Abstract
Background Cigarette smoking is associated with lower body mass index (BMI), and a commonly cited reason for unwillingness to quit smoking is a concern about weight gain. Common variation in the CHRNA5-CHRNA3-CHRNB4 gene region (chromosome 15q25) is robustly associated with smoking quantity in smokers, but its association with BMI is unknown. We hypothesized that genotype would accurately reflect smoking exposure and that, if smoking were causally related to weight, it would be associated with BMI in smokers, but not in never smokers.
Methods We stratified nine European study samples by smoking status and, in each stratum, analysed the association between genotype of the 15q25 SNP, rs1051730, and BMI. We meta-analysed the results (n = 24 198) and then tested for a genotype × smoking status interaction.
Results There was no evidence of association between BMI and genotype in the never smokers {difference per T-allele: 0.05 kg/m2 [95% confidence interval (95% CI): −0.05 to 0.18]; P = 0.25}. However, in ever smokers, each additional smoking-related T-allele was associated with a 0.23 kg/m2 (95% CI: 0.13–0.31) lower BMI (P = 8 × 10−6). The effect size was larger in current [0.33 kg/m2 lower BMI per T-allele (95% CI: 0.18–0.48); P = 6 × 10−5], than in former smokers [0.16 kg/m2 (95% CI: 0.03–0.29); P = 0.01]. There was strong evidence of genotype × smoking interaction (P = 0.0001).
Conclusions Smoking status modifies the association between the 15q25 variant and BMI, which strengthens evidence that smoking exposure is causally associated with reduced BMI. Smoking cessation initiatives might be more successful if they include support to maintain a healthy BMI.
Keywords: Smoking, BMI, SNP, genetic association, interaction
Introduction
Cigarette smoking is a major preventable cause of death and disease.1 However, while the majority of smokers report a willingness to quit, only a relatively small proportion attempt to do so in any given year.2 One reason commonly cited for being unwilling to stop smoking is a concern about weight gain following cessation,3 and weight gain following smoking cessation has also been associated with an increased risk of smoking relapse.4
There is a general consensus that smoking cessation does lead to some degree of weight gain. There is substantial evidence from experimental studies indicating that cigarette smoking exerts an appetite-suppressant effect,5 and it has long been recognized that cigarette smoking is employed as a means of weight control, particularly by women,6 possibly mediated via a lowering of the body weight set point following chronic nicotine use.7 Epidemiological evidence consistently shows that smokers have a lower body mass index (BMI) than non-smokers,8,9 but whether smoking exposure is causal to lower BMI is unclear.
Recently, a number of genome-wide association (GWA) studies have provided robust evidence that genetic variation at the CHRNA5-CHRNA3-CHRNB4 locus on chromosome 15q25 is associated with quantity of smoking10–16 in those who smoke. Each additional copy of the T-allele of the single-nucleotide polymorphism (SNP), rs1051730, is associated with an increase in smoking quantity of about one cigarette per day.10 However, the similarity of allele frequency between ever and never smokers does not support association of this variant with smoking initiation.10 The rs1051730 SNP lies within the nicotinic acetylcholine receptor-α 3 subunit gene (CHRNA3), but is in linkage disequilibrium with a large number of other SNPs across CHRNA3, CHRNA5 and CHRNB4. The functional variant at the 15q25 locus is not known. The minor allele of the mis-sense polymorphism, D398N (rs16969968), in CHRNA5, conferred a reduced response to a nicotinic agonist in vitro.17 However, a recent study, which used data from the 1000 Genomes Project to impute virtually all common variants in the region, showed that rs55853698, in the 5′-untranslated region of CHRNA5, was the most strongly associated SNP.14 Both rs16969968 and rs55853698 are highly correlated with rs1051730 in Europeans (all pairwise r2 > 0.96).14
The association between variation at the 15q25 locus and BMI is not known. Since genotypes are (i) assigned at conception and (ii) not generally associated with the wide range of environmental characteristics that confound conventional epidemiological association studies,18 genetic variation at 15q25 should serve as an unconfounded measure of smoking exposure in smokers, unbiased by reverse causality. If cigarette smoking is causally associated with lower BMI, we would expect the allele that predisposes to heavier smoking to be associated with a lower BMI in those who smoke, but not in those who have never smoked. This would constitute a gene × environment interaction, and would provide strong evidence of causality.19 To investigate this, we performed a meta-analysis (n = 24 198 participants; k = 9 study samples) to examine the association between the rs1051730 variant and BMI. We stratified our analyses by smoking status, and tested for a genotype × smoking status interaction.
Methods
Study participants
We analysed data on a total of 24 198 participants from nine study samples. All participants gave informed consent and ethical approval was obtained from the relevant local review committees. In each study, BMI was calculated as weight (kg) divided by height squared (m2).
The Avon Longitudinal Study of Parents and Children
This is a prospective study, which recruited pregnant women from Bristol, UK, with expected delivery dates between April 1991 and December 1992. Ethical approval was obtained from the Avon Longitudinal Study of Parents and Children (ALSPAC)20 Law and Ethics Committee in addition to the local review committee. Data collection on smoking behaviour have been described previously.21 Women were grouped as never smokers, former smokers or current smokers on the basis of a questionnaire administered in the 18th week of gestation. The latter two categories were combined in analyses that compared strata of never with ever smokers. Pre-pregnancy BMI was calculated from self-reported pre-pregnancy weight and height. We included a total of 6148 women of European ancestry, with rs1051730 genotype, BMI and smoking data available.
The British Regional Heart Study
The study recruited 7735 men aged 40–59 years in 1978–80; full details are reported elsewhere.22 Men were recruited from 24 medium-sized British towns; at that time, very few eligible subjects were of non-European ancestry. Twenty years later, when aged 60–79 years, n = 4252 participants were re-measured and provided a whole-blood sample from which DNA was extracted. The physical examination included weight and height. Height without shoes and weight in trousers and socks were measured, to the nearest millimetre and 0.1 kg, respectively. Participants were asked whether they had ever smoked cigarettes regularly and, if yes, whether they smoked cigarettes now; from these two questions, we defined current smokers, former smokers and never smokers. Cotinine was assayed by a widely applied gas chromatographic method with a detection limit of 0.1 ng/ml.23 Regular internal quality controls were run to ensure comparability and reliability of results over time. For non-smokers, cotinine levels were further defined using a liquid chromatography tandem mass spectrometry (LC–MS/MS) assay with a lower limit of detection of 0.02 ng/ml and with a limit of quantification of 0.1 ng/ml. Cotinine values at the limit of quantification (0.1 ng/ml) were assigned a value of 0.05 ng/ml. Further details of the assay can be obtained from abslabs@biopark.org.uk. In the current analyses, we included 3870 participants with rs1051730 genotype, BMI and smoking data available.
