameters, which showed that some CpGs could harbor wider inter-individual differences than other loci which are more stable in their methylation among the population (Fig. 1B and Supplementary Material, Table S1). The reason why only few loci showed large variability so that we defined them as hyper-variable loci with a variance of above 100, while others are more stable, is not known. Notably, local C to T polymorphisms are absent from these loci (at least from three regions that we sequenced in all individuals: C2Hs, C10Hs and C11Hs). Hence, such single nucleotide polymorphisms (SNPs) presence/absence does neither cause nor contribute to these inter-individual differences at the hyper-variable loci. However, polymorphisms in the surrounding sequences that would affect the methylation levels at these loci, as previously observed at non-repetitive elements, cannot be excluded (25,26). Thus, we distinguish C2Hs on autosomes as the hyper-variable region and CXHs1, CXP4, CXP9, CXP13, CXM6 and CXM7 on the X chromosome (Fig. 1B and Supplementary Material, Table S1). In summary, variability of methylation levels was observed in two dimensions, namely between different loci and between individuals.
LINE-1 promoters at Xi are less methylated at L1Hs, equally methylated at both Xi and Xa at L1P and variable at L1M
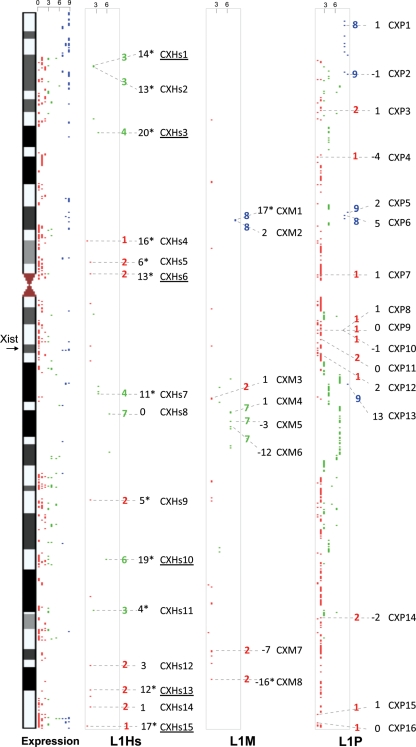
We studied three major LINE-1 subtypes on the X chromosome in two groups of healthy males and females: namely L1Hs, L1M and L1P. We restricted our analysis to full-length LINE-1 (>6 kb). Thus, we could find 25 L1Hs loci on the X chromosome (UCSC RepeatMasker table). We designed specific assays for all of the 25 loci, However, 10 of them were either impossible to analyze specifically because of the absence of loci-specific flanking sequences in close range of 500 bases upstream of the start of the repeat sequence or because the quality and sequence specificity did not pass our quality criteria. The remaining 15 loci were distributed as follows: 8 in directly strongly inactivated gene/loci (escaping score of 1 or 2) and 7 in moderately inactivated loci (inactivation score 3–7). No full-length L1Hs were present in strong non-inactivated (escaping) genes/loci (Fig. 2 and Supplementary Material, Table S2). At 14 of these loci, male samples were methylated higher than female samples, of which 12 are statistically significant. At only one loci, CXHs8, which has the highest escaping score of 7, equal methylations between male and female were observed. As women have one inactivated X, we supposed that this reduction in methylation in female samples is derived from this inactive X. Thus, we assumed that these loci are hypomethyled at the inactive X.
Figure 2.
Relative positions of the full-length (>6 kb) L1Hs, L1P and L1M on the X chromosome. The studied regions in this study are labeled with dashed line. Expression data from Carrel and Willard (57) are presented in the first column, where scale of expression is 0–9 (number of cell lines with inactive X that were expressing a given gene); we empirically considered score of 0–2 to be inactivated (red), 3–6 to be middle inactivated (green) and 8–9 escaping inactivation (blue). In the second, third and forth columns are the L1Hs, L1M and L1P full-length copies (>6 kb) represented by dots. An inactivation score for every locus was empirically calculated according to the position of the LINE-1 sequence relative to the studied transcripts represented in column 1, thus a 1 score is given for a locus in a gene that is inactivated, 2: between two inactivated genes, 3: between one inactivated gene and one middle inactivated gene, 4: between one inactivated gene and one non-inactivated gene, 5: between two middle inactivated genes, 6: in a middle inactivated gene, 7: between one middle inactivated gene and non-inactivated gene, 8: between two non-inactivated gene, 9: in a gene that is non-inactivated. The inactivation score is given right to the given studied L1 dot, followed by the male–female average methylation difference, followed by the name of the loci. The statistically significant difference between the males and females is labeled with a star. The underlined loci represent the ones studied in all four groups of samples.
Among the second category of LINE-1, L1M, we identified 50 full-length copies and we designed assays for 14 of them and 8 of these passed our quality control. Two of the loci were in escaping gene/loci (escaping score 8) and three in inactivated gene/loci (escaping score 2), while the rest were in moderately inactivated gene/loci (escaping score 7). Four of the loci were highly methylated in males, one being statistically significant, and equally, four loci were highly methylated in females and, again, only one was statistically significant. Thus, at the studied L1M loci, we found a mixed picture of higher methylation at the Xi or Xa with no clear correlation to the inactivation status of the underlying locus.
At the third category, which is the most abundant at the X chromosome with 737 full-length copies, we designed assays for 24 different regions, 16 of which passed our quality control. These loci were divided into two groups: the first group includes 11 L1P loci in inactivated gene/loci (escaping score of 1–2), while the second group includes six loci in escaping gene/loci. We found that, on average, most loci are slightly more methylated in males, while four are slightly more methylated in females. The difference between male/female methylation was significant only at CXP5. Therefore, from the studied loci, it seems that L1P sequences are not largely differentially methylated between Xa and Xi as observed at the L1Hs sequences. Therefore, the relatively strong hypomethylation on the inactivated regions may be more restricted to L1Hs promoter regions.