The British Women’s Heart and Health Study
The study randomly selected and recruited 4286 women aged 60–79 years, from 23 British towns, between 1999 and 2001. The women were interviewed, examined, completed medical questionnaires and had detailed reviews of their medical records. Full selection and baseline measurement details of the study, including measurement of height and weight (assessed without shoes and in light clothing to the nearest millimetre and 0.1 kg, respectively), have been previously reported.24,25 Smoking history (never, former and current, including the amount and details of starting and quitting) were self-reported at baseline, either at the research nurse interview or in the mailed questionnaires. Serum samples collected at baseline were assayed for cotinine levels in exactly the same way as in the British Regional Heart Study (BRHS).23 After excluding participants of known non-White ethnicity, or who had missing rs1051730 genotype, smoking or BMI data, there were 3634 women available for analysis.
The CoLaus Study
This is a population-based study of 6188 individuals, aged 35–75 years, randomly selected from the list of residents in Lausanne, Switzerland, between 2003 and 2006.26 Risk factors for cardiovascular diseases were assessed and DNA and plasma samples were collected for the study of genetic variants and biomarkers. Between 2004 and 2008, all 35- to 66-year-old individuals of the CoLaus sample were invited to participate in a psychiatric sub-study (PsyCoLaus).27 Detailed descriptions of recruitment procedures and assessments have been provided previously.26,27 The physical assessment within the CoLaus study included measurement of body weight and height, from which BMI was calculated. Ever smokers were those who had smoked regularly at some point in their life. Of these, individuals who smoked at least one cigarette daily at the time of interview were classified as current smokers, whereas the rest were classified as former smokers. For the purpose of genetic analysis, only individuals with European origin were included (n = 5692), and of these, a total of 5426 participants had the genotype and phenotype data necessary for inclusion in the current study.
The Exeter Family Study of Childhood Health
This is a prospective study of children born between 2000 and 2004, and their parents, from a geographically defined region of Exeter, UK.28 Data collection on smoking behaviour have been described previously.21 In the current study, we included 811 pregnant women and 762 men of European ancestry. We grouped them as never, former or current smokers on the basis of questionnaire data collected during the 28th week of gestation. Height and weight were measured at that time by research midwives and used to calculate BMI. Since women were pregnant at the time of data collection, we analysed men and women separately in the current study. For the women-only analysis, we used pre-pregnancy BMI (calculated from self-reported pre-pregnancy weight).
The Danish GOYA Male Study
Among 362 200 Caucasian men examined at the mean age of 20 years at the draft boards in Copenhagen and its surroundings during 1943–77, a randomly selected control group of 1 in every 100 men (n = 3601) and all obese men (n = 1930) were manually identified. Obesity was defined as 35% overweight relative to a local standard in use at the time, and this corresponds to a BMI ≥ 31.0 kg/m2, which proved to be above the 99th percentile. All obese men and half of the random sample, still living in the region, were invited to a follow-up survey in 1992–94 at the mean age of 46 years. The criteria for invitation to the follow-up surveys and the participation have been described previously.29–31 A total of 1441 men (661 obese and 780 randomly selected) were included in the present study with BMI, smoking status and rs1051730 genotype data available.
The MIDSPAN Family Study
This is one of the four MIDSPAN population cohort studies based in Scotland.32 The three original studies took place between 1964 and 1976. Twenty years later, in 1996, the next generation was studied when offspring of couples in the original Renfrew/Paisley Study were recruited into the Family Study. This latter group is the subject of the present analysis. The offspring were ascertained (by self-report) to be from full-sibling families with no step children, adoptees or half-siblings. All were White and living in the west of Scotland. Details of the study have been described previously.33 Standing height was measured without shoes. A single measurement (to the nearest 1 mm) was made after participants inhaled and stretched to reach their maximum height. Weight was measured to the nearest 0.1 kg without shoes and wearing indoor clothes. Data on smoking habit (never, current or former) were collected via questionnaires completed by the participants. Serum samples were assayed for cotinine at ABS Laboratories Ltd, London, UK. For smokers and non-smokers, capillary column gas–liquid chromatography with a nitrogen detector was used. The lower limit of detection was 0.1 ng/ml.23 In the current analyses, we included 2106 individuals of European ancestry with rs1051730 genotype, BMI and smoking data available.
Genotyping
The genotyping of the rs1051730 variant in the ALSPAC and The Exeter Family Study of Childhood Health (EFSOCH) studies has been described previously.21 In the British Women’s Heart and Health Study (BWHHS) and BRHS studies, genotyping of rs1051730 was performed by KBioscience (http://www.kbioscience.co.uk), using KASPar chemistry, which is a competitive allele-specific PCR SNP genotyping system. The respective genotyping call rates were 98.2 and 98.7%, and there was no evidence of deviation from Hardy–Weinberg Equilibrium (HWE; BWHHS P = 0.58; BRHS P = 0.29). In the BWHHS, blind duplicates (concordance 100%) were used as additional quality control tests. In the MIDSPAN study, genotyping was performed on an ABI PRISM 7900HT sequence detection system using a Taqman assay (Assay ID: C_9510307_20, Applied Biosystems), followed by allelic discrimination using software from Applied Biosystems (SDS V2.0).34,35 All genotyping errors were manually resolved by checking raw genotype data, individuals were either blanked (zeroed) or corrected prior to analysis. The call rate was 94.5% and there was no evidence of deviation from HWE (P = 0.92), which was assessed using the eldest sibling from each family to ensure a sample of unrelated individuals (n = 1386).
In the CoLaus study, genome-wide SNP genotyping was performed using the Affymetrix 500 K SNP chip. Samples were excluded if they showed gender inconsistencies or had an efficiency <90%. Marker quality control resulted in exclusion of SNPs that were monomorphic (4052), had a call rate <95% (30 873), deviated (P < 10−3) from HWE (35 417) or had minor allele frequencies <0.01 (61 654), leaving a total number of 370 162 genotyped markers for analysis. Imputation of SNP genotypes was performed using the software IMPUTE v.0.5.036 based on the HapMap CEU II samples. Approximately 2.5 million markers were imputed. A total of 89 SNPs (34 genotyped and 55 imputed) within the CHRNA5-CHRNA3-CHRNB4 gene region were obtained. The SNP rs1051730 was imputed in the study sample with a posterior call rate of 0.97.