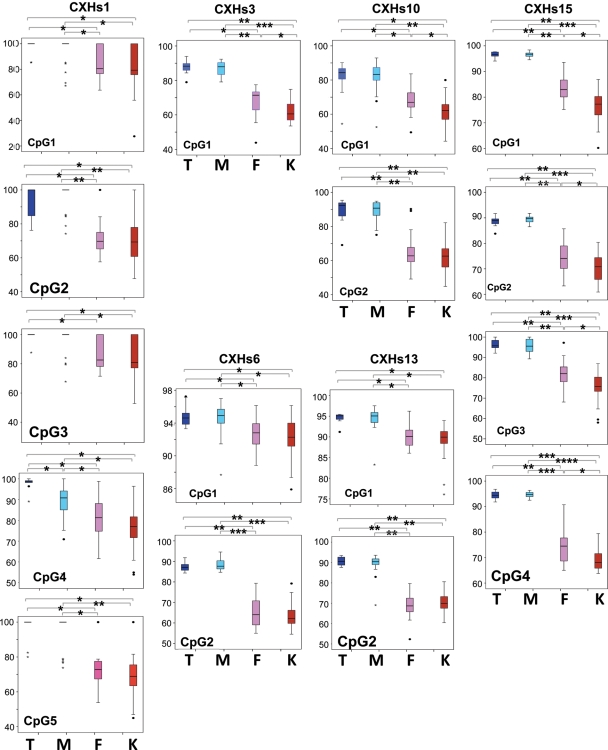
Next, we wanted to investigate whether the differences between males and females at the L1Hs loci are indeed stemming from the inactivated X or whether these are gender-specific differences induced by sex differences such as hormones. Thus, we analyzed six X-linked L1Hs (Fig. 2, underlined) that showed the highest differences between males and females in a group of Turner and Klinefelter individuals who are physiologically and phenotypically female and male, respectively. However, Turners have only one active X, while Klinefelters have two Xs with one active and one inactive like a typical female. Indeed, a pair-wise comparison of a group of samples with one X chromosome (46,XY male samples and 45,X Turner samples) and samples with two X chromosomes and one inactivated X (46,XX female samples and 47,XXY Klinefelter samples) shows a significant drop of methylation at all loci in genomes with two X chromosomes (Fig. 3 and Supplementary Material, Table S1). Therefore, we assumed that this difference is not specific for female versus male samples since Klinefelter versus Turner showed a similar significance with the Klinefelter group being hypomethylated. Moreover, the differences are specific for the X chromosome as all studied LINE-1 CpG sites at autosomes were not differentially methylated between different groups of samples (Supplementary Material, Table S1).
Figure 3.
Box plots summarizing methylation levels at six L1Hs X-linked loci in inactivated regions and middle inactivated regions. Blue, light blue, pink and red boxes represent Turner (T), male (M), female (F) and Klinefelter (K) samples, respectively. Statistical significance is represented above each box with stars and connecting lines; each star corresponds to a significant P-value of E-10.
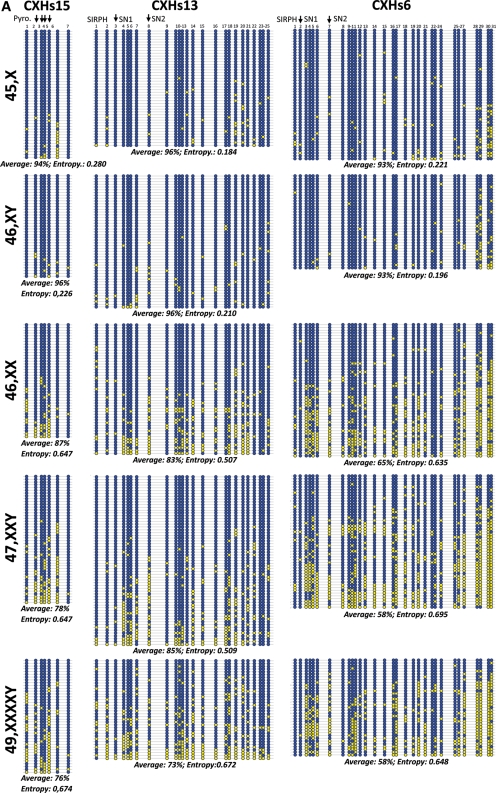
Bisulfite sequencing of individual PCR products from three regions (CXHs15, CXHs13 and CXHs6) clearly confirmed the quantitative data and provided a clear picture on the nature of the difference between the active and inactive X chromosome at such regions (Fig. 4A). Two major facts can be observed: (i) the presence of a second X chromosome (supposedly inactivated) clearly reduces average methylation at all three loci indicating that the inactive X is hypomethylated at these repeats. (ii) This hypomethylation is largely non-allelic in its nature as the methylated and non-methylated CpG sites do not cluster in nearly completely methylated and non-methylated clones as observed at differentially methylated regions associated with imprinted genes. The difference in occurrence of methylation between the two groups of samples (with one X and with two Xs) as tested by the non-parametric Wilcoxon test was significant with P-values < 0.0001 for all three regions. Additionally, the methylation entropy was calculated for all samples at every region, revealing a clear-cut between the samples with one X (entropy <0.280) and those with multiple Xs (entropy >0.507) (Fig. 4A). Thus, the effect of the presence of inactivated X on the entropy was even more pronounced than what is seen with the average methylation differences.
Figure 4.
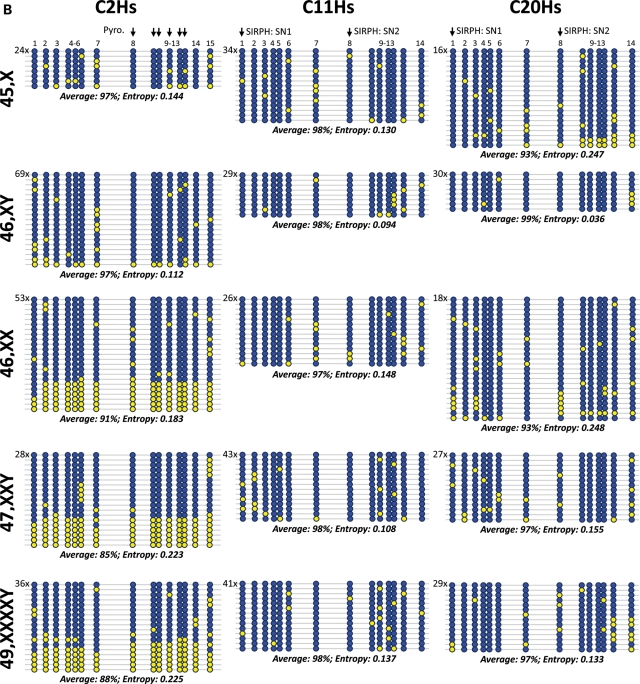
Summary of bisulfite sequencing results. (A) Three X-linked loci (CXHs15, CXHs13 and CXHs6) and (B) three autosomal loci (C2Hs, C11Hs and C20Hs) in Turner (45,X), male (46,XY), female (46,XX), Klinefelter (47,XXY) samples and one Klinefelter sample with a 49,XXXXY karyotype. Equal amounts of DNA from three individuals were pooled together prior to bisulfite treatment, PCR products were cloned into a TA cloning vector and large numbers of individual clones were sequenced. Individual CpGs are represented as blue filled and yellow filled circles representing methylated and unmethylated CpGs, respectively. Average methylation of all CpGs in all sequenced clones in a given region is provided under each group of clones. Vertical arrows indicate the positions of the CpGs studied by SIRPH or pyrosequencing reaction.
We further investigated selected loci on X-linked inactivated regions (CXHs15, CXHs3 and CXHs10) in lymphoblastoid and fibroblastoid cell lines with different X chromosome counts (Supplementary Material, Table S3). Clear negative correlations of X chromosome number and methylation levels in each cell line were observed at CXHs15 and CXHs10. This indicates that the hypomethylation phenomenon is not only restricted to blood cells, but that it is a more general phenomenon present in lymphoblastoid as well as fibroblastoid cell lines.
Methylation levels on the X chromosomes in 47,XXY Klinefelter samples are reduced in comparison to 46,XX female samples
A direct comparison between female and Klinefelter samples revealed distinctly higher methylation in female samples at several X-linked LINE-1-inactivated regions (Fig. 3 and Supplementary Material, Table S1). This higher methylation was observed at all CpG sites except CXHs13-CpG2 and it was statistically significant at all CpG sites of CXHs15 and CXHs3 as well as CpG1 of CXHs10. Cloning and sequencing data largely confirm the existence of this phenomenon at CXHs15 and CXHs6 loci but not at CXHs13 (Fig. 4A), which is consistent with the quantitative data. Moreover, at one of the six studied CpG islands (CX0) subject to X chromosome inactivation, a statistically significantly higher methylation in female samples at three CpGs was also observed (Supplementary Material, Table S1). Several factors could explain these differences among which is the possibility that the inactivation process may differ slightly between 46,XX and 47,XXY possibly due to factors on X or Y affecting the inactivation process. A second explanation could be the larger genome size in 47,XXY cells that would leave less methylation capacity at individual LINE-1 sites, while a third possibility is the hormonal differences between females and Klinefelters, with the latter having therapeutic male levels of hormones.
A tendency for inverse correlation of methylation at autosomal LINE-1 with genome size is observed
We studied autosomal LINE-1 methylation at five different loci on chromosomes 2, 6, 10, 11 and 20 in the four groups of samples: Turners, males, females and Klinefelters. These loci cover 14 different CpG sites that occur at the promoter region of L1Hs consensus sequence. In contrast to the X-linked inactivated L1Hs regions, there was no statistically significant difference between any two groups of the studied samples at the 14 studied autosomal CpG sites (Supplementary Material, Table S2).
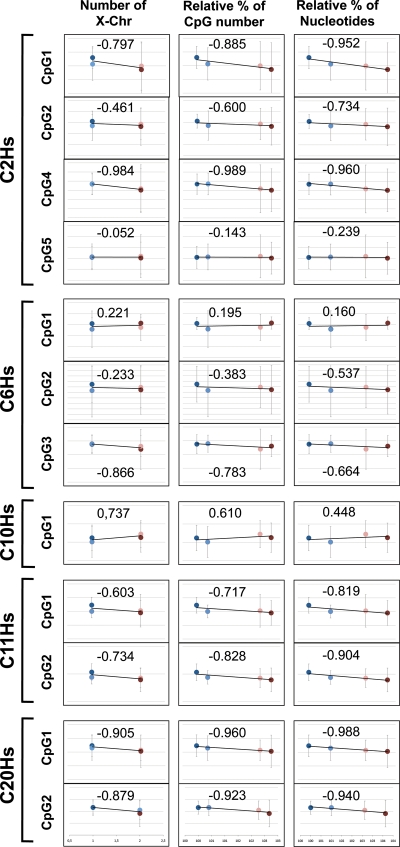
Nevertheless, an unexpected but a clear tendency was observed at the autosomal CpG sites. When we plotted the average methylation from every group in the order of increasing genome size (i.e. number of nucleotides in each genome with Turner samples as 100% reference genome size, male samples: 101.00%, female samples: 102.62% and Klinefelter samples: 103.63%), we could observe a clear correlation (Fig. 5 and Supplementary Material, Table S4). The relatively smallest genome in Turner (reference of 100%) samples showed the highest methylation. This average methylation decreased in a proportional manner towards the Klinefelter samples (size 103.62% relative to Turner samples), which harbor the biggest genome group of samples studied here. We calculated Pearson correlation of the average methylation at each site in every group with genome size as estimated from the nucleotide bases in every chromosome (UCSC). Most of the correlations (10 out of 12) were inversely proportional to genome size with r-values ranging from −0.24 (C2Hs-CpG5) to −0.99 (C20Hs-CpG1), with the exception of C6Hs-CpG1 and C10Hs-CpG1 with positive but weak correlations (Fig. 5 and Supplementary Material, Table S4). In addition, similar results were obtained when correlation with either the CpG numbers in a genome or the number of X chromosome were tested (Fig. 5). Albeit, correlations were highest with genome size (nucleotide number) followed by number of CpGs in a given genome, followed by number of present X chromosomes.
Figure 5.
Correlation of average methylation levels of autosomal LINE-1 and genomic size parameters. (A) With number of X chromosomes, (B) with relative percentage of CpG numbers, (C) with relative percentage of nucleotides. Blue, light blue, pink and red filled circles represent Turner, male, female and Klinefelter samples, respectively. Pearson correlation value is given for each correlation.
Yet, a strong positive correlation between average methylation levels of the four groups and genome size was observed at CpG3 and CpG6 of C2Hs (Supplementary Material, Table S4). One common feature between both of these CpGs is that they 3′ flank directly another CpG site. To investigate the possibility of experimental artifacts that could have been introduced by using one pyrosequencing reaction with a single sequencing primer to read all six CpGs in the C2Hs region, we designed one sequencing primer for every CpG site. We observed that CpGs 1, 2, 4 and 5 show similar tendencies with all primer sets, while CpGs 3 and 6 were not consistent with different primers. Therefore, we excluded the latter two CpGs from this evaluation (Supplementary Material, Fig. S1).
In addition to the above four sample groups, when we added methylation values of an available single DNA sample with the karyotype of 49,XXXXY [samples show a considerable drop of autosomal LINE-1 methylation at most CpG sites compared with all other groups (Supplementary Material, Table S4)] as a fifth point of measurement, stronger r-values for correlation between methylation levels and genome size at most CpG loci were observed, namely at C2Hs (CpG2, CpG4, CpG5), C6Hs (CpG1, CpG2), C10Hs (CpG1) and C11Hs (CpG1) (Supplementary Material, Table S4; Fig. 5 versus Supplementary Material, Fig. S2). This strengthens the hypothesis that increasing genome size will further reduce methylation at individual CpG sites in LINE-1 promoter regions in peripheral blood-derived DNA.
To further validate and visualize the highly quantitative SIRPH (SNuPE combined with Ion paired Reverse Phase HPLC) and pyrosequencing results, we performed extensive sequencing of individual PCR strands from three autosomal regions, C2Hs, C11Hs and C20Hs (Fig. 4B). For this analysis, we grouped three DNA samples in equal proportions from every group, with the exception of 49,XXXXY sample where only one sample is available. At the two loci C11Hs and C20Hs, where quantitative methylation results showed a 4–5% gradual decrease from Turner samples to Klinefelter sample of 49,XXXXY, only minor differences between the groups were observed [statistical comparison of the group of samples with one active X (45,X and 46,XY) and the group of samples with two Xs (46,XX and 47,XXY) by Wilcoxon test gave non-significant P-values of 0.8643 and 0.0907 for C20Hs and C11Hs, respectively]. The only statistically significant detectable differences in the average methylation were at C2Hs between Turner/male samples and female/Klinefelter samples (significant P-value of 0.0261). In fact, at the C2Hs locus, quantitative methylation data showed a distinct gradual decrease in methylation from the smaller genome to the largest (Supplementary Material, Table S1). Sequencing data also confirmed these differences. Especially female (91% methylation average) and Klinefelter samples (85 and 88%) showed a considerable drop compared with male and Turner samples (97% each) (Fig. 4B). Interestingly, differences in methylation largely stem from allelic methylation pattern, whereby some clones are completely unmethylated. This allelic methylation resulted in a relatively lower methylation entropy than if a random (non-deterministic) distribution of unmethylation were present. This was significant at several 4-CpGs windows within the three samples with two or four X chromosomes at the C2Hs locus (data not shown). This is a clear indication of an allelic unmethylation.