In the GOYA study, genome-wide SNP genotyping was performed using the Illumina 610 K Quad SNP chip. The rs1051730 SNP was genotyped on this platform. Individuals were excluded if they showed gender inconsistencies, if they had a genotyping success rate of <95%, if they had outlying heterozygosity (<0.30 or >0.35) or if they clustered with individuals of non-European ancestry on multi-dimensional scaling analysis. We also excluded one individual from each pair that appeared to be related (identity by descent > 0.2). The rs1051730 SNP had an overall call rate of 99.9% and the Hardy–Weinberg P-values were 0.21 and 0.44 in the controls and obese cases, respectively.
Statistical methods
Within-study analyses
Within each study, we verified that the association between smoking quantity and rs1051730 genotype was consistent in magnitude and direction with published data (data not shown, but the association has been previously published for the ALSPAC, CoLaus and EFSOCH women studies14,21). We then stratified by smoking status in each study (for our main analyses into never and ever strata, and for our second analyses into finer strata of never, former and current). To test for association between BMI and rs1051730 genotype within each stratum, we performed linear regression of log-transformed BMI against genotype (coded as 0, 1 or 2 T-alleles; additive genetic model). We repeated the analysis including age and sex (where appropriate) as covariables. We then analysed the association between BMI and genotype, adjusting for smoking quantity (cigarettes per day) where available, in the current and former smoker strata. In addition, we analysed the association between BMI and genotype, adjusting for measured cotinine levels (available in the BRHS, BWHHS and MIDSPAN studies only). In the MIDSPAN study, which includes related individuals, associations between BMI and SNP genotype were investigated using linear mixed effects regression models fitted using restricted maximum likelihood (REML). Family structure among the MIDSPAN offspring generation was accounted for by fitting random intercepts within sibships.
Meta-analysis
Meta-analysis statistics and plots were produced using the inverse-variance method (fixed effects), implemented in the ‘metan’ module developed for Stata (StataCorp, College Station, TX, USA).37 We pooled regression coefficients and standard errors from the linear regression analyses performed in the individual studies. We used the I2 statistic to estimate the percentage of total variation in study estimates that is because of between-study heterogeneity38 and derived 95% confidence intervals (95% CIs) for I2 using the user-written Stata command, ‘heterogi’. Conventionally, I2 values of 25, 50 and 75% represent low, moderate and high heterogeneity, respectively. In addition, we used Cochran’s Q-test to evaluate the statistical evidence for between-study heterogeneity, both within and between the strata defined by smoking status. The test of heterogeneity between strata served as a test of interaction between genotype and smoking status. As a sensitivity analysis, we repeated the meta-analysis excluding the ALSPAC and EFSOCH women studies, which analysed pre-pregnancy BMI, but which stratified by smoking status (never/former/current) using data collected during pregnancy.
Estimation of approximate overall effect sizes in kilogram per square metre
Since the outcome was natural log transformed, we obtained the percentage change in the average value of BMI per allele using 100 x [exp(beta)–1], where ‘beta’ is the effect size on the logBMI scale. We took the median study BMI within each stratum (never/former/current/ever) as the average value.
Results
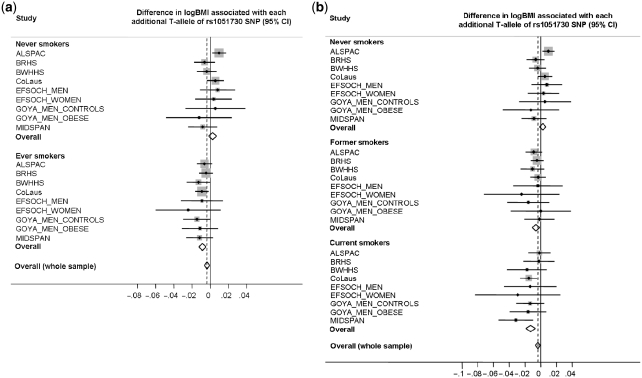
The basic characteristics of study participants are presented in Table 1. The associations between BMI and the rs1051730 variant are presented (overall and by study) in Figure 1 and Tables 2 and 3. In the ever smokers, each additional T-allele, which is associated with greater smoking quantity, was associated with a 0.9% (95% CI: 0.5–1.2) lower BMI (P = 8 × 10−6). This equates to a difference of 0.23 kg/m2 (95% CI: 0.13–0.31), relative to the median study BMI for ever smokers (25.8 kg/m2). However, in the never smokers, we observed no evidence of association between BMI and the genotype [0.2% BMI change (−0.2 to 0.7)], which equates to 0.05 kg/m2 (95% CI: −0.05 to 0.18) per T-allele (median study BMI for never smokers = 25.5 kg/m2; P = 0.25). There was low heterogeneity within each of the two strata [I2 = 21% (95% CI: 0–62), P = 0.26 for never; I2 = 0% (95% CI: 0–65), P = 0.91 for ever smokers], but strong evidence of heterogeneity between the strata (P = 0.0001), indicating an interaction between genotype and smoking status in their association with BMI.
Table 1.