Next, we investigated whether we could see this genome size effect also in methylation of autosomal LINE-1 in fibroblastoid and lymphoblastoid cell lines used in the above section. Although hypomethylation associated with the number of X chromosome was clearly seen, none of the autosomal LINE-1 loci (C2Hs, C10Hs and C11Hs) showed significant correlation with genome size (after correction for multiple testing). The discrepancy between blood-derived data and cell lines could be due to either the lower sample number of the cell lines (thus less statistical power of detection) or to the differences between in vivo and in vitro cell culture conditions. In this study, cells were cultured in Dulbecco's modified Eagle medium (DMEM) or Rosewell Park Memorial Institute (RPMI) mediums providing unrestricted supply of nutrients and methyl group donors.
Correlations of methylation levels between different CpG sites
The wide distribution of studied CpGs in different LINE-1 enabled us to investigate whether a correlation of methylation levels exists between different CpGs located at different chromosomes, possibly suggesting a genetic or environmental influence on levels of LINE-1 methylation genome-wide. Therefore, we tested the correlations at the autosomal and X-linked LINE-1 at L1Hs loci, and the X-linked CpG islands that were studied in all four groups of samples. Hence, we examined for (i) specific correlation of methylation of CpGs within one L1Hs LINE-1 locus (intra-loci correlations), (ii) correlation between CpGs of different L1Hs LINE-1 loci (inter-loci correlations), and (iii) correlations between LINE-1 CpGs and non-LINE-1 CpGs.
At many LINE-1 loci, strong positive and some sporadic negative correlations within each of the studied loci were observed in Turner, male, female and Klinefelter samples (intra-loci correlations) (data not shown). Also, positive as well as negative correlations between different loci were observed (inter-loci correlations) (P-values <0.05); after correction for multiple testing only few inter-loci correlations remained significant (data not shown). In addition, age did not correlate with methylation at any of the studied loci (data not shown).
The absence of statistically significant widespread correlations between different LINE-1 loci indicates that this variability may not be controlled by a genetic factor affecting all repeats simultaneously. Instead, they may be influenced by loci-specific factors as local nearby polymorphisms and/or environmental factors, which could control methylation levels separately at each individual locus. Also, we cannot exclude the possibility that the stringent corrections for multiple testing (i.e. relatively high number of tests performed) and the relatively low number of sample size are responsible for the low power of detection of such correlations. Moreover, the absence of correlations in the female and Klinefelter samples at the severely hypomethylated L1Hs loci indicates that this hypomethylation affects different loci at the inactive X to different degrees and thus suggests that this is a randomized process.
DISCUSSION
Repetitive elements, like the LINE-1 subfamily, are known to be hypomethylated in tumors and their methylation levels are affected by environmental factors (17,19,27,28). Hypomethylation of specific LINE-1 could also activate alternative transcripts of neighboring genes as in the case of MET oncogene in bladder tumors or different cancer cell lines (29,30). In addition, we and others have previously shown that men have slightly higher methylation levels than women, a fact that became significant when comparing about 96 samples from every group (20,21,31,32). We also recently proved that the gender difference in the methylation of LINE-1 is not induced by male/female hormone differences (21). Moreover, the involvement of the LINE-1 elements and consequently their methylation is strongly suggested in the X chromosome inactivation process either by initializing, promoting or maintaining the inactivation process (3).
Variability of methylation at individual LINE-1
Detailed data on inter-individual variability of methylation at individual LINE-1 from total blood in large number of healthy individuals have not been studied sufficiently. In fact, using a degenerate assay to measure global LINE-1 methylation at promoter regions, we identified a considerable variability in male and female blood-derived DNA (21). Moreover, tissue-specific differences have been reported (33). However, only one report described in detail loci-specific methylation patterns at 17 different LINE-1 loci in several cell types, including different tumor cell lines but only 12 samples of normal oral epithelium (22). In contrast, no report dealt with inter-individual differences in a large cohort of blood samples derived from healthy individuals or with different types of aneuploidy using a highly sensitive method to measure subtle methylation differences. Our data presented here provide evidence for three types of variability in methylation at LINE-1-specific loci. The first type is the inter-loci variability. Although most loci were heavily methylated, there were some considerable inter-loci variations. The second type of variability was observed in different CpGs on the same locus (intra-loci variations). Both of these variation types, inter- and intra-loci variation, could be induced by specificity of DNMT3A and DNMT3B, which preferentially methylate specific CpGs at non-repeats, dependent on the underlying DNA sequence (34). Yet, evidence that this could also be the case for LINE-1 repeats is still lacking.
Both of the above variations have been previously known and reported by others in several studies. However, the third and most significant of the observed variations that have not been discussed sufficiently in the literature is the inter-individual variation, which can be clearly seen by the variance parameters of each locus (Fig. 1B and Supplementary Material, Table S1). This was previously reported in a small number of normal oral epithelium samples (22) but similar data in a large cohort of healthy blood samples have been lacking. Considering the male and female groups, it was obvious that not all loci are equally variable in their methylation but that some minorities of loci are hyper-variable (defined here as variance >100) in both males and females (Fig. 1B). Only two such loci were observed at L1Hs loci (CXHs1 and C2Hs), while more hyper-variable loci were present at L1P (CXP4, CXP9, CXP13) and L1M (CXM6, CXM7) subtypes.
Polymorphisms of mainly C to T at the investigated CpG sites can cause high inter-individual variability. However, this has been excluded by sequencing all samples in three regions (C2Hs, C10Hs and C11Hs). This observed inter-individual variability could still be due to other neighboring polymorphisms that act in cis at a lower distance (25,26). We cannot exclude this possibility. Moreover, this variability could also be global, affecting several loci simultaneously or genome-wide (35) due to genetic factors that act globally in trans or as a result of environmental factors. However, inter-CpG correlation analysis did not show significant inter-loci correlation. Thus, we assume that these variations are random stochastic variations not resulting from global but largely rather locus-specific factors.
Hypomethylation of LINE-1 at X-linked L1Hs inactivated regions: cause or consequence of inactivation?
On the X chromosome, we investigated three groups of LINE-1 subtypes: 15 L1Hs, 16 L1P and 8 L1M. In comparison to males, female samples demonstrated considerable and significant hypomethylation at almost all L1Hs but not at L1P or L1M (Fig. 2). However, most L1P loci were also higher methylated in males, while the L1M loci particularly showed a mixture of correlation to gender with four loci each more methylated in males and females, respectively (Fig. 2). We assume that the high order lowering of methylation at L1Hs and—to a lesser extent—at L1Ps stems from the inactive X chromosome. This clearly explains our previous results of a significant decrease in methylation in women at genome-wide global LINE-1 5′ regions when using degenerate primers based on the L1Hs promoter consensus sequence (20,21). However, these results appear to be at odds with the observations made by Hansen (36), who concluded that both LINE-1 promoters on active as well as inactive chromosomes are hypermethylated in normal cells, while a significant hypomethylation on the inactive X in ICF cells is observed. Although the conclusions from Hansen were largely correct regarding hypermethylation of the studied loci on X chromosomes, the relatively small difference in the hypermethylation between men and women was missed because—in contrast to our analysis—the methods used were largely non-quantitative (based on southern blots and bisulfite cloning and sequencing). Yet, a close look at his data revealed clear indication of healthy female hypomethylation in comparison to healthy male samples and ∼12% lower methylation in female lymphoblastoid cells at one X-linked L1Hs [81.7% methylated sites in male samples versus 69.7% in female samples using the average of methylated sites from the cloning and sequencing data of Hansen's Figure 4 (36)].
To test whether hypomethylation of the LINE-1 regions is specific for inactivated or non-inactivated (escaping) genes/regions, we analyzed LINE-1 loci distributed on both inactivated and non-inactivated regions of the X chromosome. This was possible only for L1M and L1P loci but not for L1Hs due to the absence of full-length L1Hs loci on the non-inactivated (strongly escaping) regions. In the L1P group as well as the L1M group, there was no clear association between hypomethylation on the inactive X and the inactivation status of the host DNA (Fig. 2). Especially at the L1P, the degree of hypomethylation on the inactive X was quite similar between the inactivated and the escaping regions. Apparently, the generalized severe hypomethylation of the inactive X is not only restricted to the inactivated regions but specifically so to L1Hs promoter regions that, in contrast to other LINE-1 subtypes, contain higher density of CpGs predicted to be a CpG island.
Since the hypomethylation at the L1Hs on the inactive X chromosome could be a simple effect of hormone differences rather than a cause or consequence of the inactivation, we analyzed six L1Hs regions showing the highest differences between male and females in two cohorts of Turners (hormone-wise female, but with only one active X) and Klinefelters (hormone-wise male, but with two X chromosomes: one active and one inactive). Indeed, all samples with two X chromosomes showed this significant hypomethylation (Fig. 3), but not samples with one active X. Therefore, the L1Hs hypomethylation on the Xi is linked to the X chromosome inactivation process or it is a consequence of the inactivation status rather than a simple effect of hormonal differences.
Previous methylation profiling of the X chromosome has also shown differences between Xi and Xa in a sequence- and region-specific manner. Most CpG islands show a clear hypermethylation on the Xi as expected because of inactivation, while ∼7% showed a hypomethylation at the Xi (37). Outside CpG islands, Xi was shown to be hypomethylated in the gene bodies (38) and global hypomethylation of Xi was also reported (39). Therefore, and based on our reported results here, it seems that the methylation status of a given CpG on the inactive X depends on many factors, such as CpG density, sequence context (class of repeat) and relative position to gene expression. However, a single base resolution of methylation profiling of the Xi and Xa in different stages of X chromosome inactivation is necessary to determine the factors that rule the methylation status of a given X-linked CpG site.
Involvement of the LINE-1 repeats in the X chromosome inactivation process was suggested in 1998 by Lyon (40). The mainstream theory is that LINE-1 elements could work as boosters (or ‘way stations’ as termed by Gartler and Riggs (41), yet without naming LINE-1 ‘stations’) for the spreading of the inactivation signal or to maintain the inactivation status. Since this first hypothesis, several studies have supported involvement of LINE-1 in the process of X-inactivation, but none has provided a direct/ultimate proof to date.
Density of LINE-1 on the X chromosome was found to be double compared with autosomes (26.50 versus 13.43%), while several other repeats, such as SINE, were not enriched on the X (42). The highest density of LINE-1 enrichment is on the Xq13, which contains the center of inactivation. Moreover, density of LINE-1 was higher on inactivated regions than non-inactivated regions, escaping the inactivation process (26.5 versus 10.3%) (42). Considering in more detail Xp22, which contains both active and inactive genes, Carrel et al. (43) could show that the surroundings of inactive gene regions are particularly enriched with sequences corresponding to LINE-1 sequences which are otherwise underrepresented on Xp22 in comparison to the inactivated regions of the X chromosomes. A direct link between LINE-1 density and effectiveness of inactivation comes from the X;autosomal and autosomal;X translocations. These cases show that different autosomal regions are not effectively/equally silenced when translocated to the X (44,45). Reciprocally, when an inducible Xist transgene is inserted into different mice autosomal loci, the silencing efficiency is determined by the chromosomal domain organization of the integration locus (46). Inactivation on autosomes is more effective in gene poor and LINE-1 rich domains. Popova et al. (47) also provided evidence that a LINE-1 poor region is able to attenuate the primary inactivating Xist signal instead of failing to maintain the inactivation status.
The strongest involvement of LINE-1 in the inactivation process came from a recent mice study by Chow et al. (48) which showed that LINE-1 repeats participate in the inactivation process by creating with the Xist RNA a silent nuclear compartment where inactivated regions are recruited to. Moreover, the youngest LINE-1 element families (Tf and Gf LINE-1 in mice) are also transiently transcribed (from the to-be-inactivated X) during the early inactivation process (48). This LINE-1 expression is dependent on Xist expression as well as facultative heterochromatin formation. Additionally, the efficiency of Xist-mediated inactivation on mice autosomal loci (in autosomal Xist transgenes) was correlated to the presence of full-length young LINE-1 elements (48). Therefore, could L1Hs sequences in humans, which are exclusively hypomethylated to a larger degree than L1P or L1M sequences on the inactive X and are absent from the non-inactivated loci, correspond to Tf and Gf in mice and could they be also transiently expressed during the inactivation process where their expression is down regulating in cis the neighboring genes? And could the hypomethylation of their 5′ promoter be a mark of such earlier activity? Such data can be expected from human ES cell studies. Although our data do not provide a direct evidence for the involvement of L1Hs sequences with the X chromosome inactivation process, still we provide one more direct support for the likelihood of the involvement of LINE-1 in the inactivation process in humans, particularly L1Hs which are specifically and largely hypomethylated on the inactive regions.
Methylation levels at specific LINE-1 and size of the genome
In this study, we found an unexpected correlation between genome size and methylation at autosomal LINE-1 promoter elements. Several of the autosomal CpG sites showed a negative correlation between genome size as estimated from the number of nucleotides in a given genome and methylation levels (Fig. 5). The strongest negative correlation was observed at C20Hs and C11Hs, while a positive correlation was observed at C6Hs-CpG1 and C10Hs. The global LINE-1 assay with degenerate primers did not show this tendency (Supplementary Material, Table S1) probably due to the insensitivity of the assay to variations at specific loci or because only a subset of full-length L1Hs LINE-1 elements studied here are showing this tendency.
However, the unexpected observation of a correlation between genome size and methylation at autosomal LINE-1 promoter elements could also be explained by either presence of a factor on the X chromosome that could induce hypomethylation of the repeats or a limiting amount of a given factor required for methylation of repeats. However, since the correlation is strongest with genome size rather than X chromosome count, the first factor could be of lesser importance. What supports the second possibility is that, in a tissue culture system where the nutrient factors—particularly the methyl donor—are unlimited, no correlation between genome size and methylation levels of LINE-1 CpG sites was found. Thus, our data argue for a factor that is present in a limited amount so that its deficiency effect is seen in either rapidly dividing cells (like blood-derived cells) and/or in larger genomes. We should acknowledge here that this phenomenon could be also caused by differences in expression of gene(s) that could be involved directly or indirectly with the methylation of LINE-1 repeats. Such genes could be located at the X or even the Y chromosome, for example on the X non-inactivated regions, and thus exert a dose effect differences among different groups of samples studied. An effect on gene expression exerted by sex chromosome counts rather than sex was previously reported by Wijchers et al. (49). Therefore, it is not unexpected that sex chromosome counts affect one of the epigenetic layers that regulate gene expression.