Basic characteristics of study participants
| Study | Sample group | Participants, na (% male) | Age, median (IQR) | BMI, median (IQR) |
|---|---|---|---|---|
| ALSPACb | Never smokers | 3193 (0) | 29 (26–32) | 22.2 (20.5–24.4) |
| Former smokers | 1851 (0) | 29 (25–32) | 22.4 (20.7–24.8) | |
| Current smokers | 1104 (0) | 26 (24–30) | 22.0 (20.3–24.7) | |
| Ever smokers | 2955 (0) | 28 (25–31) | 22.3 (20.5–24.8) | |
| All | 6148 (0) | 28 (25–32) | 22.2 (20.6–24.5) | |
| BRHS | Never smokers | 1124 (100) | 67 (64–72) | 26.3 (24.3–28.5) |
| Former smokers | 2243 (100) | 69 (65–74) | 26.9 (24.9–29.4) | |
| Current smokers | 503 (100) | 68 (64–72) | 25.9 (23.4–28.1) | |
| Ever smokers | 2746 (100) | 69 (64–74) | 26.7 (24.6–29.2) | |
| All | 3870 (100) | 68 (64–73) | 26.6 (24.5–29.0) | |
| BWHHS | Never smokers | 2047 (0) | 69 (64–73) | 27.0 (24.0–30.0) |
| Former smokers | 1194 (0) | 69 (64–74) | 27.0 (25.0–31.0) | |
| Current smokers | 393 (0) | 67 (63–72) | 26.0 (23.0–29.0) | |
| Ever smokers | 1587 (0) | 69 (64–73) | 27.0 (24.0–30.0) | |
| All | 3634 (0) | 69 (64–73) | 27.0 (24.0–30.0) | |
| CoLaus | Never smokers | 2193 (38.0) | 53 (44–62) | 25.3 (22.6–28.3) |
| Former smokers | 1801 (54.9) | 56 (47–63) | 25.8 (23.1–28.8) | |
| Current smokers | 1432 (51.2) | 50 (43–60) | 24.7 (22.1–27.7) | |
| Ever smokers | 3233 (53.3) | 53 (45–62) | 25.3 (22.6–28.3) | |
| All | 5426 (47.1) | 53 (44–62) | 25.3 (22.6–28.3) | |
| EFSOCH MEN | Never smokers | 376 (100) | 32 (30–36) | 26.6 (24.6–28.7) |
| Former smokers | 181 (100) | 33 (30–37) | 26.5 (24.5–29.2) | |
| Current smokers | 205 (100) | 32 (27–25) | 26.2 (23.2–29.2) | |
| Ever smokers | 386 (100) | 32 (29–36) | 26.4 (24.0–29.2) | |
| All | 762 (100) | 32 (29–36) | 26.5 (24.2–29.0) | |
| EFSOCH_WOMENb | Never smokers | 593 (0) | 31 (29–34) | 22.9 (21.1–25.3) |
| Former smokers | 118 (0) | 29 (25–32) | 23.4 (21.0–25.5) | |
| Current smokers | 100 (0) | 29 (24–32) | 23.7 (20.9–27.7) | |
| Ever smokers | 218 (0) | 29 (25–32) | 23.5 (20.9–26.6) | |
| All | 811 (0) | 31 (27–34) | 23.0 (21.1–25.5) | |
| GOYA MEN CONTROLS | Never smokers | 174 (100) | 44 (40–49) | 25.4 (23.5–28.7) |
| Former smokers | 216 (100) | 48 (42–54) | 26.2 (24.2–28.4) | |
| Current smokers | 390 (100) | 47 (42–55) | 25.6 (23.6–27.8) | |
| Ever smokers | 606 (100) | 47 (42–55) | 25.8 (23.7–28.1) | |
| All | 780 (100) | 46 (41–53) | 25.8 (23.6–28.2) | |
| GOYA MEN OBESE | Never smokers | 163 (100) | 41 (37–45) | 35.8 (32.2–39.9) |
| Former smokers | 148 (100) | 43 (40–48) | 35.2 (32.7–40.3) | |
| Current smokers | 350 (100) | 42 (38–47) | 34.5 (31.6–38.2) | |
| Ever smokers | 498 (100) | 42 (39–47) | 34.6 (31.8–38.6) | |
| All | 661 (100) | 42 (38–46) | 34.9 (32.0–39.1) | |
| MIDSPAN | Never smokers | 996 (43.0) | 45 (40–49) | 25.5 (23.1–28.4) |
| Former smokers | 574 (49.7) | 47 (42–50) | 26.3 (24.0–29.2) | |
| Current smokers | 536 (44.2) | 46 (40–50) | 24.6 (22.1–28.1) | |
| Ever smokers | 1110 (47.0) | 46 (41–50) | 25.6 (23.0–28.8) | |
| All | 2106 (45.1) | 45 (41–49) | 25.5 (23.0–28.6) |
Ever smokers: former plus current. IQR: inter-quartile range.
aN included in main analysis.
bPre-pregnancy BMI was analysed in ALSPAC and EFSOCH_WOMEN.
Figure 1.
Meta-analysis plots of the association between the rs1051730 variant and BMI, stratified by smoking status. (a) Never/ever smokers. There was strong evidence of heterogeneity between strata (P = 0.0001). In the ever smokers, the effect size equates to a 0.23 kg/m2 (95% CI: 0.13−0.31) lower BMI per T-allele. In the never smokers, there was no evidence of association [BMI difference per T-allele: 0.05 kg/m2 (95% CI: −0.05 to 0.18)]. (b) Never/former/current smokers. There was strong evidence of heterogeneity among the three strata (P = 0.0002). In the former and current smokers, there was a 0.16 kg/m2 (95% CI: 0.03–0.29) and 0.33 kg/m2 (95% CI: 0.18–0.48) lower BMI per T-allele, respectively. Overall, across all smoking status strata, each additional T-allele was associated with a 0.10 kg/m2 (95% CI: 0.03–0.18) lower BMI
Table 2.