In this study, using highly quantitative methylation analysis of individual LINE-1 repeats, we showed that methylation is not the same in all individuals and that it does not have the same levels on all loci even when comparing the same category of loci (autosomal or X-linked). In addition, we report on a strong difference in methylation levels between active and inactive X chromosomes with inactive ones considerably hypomethylated (in the inactivated region) at the promoters of full-length L1H regions. Finally, genome size affected methylation levels at most autosomal loci with the lowest methylation observed in the bigger genome.
MATERIALS AND METHODS
Blood samples and cell lines
Blood samples were collected from 22 female patients with Turner syndrome (expected karyotype 45,X) and were obtained from the Pediatric Endocrinology Division, University Children's Hospital, Bonn, Germany. Forty men with Klinefelter syndrome (expected karyotype 47,XXY) were recruited to this study through the German Klinefelter association, Falkenstein, Germany. In the latter group, two men were reported to have 48,XXYY and 49,XXXXY karyotypes each. In addition, control blood samples, matching in age with Turner and Klinefelter samples, from 28 healthy men and 28 healthy women with a normal X chromosome count, matching in age with Turner and Klinefelter samples, were collected from blood donors at the Institute of Experimental Hematology and Transfusion Medicine, Bonn, Germany. Furthermore, a large sample group of young healthy men and women (96 each) with an average age of 23 ± 3.46 years was also available to study inter-individual and inter/intra-loci variations. All samples from all subjects were obtained upon written informed consent. The local Ethics Committee of the University Clinics of Bonn approved the study (approval numbers: 106/05 and 121/06).
Several lymphoblastoid and fibroblastoid cell lines with essentially different X chromosome counts were used in this study. These included: 45,X; 46,XY; 46,XX; 47,XXX; 48,XXXX; 49,XXXXY and 49,XXXXX lymphoblastoid cell lines and 45,X; 47,XXX and 49,XXXXY fibroblastoid cell lines. All cell lines were in their early 20 passages. The fibroblastoid cell lines were maintained in DMEM, 10% FCS, 2 mm glutamine and Pen/Strep, while the lymphoblastoid cell lines were incubated in RPMI, 10% FCS, 2 mm glutamine and Pen/Strep. DNA from cell lines was extracted using the DNeasy blood and Tissue kit from Qiagen (Hilden, Germany).
Chromosome copy number verification
All Klinefelter and Turner blood samples as well as the cell lines that we received were documented for their karyotypes (especially X chromosome copy number). However, to verify X chromosome copy number in Klinefelter and Turner samples as well as in the cell lines and to determine the mosaics rates (if any), we first sequenced all samples at six X-linked regions distributed over the whole X chromosome containing SNPs with a predicted high heterozygosity in Caucasian populations (as reported in HapMap) (Supplementary Material, Table S5, primers used to amplify these regions are listed in Supplementary Material, Table S6). Loss of heterozygosity at all markers would predict one X and at least one heterozygous site would predict two X. To predict the degree of cellular mosaicism, we applied a quantitative pyrosequencing assay for five of the SNPs (primers are listed in Supplementary Material, Table S6).
As a second method to quantitatively determine the copy number of X chromosomes, a copy number assay from Applied Biosystems was used. Briefly, the assay consists of a TaqMan minor groove binding probe labeled with FAM dye and unlabeled PCR primers designed for the X-linked region TCEAL4 not containing any copy number variations. The assay was run simultaneously with a TaqMan® Copy Number Reference Assay (RNaseP using VIC® dye-labeled TAMRA™ probes) in a duplex real-time PCR. Relative quantitation analysis of the target to the reference sequences was performed by CopyCaller™ software, using a known calibrator sample with two X chromosomes. Twenty nanograms of DNA was used in a total reaction volume of 16 μl. The reaction mix was prepared with the TaqMan Universal Master Mix. The assay was performed in four replicates for all samples.
Klinefelter samples showed heterozygosity in at least one of six markers in all samples except one (11 K). However, the TaqMan-based copy number assay clearly confirmed the presence of two X chromosome copies in all samples except one that was known from karyotype analysis to have four X chromosomes. Based on the results of the two methods used, Turner samples were divided into two groups: 13 samples lacking heterozygosity at all six SNPs and 8 samples that are heterozygous at least at one of the six studied loci. From the latter group, the TaqMan copy number assay showed two samples, to be mosaic for the presence of an additional X chromosome, while the rest are supposed to have a normal female karyotype (Supplementary Material, Table S5).
The fibroblastoid and lymphoblastoid cell lines were also tested with the TaqMan-based copy number assay for the expected X chromosome numbers. All four fibroblastoid lines shows the expected X copy numbers. However, the lymphoblastoid cell lines with the expected 4X (375-99 LCL) and 5X (377-99 LCL) showed a considerable decrease from the expected X chromosome numbers with 3.25 and 4.12 predicted X copy numbers (Supplementary Material, Table S5). This could be due to recombination events occurring during different passaging in cell culture or to a decrease in sensitivity of the used assay with increasing copy number of target sequences. It would require much larger numbers of replicates to achieve accurate copy numbers determination as previously suggested (50). However, we assumed that the quantitative TaqMan-based assay is reflecting the true X chromosome copy number.
In summary, the samples in this study include peripheral blood samples from 13 Turner syndrome patients (45,X), 28 men (46,XY), 28 women (46,XX) and 39 Klinefelter syndrome patients (47,XXY). Moreover, one Klinefelter sample with 49,XXXXY karyotype was also available. In addition, 10 lymphoblastoid and 3 fibroblastoid cell lines were included in the study.
Exclusion of C to T polymorphisms at the investigated autosomal CpG sites
Since C to T polymorphisms are common at CpG sites, such C to T changes would be detected after bisulfite as non-methylated cytosine, thus interfering with the average methylation values of CpG sites. To exclude this influence, we sequenced three autosomal regions (C2Hs, C10Hs, C11Hs) in all studied samples from non-bisulfite-treated DNA. No detectable C to T transition was found in any of the samples. As individual methylation values at other regions do not harbor considerable variations >50% (which could be an indication for the presence of a C to T polymorphism), we assumed that all of the CpG sites included in this study are not overlapping with polymorphisms that could interfere with the quantitative methylation data.
Methylation analysis
Bisulfite modifications and amplification of specific repeats
About 1000 ng input DNA was used for the bisulfite conversion; this was done using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's protocol. To minimize assay variability or batch variability, we distributed the different samples of the Turner, male, female and Klinefelter groups over two plates, whereby each plate contained an approximately similar number of samples from every group.
A semi-nested PCR approach was used to amplify specific LINE-1 repeats. Primer sequences used for amplifications in this study are listed in Supplementary Material, Table S6. To select LINE-1 regions for amplifications, we used a Blat search on the UCSC genome browser (http://genome.ucsc.edu; 51) of the LINE-1 promoter region (L1Hs; NCBI accession number: X58075). We selected LINE-1 on autosomes that were >6 kb in length, with the highest homology to the consensus sequence, flanked by a unique genomic DNA sequence and covering the highest possible numbers of CpGs. Using this strategy, we were able to specifically amplify five autosomal LINE-1 (L1Hs) promoter regions with the highest homology to the consensus sequence. We then localized all 25 L1Hs on the X chromosome and were able to specifically amplify 15 of them, while the remaining 10 were either not flanked by unique DNA sequence or unable to verify their specific amplification and thus were discarded. Moreover, 14 L1M and 22 L1P on the X chromosome were chosen to be imbedded in genes or loci that are known either to be inactivated or to escape inactivation. Therefore, we were able to specifically amplify 8 L1M and 16 L1P loci. All PCR amplifications were done using hot-start Taq polymerase (HOT FIREPol from Solis BioDyne, Tartu, Estonia). The success of locus-specific amplification was confirmed by direct sequencing of each region (data not shown). In addition, we targeted six X-linked CpG islands subject to X chromosome inactivation, Information regarding their methylation levels in male and female samples was established in our MeDIP experiments on male and female peripheral blood-derived DNA as previously reported (35).
Quantitative methylation analysis
Quantitative methylation at specific CpG sites was done either by SIRPH protocol as previously described (52,53) or by pyrosequencing performed according to Tost et al. (54). The latter was performed using Pyro Gold reagents from Biotage (Uppsala, Sweden) on a PyroMark ID system. SNuPE and pyrosequencing primers are listed in Supplementary Material, Table S6.
Cloning and sequencing and calculation of methylation entropy
Individual PCR products were cleaned using the QIAquick PCR purification kit (order no. 28106; Qiagen). Next, the purified PCR product was ligated to a pGEM-T cloning vector (pGEM-T Vector System 1, order no. A3600, Promega, Mannheim, Germany). Individual positive clones were isolated by PCR using M13 primers and sequenced using ABI standard protocols. The methylation entropy that reflects on the allelic or non-allelic distribution of methylation at a given locus was calculated according to Xie et al. (55). A sliding window of four CpGs was used at one time, and next, the average of all windows was used as an estimate of the methylation entropy in a given sample at a given region.
Global genomic methylation contents by the LUMA method (methylation levels at CCGG sites)
Luminescence methylation assay (LUMA) was essentially performed as previously described by Karimi et al. (56). Briefly, 500 ng of DNA to be measured was cleaved with HpaII + EcoR1 or MspI + EcoR1 (five units from each enzyme) in two separate tubes. The total volume of reaction was 20 µl. The tubes were incubated at 37°C for exactly 4h. Next, 20 µl annealing buffer (20 mm Tris-acetate, 2 mm Mg-acetate pH 7.6) was added and samples were measured in a PSQ96™ MA pyrosequencing machine (Biotage AB). EcoR1 leaves 5′-TTAA sticky ends on the DNA that can be extended by As and Ts, while HpaII and MspI leaves 5′-GC that can be extended by Cs and Gs. Therefore, the machine was programmed to add As, next Gs and Cs. The first would generate peaks corresponding to the EcoRI digestion, which can be used as internal control for the quantity of DNA and the efficiency of cleavages, while the second would give peaks corresponding to all CCGG sites (for MspI) or for the unmethylated portion of CCGG (for HpaII). Next, the percentage of methylated CCGG sites was calculated according to the formula: Meth% = [1-(peak HpaII/peak EcoRI)/(peak MspI/peak EcoRI)]*100.
Statistical analysis
T-test and Bonferroni–Holmes corrections
T-test was applied to investigate at a specific CpG site whether the methylation levels between two groups (Turner, male, female and Klinefelter samples, each two at a time) are significantly different. The Bonferroni–Holmes correction was applied to correct for multiple testing.
Correlation between different CpG sites within one group
To investigate a possible correlation between different CpG sites within one studied locus and across different loci in one group of individuals with the same karyotypes, we applied Pearson correlation followed by Bonferroni–Holmes correction for multiple testing.
Statistical significance of sequencing data was assesses by the non-parametric Wilcoxon test. Comparison of the two groups, the first derived from 45,X and 46,XY (one active X) and the second included the clones derived from 46,XX and 47,XXY (containing active and inactive X), at each of the six loci was performed.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
The study was supported by institutional fund of the Institute of Experimental Hematology and Transfusion Medicine, University Clinic of Bonn. Additionally, work in the lab of OEM is supported by a German Research Council grant (DFG EL 499/2-1). Funding to pay the Open Access publication charges for this article was provided by the Institute of Experimental Haematology and Transfusion Medicine, University of Bonn.
Supplementary Material
ACKNOWLEDGEMENTS
The authors like to thank all the participants in this study and the German Klinefelter association for help in sample recruitment and to Hehuang Xie for the methylation entropy calculations.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bailey J.A., Laura C., A C., E E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. PNAS. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyon M. LINE-1 elements and X chromosome inactivation: a function for Junk DNA. PNAS. 2000;97:6248–6249. doi: 10.1073/pnas.97.12.6248. doi:10.1073/pnas.97.12.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohjoh H., Singer M.F. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. doi:10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. doi:10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 6.Mathias S.L., Scott A.F., Kazazian H.H., Jr, Boeke J.D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. doi:10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 7.Babushok D.V., Kazazian H.H., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. doi:10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 8.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. doi:10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. doi:10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 10.Aporntewan C., Phokaew C., Piriyapongsa J., Ngamphiw C., Ittiwut C., Tongsima S., Mutirangura A. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PloS ONE. 2011;6:e17934. doi: 10.1371/journal.pone.0017934. doi:10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C.R., Schneider A.M., Lu Y., Niranjan T., Shen P., Robinson M.A., Steranka J.P., Valle D., Civin C.I., Wang T., et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. doi:10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. doi:10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. doi:10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing A.D., Kazazian H.H., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. doi:10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thayer R.E., Singer M.F., Fanning T.G. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–277. doi: 10.1016/0378-1119(93)90651-i. doi:10.1016/0378-1119(93)90651-I. [DOI] [PubMed] [Google Scholar]