Associations between BMI and rs1051730 genotype in all study participants, stratified by smoking status (never/ever) and by study
| No. and BMI (95% CI) by rs1051730 genotype |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Study | Total no. of participants | CC | CT | TT | Per-T allele change in ln(BMI) (95% CI)a | P-valuea | |||
| Never smokers | ALSPAC | 3193 | 1412 | 22.5 (22.3 to 22.7) | 1403 | 22.8 (22.6 to 23.0) | 378 | 22.8 (22.5 to 23.2) | 0.010 (0.002 to 0.017) | 0.01 |
| BRHS | 1124 | 475 | 26.6 (26.2 to 26.9) | 514 | 26.2 (26.0 to 26.5) | 135 | 26.3 (25.7 to 27.0) | −0.006 (−0.018 to 0.005) | 0.27 | |
| BWHHS | 2047 | 898 | 27.1 (26.8 to 27.4) | 910 | 27.2 (26.9 to 27.5) | 239 | 26.7 (26.1 to 27.3) | −0.004 (−0.015 to 0.007) | 0.49 | |
| CoLaus | 2193 | 884 | 25.5 (25.2 to 25.7) | 1016 | 25.6 (25.4 to 25.9) | 293 | 25.7 (25.3 to 26.2) | 0.005 (−0.004 to 0.015) | 0.25 | |
| EFSOCH_M | 376 | 179 | 26.5 (26.0 to 27.0) | 160 | 26.5 (26.0 to 27.0) | 37 | 27.1 (26.0 to 28.4) | 0.008 (−0.012 to 0.027) | 0.43 | |
| EFSOCH_W | 593 | 264 | 23.4 (23.0 to 23.9) | 272 | 23.7 (23.2 to 24.1) | 57 | 23.4 (22.3 to 24.5) | 0.004 (−0.017 to 0.024) | 0.73 | |
| GOYA_MC | 174 | 73 | 25.7 (24.8 to 26.6) | 83 | 26.2 (25.5 to 27.0) | 18 | 25.5 (23.8 to 27.4) | 0.005 (−0.027 to 0.038) | 0.75 | |
| GOYA_MO | 163 | 74 | 36.2 (35.0 to 37.5) | 68 | 36.0 (34.6 to 37.4) | 21 | 35.2 (32.5 to 38.1) | −0.012 (−0.048 to 0.024) | 0.50 | |
| MIDSPAN | 996 | 447 | 25.9 (25.5 to 26.3) | 455 | 25.9 (25.5 to 26.3) | 94 | 25.1 (24.4 to 25.9) | −0.008 (−0.024 to 0.007) | 0.30 | |
| Overall | 10 859 | 0.002 (−0.002 to 0.007) | 0.25 | |||||||
| Overallb | 10 859 | 0.002 (−0.002 to 0.007) | 0.25 | |||||||
| Ever smokers | ALSPAC | 2955 | 1323 | 22.9 (22.7 to 23.1) | 1308 | 22.7 (22.5 to 22.9) | 324 | 22.7 (22.3 to 23.1) | −0.006 (−0.015 to 0.002) | 0.13 |
| BRHS | 2746 | 1290 | 26.9 (26.7 to 29.1) | 1162 | 26.7 (26.4 to 26.9) | 294 | 26.7 (26.3 to 27.1) | −0.005 (−0.012 to 0.003) | 0.23 | |
| BWHHS | 1587 | 700 | 27.4 (27.0 to 27.7) | 723 | 27.1 (26.8 to 27.5) | 164 | 26.6 (25.9 to 27.3) | −0.013 (−0.026 to 0.000) | 0.05 | |
| CoLaus | 3233 | 1302 | 25.6 (25.4 to 25.8) | 1509 | 25.3 (25.1 to 25.5) | 422 | 25.2 (24.8 to 25.5) | −0.009 (−0.017 to −0.002) | 0.01 | |
| EFSOCH_M | 386 | 182 | 26.4 (25.8 to 27.0) | 162 | 26.4 (25.8 to 27.1) | 42 | 26.4 (25.8 to 27.1) | −0.009 (−0.032 to 0.014) | 0.43 | |
| EFSOCH_W | 218 | 103 | 24.2 (23.3 to 25.1) | 91 | 24.2 (23.3 to 25.0) | 24 | 22.5 (21.2 to 24.0) | −0.024 (−0.060 to 0.011) | 0.18 | |
| GOYA_MC | 606 | 276 | 26.1 (25.8 to 26.5) | 250 | 25.8 (25.4 to 26.2) | 80 | 25.4 (24.6 to 26.1) | −0.015 (−0.030 to 0.000) | 0.06 | |
| GOYA_MO | 498 | 220 | 35.2 (34.5 to 35.9) | 219 | 34.7 (34.0 to 35.4) | 59 | 34.5 (33.1 to 35.9) | −0.011 (−0.031 to 0.009) | 0.26 | |
| MIDSPAN | 1110 | 510 | 26.0 (25.6 to 26.4) | 479 | 25.6 (25.3 to 26.0) | 121 | 25.2 (24.5 to 26.0) | −0.012 (−0.027 to 0.003) | 0.12 | |
| Overall | 13 339 | −0.009 (−0.012 to −0.005) | 8 × 10−6 | |||||||
| Overallb | 13 339 | −0.008 (−0.012 to −0.004) | 3 × 10−5 | |||||||
BMI values are geometric means and 95% CIs.
aFrom linear regression of ln(BMI) against genotype, assuming an additive genetic model.
bAdjusted for age and sex. Meta-analysis results for per-allele changes in logBMI (95% CI), excluding the pregnant women from the ALSPAC and EFSOCH_WOMEN studies were: −0.001 (−0.006 to 0.004) for never smokers and −0.009 (−0.013 to −0.005) for ever smokers.
EFSOCH_M, EFSOCH_MEN; EFSOCH_W, EFSOCH_WOMEN; GOYA_MC, GOYA_MEN_CONTROLS; GOYA_MO, GOYA_MEN_OBESE.
Table 3.
Associations between BMI and rs1051730 genotype in the ever smokers, stratified by smoking status (former/current) and by study
| No. and BMI (95% CI) by rs1051730 genotype |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Study | Total no. of participants | CC | CT | TT | Per-T allele change in ln(BMI) (95% CI)a | P-valuea | |||
| Former smokers | ALSPAC | 1851 | 858 | 23.0 (22.8 to 23.3) | 806 | 22.8 (22.6 to 23.0) | 187 | 22.6 (22.2 to 23.1) | −0.009 (−0.019 to 0.002) | 0.10 |
| BRHS | 2243 | 1067 | 27.1 (26.9 to 27.3) | 934 | 27.0 (26.8 to 27.2) | 242 | 26.8 (26.3 to 27.2) | −0.005 (−0.013 to 0.003) | 0.26 | |
| BWHHS | 1194 | 532 | 27.7 (27.3 to 28.2) | 542 | 27.6 (27.2 to 28.0) | 120 | 26.9 (26.2 to 27.7) | −0.010 (−0.025 to 0.005) | 0.18 | |
| CoLaus | 1801 | 738 | 26.0 (25.7 to 26.2) | 838 | 25.9 (25.7 to 26.2) | 225 | 25.8 (25.3 to 26.3) | −0.003 (−0.013 to 0.007) | 0.51 | |
| EFSOCH_M | 181 | 88 | 26.5 (25.7 to 27.3) | 73 | 27.5 (26.7 to 28.4) | 20 | 25.2 (23.8 to 26.6) | −0.004 (−0.035 to 0.028) | 0.82 | |
| EFSOCH_W | 118 | 61 | 23.9 (22.9 to 25.0) | 48 | 23.9 (22.9 to 25.1) | 9 | 21.9 (20.0 to 24.0) | −0.024 (−0.072 to 0.024) | 0.33 | |