- 16.Hata K., Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. doi:10.1016/S0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 17.Schulz W.A., Steinhoff C., Florl A.R. Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. doi:10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 18.Pilsner J.R., Hu H., Ettinger A., Sanchez B.N., Wright R.O., Cantonwine D., Lazarus A., Lamadrid-Figueroa H., Mercado-Garcia A., Tellez-Rojo M.M., et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ. Health Perspect. 2009;117:1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollati V., Baccarelli A., Hou L., Bonzini M., Fustinoni S., Cavallo D., Byun H.M., Jiang J., Marinelli B., Pesatori A.C., et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. doi:10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 20.El-Maarri O., Becker T., Junen J., Manzoor S.S., Diaz-Lacava A., Schwaab R., Wienker T., Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. doi:10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 21.El-Maarri O., Walier M., Behne F., van Üüm J., Singer H., Diaz-Lacava A., Nüsgen N., Niemann B., Watzka M., Reinsberg J., et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PloS ONE. 2011;6:e16252. doi: 10.1371/journal.pone.0016252. doi:10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phokaew C., Kowudtitham S., Subbalekha K., Shuangshoti S., Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36:5704–5712. doi: 10.1093/nar/gkn571. doi:10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popp C., Dean W., Feng S., Cokus S.J., Andrews S., Pellegrini M., Jacobsen S.E., Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. doi:10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J., et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. doi:10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Rohde C., Reinhardt R., Voelcker-Rehage C., Jeltsch A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 2009;10:R138. doi: 10.1186/gb-2009-10-12-r138. doi:10.1186/gb-2009-10-12-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N., Plomin R., Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. doi:10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baccarelli A., Wright R.O., Bollati V., Tarantini L., Litonjua A.A., Suh H.H., Zanobetti A., Sparrow D., Vokonas P.S., Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am. J. Resp. Crit. Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. doi:10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavanello S., Bollati V., Pesatori A.C., Kapka L., Bolognesi C., Bertazzi P.A., Baccarelli A. Global and gene-specific promoter methylation changes are related toanti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int. J. Cancer. 2009;125:1692–1697. doi: 10.1002/ijc.24492. doi:10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 29.Wolff E.M., Byun H.-M., Han H.F., Sharma S., Nichols P.W., Siegmund K.D., Yang A.S., Jones P.A., Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PloS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. doi:10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber B., Kimhi S., Howard G., Eden A., Lyko F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene. 2010;29:5775–5784. doi: 10.1038/onc.2010.227. doi:10.1038/onc.2010.227. [DOI] [PubMed] [Google Scholar]
- 31.Wilhelm C.S., Kelsey K.T., Butler R., Plaza S., Gagne L., Zens M.S., Andrew A.S., Morris S., Nelson H.H., Schned A.R., et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin. Cancer Res. 2010;16:1682–1689. doi: 10.1158/1078-0432.CCR-09-2983. doi:10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z.Z., Hou L., Bollati V., Tarantini L., Marinelli B., Cantone L., Yang A.S., Vokonas P., Lissowska J., Fustinoni S., et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int. J. Epidemiol. 2010;39:1–14. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalitchagorn K., Shuangshoti S., Hourpai N., Kongruttanachok N., Tangkijvanich P., Thong-ngam D., Voravud N., Sriuranpong V., Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. doi:10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 34.Wienholz B.L., Kareta M.S., Moarefi A.H., Gordon C.A., Ginno P.A., Chédin F. DNMT3L modulates significant and distinct flanking sequence preference for DNA Methylation by DNMT3A and DNMT3B in vivo. PloS Genet. 2010;6:e1001106. doi: 10.1371/journal.pgen.1001106. doi:10.1371/journal.pgen.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Maarri O., Kareta M.S., Mikeska T., Becker T., Diaz-Lacava A., Junen J., Nusgen N., Behne F., Wienker T., Waha A., et al. A systematic search for DNA methyltransferase polymorphisms reveals a rare DNMT3L variant associated with subtelomeric hypomethylation. Hum. Mol. Genet. 2009;18:1755–1768. doi: 10.1093/hmg/ddp088. doi:10.1093/hmg/ddp088. [DOI] [PubMed] [Google Scholar]
- 36.Hansen R.S. X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum. Mol. Genet. 2003;12:2559–2567. doi: 10.1093/hmg/ddg268. doi:10.1093/hmg/ddg268. [DOI] [PubMed] [Google Scholar]
- 37.Sharp A.J., Stathaki E., Migliavacca E., Brahmachary M., Montgomery S.B., Dupre Y., Antonarakis S.E. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;10:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. doi:10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 39.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. doi:10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 40.Lyon M.F. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. doi:10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 41.Gartler S.M., Riggs A.D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. doi:10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 42.Bailey J., Carrel L., Chakravarti A., Eichler E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation the lyon repeat hypothesis. PNAS. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. doi:10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrel L., Park C., Tyekucheva S., Dunn J., Chiaromonte F., Makova K. Genomic environment predicts expression patterns on the human inactive X chromosome. PloS Genet. 2005;2:e151. doi: 10.1371/journal.pgen.0020151. doi:10.1371/journal.pgen.0020151.eor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cattanach B.M. Position effect variegation in the mouse. Genet. Res. 1974;23:291–306. doi: 10.1017/s0016672300014932. doi:10.1017/S0016672300014932. [DOI] [PubMed] [Google Scholar]
- 45.Sharp A.J., Spotswood H.T., Robinson D.O., Turner B.M., Jacobs P.A. Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum. Mol. Genet. 2002;11:3145–3156. doi: 10.1093/hmg/11.25.3145. doi:10.1093/hmg/11.25.3145. [DOI] [PubMed] [Google Scholar]
- 46.Tang Y.A., Huntley D., Montana G., Cerase A., Nesterova T.B., Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. doi:10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popova B.C., Tada T., Takagi N., Brockdorff N., Nesterova T.B. Attenuated spread of X-inactivation in an X;autosome translocation. Proc. Natl Acad. Sci. USA. 2006;103:7706–7711. doi: 10.1073/pnas.0602021103. doi:10.1073/pnas.0602021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chow J.C., Ciaudo C., Fazzari M.J., Mise N., Servant N., Glass J.L., Attreed M., Avner P., Wutz A., Barillot E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. doi:10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 49.Wijchers P.J., Yandim C., Panousopoulou E., Ahmad M., Harker N., Saveliev A., Burgoyne P.S., Festenstein R. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by sry but by sex chromosome complement as well. Dev. Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. doi:10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Weaver S., Dube S., Mir A., Qin J., Sun G., Ramakrishnan R., Jones R.C., Livak K.J. Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods. 2010;50:271–276. doi: 10.1016/j.ymeth.2010.01.003. doi:10.1016/j.ymeth.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El-Maarri O., Herbiniaux U., Walter J., Oldenburg J. A rapid, quantitative, non-radioactive bisulfite-SNuPE- IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res. 2002;30:e25. doi: 10.1093/nar/30.6.e25. doi:10.1093/nar/30.6.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Maarri O. SIRPH analysis: SNuPE with IP-RP-HPLC for quantitative measurements of DNA methylation at specific CpG sites. Methods Mol. Biol. 2004;287:195–205. doi: 10.1385/1-59259-828-5:195. [DOI] [PubMed] [Google Scholar]
- 54.Tost J., Dunker J., Gut I.G. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 55.Xie H., Wang M., de Andrade A., Bonaldo M.F., Galat V., Arndt K., Rajaram V., Goldman S., Tomita T., Soares M.B. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011;39:4099–4108. doi: 10.1093/nar/gkr017. doi:10.1093/nar/gkr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karimi M., Johansson S., Stach D., Corcoran M., Grander D., Schalling M., Bakalkin G., Lyko F., Larsson C., Ekstrom T.J. LUMA (LUminometric Methylation Assay)—a high throughput method to the analysis of genomic DNA methylation. Exp. Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. doi:10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 57.Carrel L., Willard H.F. X inactivation profile reveals extensive variability in X linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. doi:10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.