| GOYA_MC | 216 | 100 | 26.7 (24.8 to 27.4) | 92 | 26.3 (25.6 to 27.0) | 24 | 25.8 (24.3 to 27.5) | −0.016 (−0.043 to 0.011) | 0.25 | |
| GOYA_MO | 148 | 63 | 35.5 (34.3 to 36.8) | 69 | 35.8 (34.5 to 37.3) | 16 | 35.2 (32.1 to 38.4) | 0.000 (−0.038 to 0.038) | 1.00 | |
| MIDSPAN | 574 | 251 | 26.7 (26.2 to 27.2) | 260 | 26.3 (25.9 to 26.8) | 63 | 26.9 (25.9 to 27.8) | −0.002 (−0.021 to 0.017) | 0.86 | |
| Overall | 8326 | −0.006 (−0.011 to −0.001) | 0.01 | |||||||
| Overallb | 8326 | −0.006 (−0.010 to −0.001) | 0.02 | |||||||
| Overallc | 4169 | −0.010 (−0.017 to −0.003) | 0.008 | |||||||
| Current smokers | ALSPAC | 1104 | 465 | 22.7 (22.4 to 23.0) | 502 | 22.5 (22.2 to 22.8) | 137 | 22.8 (22.2 to 23.4) | −0.002 (−0.016 to 0.013) | 0.82 |
| BRHS | 503 | 223 | 25.9 (25.4 to 26.4) | 228 | 25.2 (24.7 to 25.7) | 52 | 26.4 (25.4 to 27.5) | −0.002 (−0.022 to 0.017) | 0.82 | |
| BWHHS | 393 | 168 | 26.2 (25.6 to 26.9) | 181 | 25.6 (25.0 to 26.3) | 44 | 25.5 (24.1 to 27.0) | −0.017 (−0.043 to 0.008) | 0.18 | |
| CoLaus | 1432 | 564 | 25.2 (24.9 to 25.5) | 671 | 24.6 (24.3 to 24.8) | 197 | 24.6 (24.1 to 25.1) | −0.015 (−0.026 to −0.004) | 0.006 | |
| EFSOCH_M | 205 | 94 | 26.3 (25.5 to 27.1) | 89 | 25.5 (24.7 to 26.5) | 22 | 26.0 (24.2 to 28.0) | −0.014 (−0.047 to 0.020) | 0.43 | |
| EFSOCH_W | 100 | 42 | 24.6 (23.0 to 26.3) | 43 | 24.4 (23.2 to 25.7) | 15 | 22.9 (21.0 to 24.9) | −0.029 (−0.084 to 0.025) | 0.30 | |
| GOYA_MC | 390 | 176 | 25.8 (25.4 to 26.3) | 158 | 25.5 (25.0 to 26.1) | 56 | 25.2 (24.3 to 36.0) | −0.013 (−0.031 to 0.005) | 0.15 | |
| GOYA_MO | 350 | 157 | 35.1 (34.2 to 35.9) | 150 | 34.2 (33.4 to 35.0) | 43 | 34.2 (32.8 to 35.8) | −0.016 (−0.039 to 0.007) | 0.18 | |
| MIDSPAN | 536 | 259 | 25.3 (24.8 to 25.9) | 219 | 24.8 (24.2 to 25.4) | 58 | 23.5 (22.6 to 24.6) | −0.032 (−0.054 to −0.009) | 0.007 | |
| Overall | 5013 | −0.013 (−0.019 to −0.007) | 6 × 10−5 | |||||||
| Overallb | 5013 | −0.012 (−0.018 to −0.006) | 0.0001 | |||||||
| Overallc | 4084 | −0.014 (−0.021 to −0.007) | 7 × 10−5 | |||||||
| Overalld | 1364 | −0.010 (−0.023 to 0.006) | 0.13 | |||||||
BMI values are geometric means and 95% CIs.
aFrom linear regression of ln(BMI) against genotype, assuming an additive genetic model.
bAdjusted for age and sex.
cAdjusted for smoking quantity.
dAdjusted for cotinine levels.
Current smoking quantity was available in all nine study samples; former smoking quantity was available in ALSPAC, BWHHS, CoLaus and MIDSPAN only. Cotinine levels were available in BRHS, BWHHS and MIDSPAN current smokers only [for comparison, the pooled per allele change in logBMI (95% CI), adjusted for smoking quantity, in BRHS, BWHHS and MIDSPAN was −0.020 (−0.033 to −0.006)].
Meta-analysis results for per-allele changes in logBMI (95% CI), excluding the pregnant women from the ALSPAC and EFSOCH studies were: −0.005 (−0.010 to 0.000) for former smokers and −0.015 (−0.022 to −0.008) for current smokers.
EFSOCH_M, EFSOCH_MEN; EFSOCH_W, EFSOCH_WOMEN; GOYA_MC, GOYA_MEN_CONTROLS; GOYA_MO, GOYA_MEN_OBESE.
When dividing the ever-smoker group into current and former smokers, we observed evidence of association in both strata (Figure 1b and Table 3), but with a larger effect size in current smokers. In the former and current smokers, respectively, there was a 0.6% (95% CI: 0.1–1.1) and 1.3% (95% CI: 0.7–1.9) lower BMI per T-allele. These effect sizes equate, respectively, to differences of 0.16 kg/m2 (95% CI: 0.03–0.29) and 0.33 kg/m2 (95% CI: 0.18–0.48), relative to the median study BMIs in these groups (26.3 kg/m2 former and 25.6 kg/m2 current). There was no detectable heterogeneity within these strata [I2 = 0% (95% CI: 0–65), P = 0.97 for former; I2 = 0% (95% CI: 0–65), P = 0.55 for current smokers], but strong evidence of heterogeneity among the never, former and current smoker strata (P = 0.0002).
Overall, across all smoking status strata, each additional T-allele was associated with a 0.4% (95% CI: 0.1–0.7) lower BMI (P = 0.007; Figure 1), which equates to a difference of 0.10 kg/m2 (95% CI: 0.03–0.18) (median overall study BMI = 25.8 kg/m2).
The results were not materially altered on adjustment for age and sex (Tables 2 and 3). A sensitivity analysis excluding the ALSPAC and EFSOCH pregnant women produced very similar effect size estimates (Tables 2 and 3).
Adjustment for smoking quantity, where available, in the current and former smokers also did not substantially change the results (Table 3). Measured cotinine levels were available in the current smokers of the BRHS, BWHHS and MIDSPAN studies. Adjustment for cotinine levels resulted in attenuation of the association between genotype and BMI [1.0% (95% CI: –0.3 to 2.3)] lower per T-allele (P = 0.13), relative to the association adjusted for reported smoking quantity in BRHS, BWHHS and MIDSPAN only: 2.0% lower BMI (95% CI: 0.6–3.2, P = 0.005; Table 3).
Discussion
In a meta-analysis of 24 198 individuals from nine study samples, we have shown that there is an interaction between smoking status and genotype of the 15q25 variant, rs1051730, in relation to BMI. In never smokers, we observed no evidence of association between rs1051730 genotype and BMI. However, in ever smokers, there was strong evidence that each additional copy of the T-allele, which is associated with higher smoking quantity, was associated with a 0.23 kg/m2 (95% CI: 0.13–0.31) lower BMI (interaction P = 0.0001). The association in the ever smokers was consistent across studies (heterogeneity P = 0.91), despite variation in mean BMI because of age and ascertainment differences.
Our results support the hypothesis that exposure to cigarette smoking is causally associated with lower BMI. Genotype–phenotype associations are not biased by reverse causality, and genotypes are generally randomized to factors that usually confound epidemiological studies.18 We have shown previously that the rs1051730 variant, which is robustly associated with smoking quantity,10–16 is not associated with covariates of smoking behaviour including age, education level and occupational position.21 We can, therefore, be confident that the association we have observed between rs1051730 genotype and BMI in ever smokers, together with the null association in never smokers, reflects a causal relationship between smoking exposure and lower BMI. Our data add support to evidence from previous epidemiological, experimental and physiological studies.5–7,9,39
Subdivision of the ever-smoker group showed that the effect size was smaller in former smokers [0.16 kg/m2 (95% CI: 0.03–0.29) lower BMI per T-allele] than in current smokers [0.33 kg/m2 (95% CI: 0.18–0.48) lower BMI per T-allele]. This suggests that the association with BMI weakens over time after cessation. It is consistent with the observation that cigarette smokers who attempt to quit experience a short-term rebound effect, whereby a marked initial weight gain following smoking cessation is a result of the removal of the lowered body weight set point induced by chronic nicotine use,7 and reduced sympathovagal ratio and resensitization of nicotinic receptors.39 Smoking cessation is associated with a corresponding average increase in BMI,9 and comparisons of BMI over time suggest that when cigarette smokers achieve long-term abstinence from tobacco they revert to a mean BMI roughly equivalent to that of never smokers. Weight gain on cessation of smoking is associated with increased risk of relapse into smoking.4 Taken together, these observations suggest that smoking cessation initiatives may be more successful if accompanied by support to maintain a healthy BMI.
We did not observe attenuation of the associations between genotype and BMI upon adjustment for smoking quantity in the former and current smokers. We would expect these associations to get weaker if, as we hypothesize, the genotype–BMI association is mediated by smoking quantity. However, since data on smoking quantity were self-reported, they were unlikely to have captured the exposure fully. Moreover, adjustment for cotinine levels in the available samples did result in attenuation, and our observation that there was no association between genotype and BMI in the never smokers is consistent with the associations observed being mediated by smoking. The observation that the effect of the rs1051730 variant on BMI is robust to adjustment for smoking quantity suggests that, even within heaviness of smoking strata, the rs1051730 variant exerts an influence on exposure, possibly via individual differences in smoking topography (e.g. depth of puff inhalation, number of puffs). This further supports a causal association between exposure and BMI, both between and within heaviness of smoking strata, and suggests that simple measures of heaviness of smoking based on reported quantity may mask important inter-individual variation in behaviour and subsequent exposure.
Although the association between rs1051730 genotype and smoking quantity has mostly been described in Europeans, it has also been observed in populations of African–American and Korean descent.40–42 Further work is needed to investigate the relationships between genotype, smoking and BMI in non-European populations.
We acknowledge some limitations to our study. First, in the ALSPAC and EFSOCH studies, pre-pregnancy BMI was analysed, whereas smoking data were collected during pregnancy. Therefore, the temporal distinction between former and current smokers may have been different from that in the other studies. However, a sensitivity analysis excluding these studies produced very similar effect size estimates of the associations within each smoking status stratum. Secondly, different methods and instruments were used for assessing weight and height, as well as smoking in the studies that were pooled here. However, we found little evidence for marked heterogeneity between studies within strata. Thirdly, our statistical evidence for association with BMI in the ever smokers (P = 8 × 10−6), and for the genotype × smoking status interaction (P = 0.0001), although strong, is modest in relation to levels now necessary for robust candidate gene association studies. However, the fact that the 15q25 variant is now incontrovertibly associated with smoking quantity greatly increases the prior odds of association.
To conclude, the 15q25 smoking quantity variant is associated with a lower BMI in former and current smokers, but not in individuals who have never smoked. This is a clear example of a gene × environment interaction because the association between genotype and BMI is dependent upon smoking status. Our observations support the hypothesis that smoking is causally associated with reduced BMI. Smoking cessation initiatives might be more successful if they include support to maintain a healthy BMI at the same time as quitting smoking.
Funding
The UK Medical Research Council, the Wellcome Trust and the University of Bristol (to ALSPAC); BRHS is a British Heart Foundation research group (current programme grant RG/08/013/25942); British Heart Foundation Senior Research Fellowship (award FS05/125, partial, to BRHS for DNA extraction); Department of Health (England) Policy Research Programme and the British Heart Foundation (to BWHHS); GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, Switzerland, and the Swiss National Science Foundation (33CSCO-122661) (to The CoLaus Study); Swiss National Science Foundation (#3200B0–105993, #3200B0-1180308, 33CSC0122661) and GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Drug Discovery—Verona, R&D) (to The PsyCoLaus Study); South West NHS Research and Development, Exeter NHS Research and Development, the Darlington Trust, and the Peninsula NIHR Clinical Research Facility at the University of Exeter (to EFSOCH); Wellcome Trust (grant WT084762) (to The Danish GOYA male study); Wellcome Trust and the NHS Research and Development Programme (to the offspring study in MIDSPAN); Sir Henry Wellcome Postdoctoral Fellowship (Wellcome Trust grant 085541/Z/08/Z) (to R.M.F.); UK Medical Research Council (G0600705) and the University of Bristol (to G.D.S. and D.A.L.).
Acknowledgements
The authors are extremely grateful to the participants and families who contributed to all of the studies and the teams of investigators involved in each one. These include interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, nurses and, for the CoLaus study, many GlaxoSmithKline (GSK) employees (especially Emiliangelo Ratti for supporting this work within GSK Drug Discovery). They are grateful to Colin Feyerabend for performing the cotinine assays in the MIDSPAN study.
Conflict of interest: The CoLaus study was partially funded by GSK. F.T., G.R.K. and V.M. are (or were at the time that the study was performed) full-time employees of GSK. P.V. and G.W. received unrestricted grant from GSK to build the CoLaus study. M.P. received an unrestricted grant from GSK to conduct the PsyCoLaus substudy.
KEY MESSAGES.
A genetic variant on chromosome 15 is known to be associated with a higher smoking quantity in smokers. We show that the higher smoking quantity genotypes are also associated with a lower BMI in current and former smokers.
There is no association between genotype and BMI in never smokers.
Since genotype is assigned at conception and reflects smoking exposure in smokers, our results strengthen evidence that smoking is causally related to lower BMI.
References
- 1.Doll R, Peto R, Boreham J, Sutherland I. Mortality in relation to smoking: 50 years' observations on male British doctors. BMJ. 2004;328:1519. doi: 10.1136/bmj.38142.554479.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes JR. Motivating and helping smokers to stop smoking. J Gen Intern Med. 2003;18:1053–57. doi: 10.1111/j.1525-1497.2003.20640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomerleau CS, Zucker AN, Stewart AJ. Characterizing concerns about post-cessation weight gain: results from a national survey of women smokers. Nicotine Tob Res. 2001;3:51–60. doi: 10.1080/14622200020032105. [DOI] [PubMed] [Google Scholar]
- 4.Borrelli B, Spring B, Niaura R, Hitsman B, Papandonatos G. Influences of gender and weight gain on short-term relapse to smoking in a cessation trial. J Consult Clin Psychol. 2001;69:511–15. doi: 10.1037//0022-006x.69.3.511. [DOI] [PubMed] [Google Scholar]
- 5.Jo YH, Talmage DA, Role LW. Nicotinic receptor-mediated effects on appetite and food intake. J Neurobiol. 2002;53:618–32. doi: 10.1002/neu.10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li MD, Kane JK, Konu O. Nicotine, body weight and potential implications in the treatment of obesity. Curr Top Med Chem. 2003;3:899–919. doi: 10.2174/1568026033452203. [DOI] [PubMed] [Google Scholar]
- 7.Cabanac M, Frankham P. Evidence that transient nicotine lowers the body weight set point. Physiol Behav. 2002;76:539–42. doi: 10.1016/s0031-9384(02)00783-7. [DOI] [PubMed] [Google Scholar]
- 8.Sneve M, Jorde R. Cross-sectional study on the relationship between body mass index and smoking, and longitudinal changes in body mass index in relation to change in smoking status: the Tromso Study. Scand J Public Health. 2008;36:397–407. doi: 10.1177/1403494807088453. [DOI] [PubMed] [Google Scholar]
- 9.Munafo MR, Tilling K, Ben-Shlomo Y. Smoking status and body mass index: a longitudinal study. Nicotine Tob Res. 2009;11:765–71. doi: 10.1093/ntr/ntp062. [DOI] [PubMed] [Google Scholar]
- 10.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berrettini W, Yuan X, Tozzi F, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–47. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey Smith G, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey Smith G. Mendelian randomization for strengthening causal inference in observational studies: application to gene by environment interaction. Perspectives on Psychol Sci. 2010;5:527–45. doi: 10.1177/1745691610383505. [DOI] [PubMed] [Google Scholar]
- 20.Golding J, Pembrey M, Jones R. ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 21.Freathy RM, Ring SM, Shields B, et al. A common genetic variant in the 15q24 nicotinic acetylcholine receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) is associated with a reduced ability of women to quit smoking in pregnancy. Hum Mol Genet. 2009;18:2922–27. doi: 10.1093/hmg/ddp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker M, Whincup PH, Shaper AG. The British Regional Heart Study 1975–2004. Int J Epidemiol. 2004;33:1185–92. doi: 10.1093/ije/dyh295. [DOI] [PubMed] [Google Scholar]
- 23.Feyerabend C, Russell MA. A rapid gas–liquid chromatographic method for the determination of cotinine and nicotine in biological fluids. J Pharm Pharmacol. 1990;42:450–52. doi: 10.1111/j.2042-7158.1990.tb06592.x. [DOI] [PubMed] [Google Scholar]
- 24.Lawlor DA, Bedford C, Taylor M, Ebrahim S. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women's Heart and Health Study. J Epidemiol Community Health. 2003;57:134–40. doi: 10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawlor DA, Ebrahim S, Davey Smith G. Association of birth weight with adult lung function: findings from the British Women's Heart and Health Study and a meta-analysis. Thorax. 2005;60:851–58. doi: 10.1136/thx.2005.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firmann M, Mayor V, Vidal PM, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preisig M, Waeber G, Vollenweider P, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight B, Shields BM, Hattersley AT. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol. 2006;20:172–79. doi: 10.1111/j.1365-3016.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 29.Black E, Holst C, Astrup A, et al. Long-term influences of body-weight changes, independent of the attained weight, on risk of impaired glucose tolerance and Type 2 diabetes. Diabet Med. 2005;22:1199–205. doi: 10.1111/j.1464-5491.2005.01615.x. [DOI] [PubMed] [Google Scholar]
- 30.Kring SI, Larsen LH, Holst C, et al. Genotype–phenotype associations in obesity dependent on definition of the obesity phenotype. Obes Facts. 2008;1:138–45. doi: 10.1159/000137665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonne-Holm S, Sorensen TI, Jensen G, Schnohr P. Independent effects of weight change and attained body weight on prevalence of arterial hypertension in obese and non-obese men. BMJ. 1989;299:767–70. doi: 10.1136/bmj.299.6702.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart CL, MacKinnon PL, Watt GC, et al. The Midspan studies. Int J Epidemiol. 2005;34:28–34. doi: 10.1093/ije/dyh348. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Rmeileh NM, Hart CL, McConnachie A, Upton MN, Lean ME, Watt GC. Contribution of Midparental BMI and other determinants of obesity in adult offspring. Obesity. 2008;16:1388–93. doi: 10.1038/oby.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–80. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–49. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 36.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 37.Harris R, Bradburn M, Deeks J, et al. METAN: Stata Module for Fixed and Random Effects Meta-Analysis. Statistical Software Components S456798, Boston College Department of Economics, revised 19 Feb 2007. http://ideasrepecorg/c/boc/bocode/s456798html (22 July 2010, date last accessed) [Google Scholar]
- 38.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun AJ, Bazar KA, Lee PY, Gerber A, Daniel SM. The smoking gun: many conditions associated with tobacco exposure may be attributable to paradoxical compensatory autonomic responses to nicotine. Med Hypotheses. 2005;64:1073–79. doi: 10.1016/j.mehy.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 40.Saccone NL, Wang JC, Breslau N, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African–Americans and in European–Americans. Cancer Res. 2009;69:6848–56. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–56. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li MD, Yoon D, Lee JY, et al. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010;5:e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]