Abstract
Aim
Tamoxifen biotransformation to endoxifen, a potent antiestrogen, is catalyzed by CYP2D6. In addition, CYP2C19 and SULT1A1 have also been implicated in the metabolism of tamoxifen. We sought to evaluate the importance of SULT1A1 copy number and CYP2C19*17 on disease-free survival (DFS) in postmenopausal women randomized to tamoxifen monotherapy in North Central Cancer Treatment Group 89-30-52 from January 1991 to April 1995.
Materials & methods
We extracted DNA from paraffin-embedded tumors and determined tumor SULT1A1 copy number and CYP2C19*17 genotype. The association of genotype with DFS was determined using the log-rank test. Multivariate cox modeling was performed using traditional prognostic factors, as well as CYP2D6 genotype. SULT1A1 copy number and CYP2C19*17 genotype was determined in 190 out of 256 patients (95% Caucasian).
Results
The median follow-up for living patients was 14 years. DFS did not differ according to SULT1A1 copy number (p = 0.482) or CYP2C19*17 genotype (p = 0.667). Neither SULT1A1 copy number or CYP2C19*17 genotype was associated with disease recurrence in this cohort.
Conclusion
Future studies are needed to identify whether other genetic and environmental factors which affect tamoxifen metabolism are associated with tamoxifen clinical outcomes.
Keywords: breast cancer, copy number polymorphism, CYP2C19, pharmacogenomic, polymorphism, single nucleotide, SULT1A1, tamoxifen
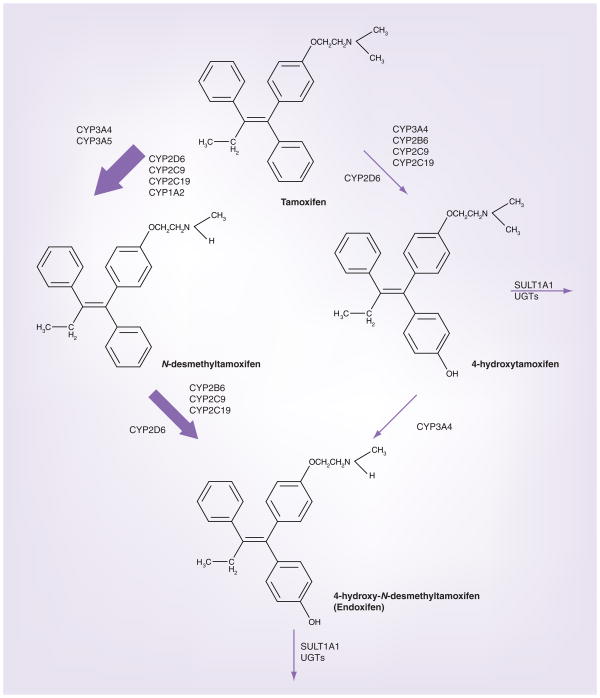
Tamoxifen is a selective estrogen receptor (ER) modulator utilized for the treatment and prevention of ER-positive breast cancer. The risk of recurrence and mortality of breast cancer is reduced by nearly a half and a third, respectively, when women with ER-positive breast cancer are treated with tamoxifen [1]. Many enzymes are important in tamoxifen metabolism. The parent drug, tamoxifen, is converted in to N-desmethyl-tamoxifen, 4-hydroxy (4-OH) tamoxifen, and endoxifen (4-hydroxy-N-desmethyl tamoxifen) by the action of several CYP enzymes, including CYP2D6 (Figure 1) [2–6]. Tamoxifen is inactivated by glucuronidation through UGT2B15 and UGT1A4, and through sulfation, SULT1A1 (Figure 1) [7,8].
Figure 1. In vitro biotransformation pathways of tamoxifen.
The relative contribution of each pathway to the overall oxidation of tamoxifen is shown by the thickness of the arrow.
Endoxifen is the most abundant active metabolite of tamoxifen. Its steady-state plasma concentrations are five- to ten-fold higher than 4-OH tamoxifen, yet it is identical to 4-OH tamoxifen in its ER-binding affinity and ability to suppress estradiol-stimulated cell proliferation [2,6,9,10]. However, recent data demonstrated that the mechanism of action of these two selective estrogen receptor modulators may differ, given that unlike 4-OH tamoxifen or the parent drug tamoxifen, endoxifen uniquely targets ERα for proteasomal degradation [11].
Genetic and environmental variation in drug metabolizing enzymes, particularly CYP2D6, has an effect on tamoxifen biotransformation and efficacy. Compared with women with normal or increased CYP2D6 activity, endoxifen concentrations are significantly reduced in women with low CYP2D6 activity [6,10]. Considering the adjuvant therapy setting, there have been conflicting data regarding the association between impaired CYP2D6 metabolism (either genotype and/or drug inhibitor use) and tamoxifen treatment outcome [12–21]. This heterogeneity is well illustrated in the data recently presented from three large adjuvant clinical breast cancer trials. While no association of CYP2D6 genotype with breast cancer recurrence was observed in the Arimidex, Tamoxifen, Alone or in Combination (ATAC) and the Breast International Group (BIG) 1–98 clinical trials [14,15], CYP2D6 genotype was associated with an increase in the odds of breast cancer recurrence in the Austrian Breast and Colorectal Cancer Study Group (ABCCSG) 8 trial [22]. However, this was only observed in patients that received tamoxifen monotherapy, and not in those that received anastrozole following tamoxifen [22]. The latter finding suggests that crossover to an active drug not metabolized by CYP2D6 (anastrozole) may affect the association between CYP2D6 genetic variation and tamoxifen clinical outcome. In addition, it should be noted that there are substantial differences in many of the published studies with regard to study population, study design or variability in assignment of CYP2D6 genotype and/or metabolizer status.
Although many studies have focused on the impact of CYP2D6 genetic variation and clinical outcome, few have considered the role of genetic variation in other enzymes mediating either endoxifen generation or inactivation. Recent studies have suggested that, in addition to CYP2D6, genetic variation in genes encoding drug-metabolizing enzymes, such as CYP2C19 and SULT1A1, may be associated with treatment outcome [16,23]. Recently, new variants of CYP2C19 have been identified, including CYP2C19*17 [24]. This variant is characterized by two SNPs in the 5′-flanking region of the gene, which are in tight linkage disequilibrium, resulting in higher enzyme activity. The extent of the increased activity and the clinical impact of this variant has been controversial [25]. Several studies have observed an increase in tamoxifen metabolites and better clinical outcomes associated with the variant allele, and some studies have found no difference from the wild-type [16,18,26]. In the case of patients with decreased CYP2D6 activity, in principle, increased CYP2C19 activity with the *17 variant may be able to compensate; however, studies investigating the combination of CYP2D6 and CYP2C19*17 genotype in tamoxifen metabolism and testing this hypothesis are scant presently.
We recently discovered that the SULT1A1 liver enzyme activity was highly correlated with SULT1A1 copy number, which explained the majority of the variation in SULT1A1 in vitro activity after considering all known sources of SULT1A1 genetic variation (e.g., SULT1A1*2) [27]. Previous studies have not identified an association between SULT1A1 SNPs and clinical outcome; therefore, we sought to evaluate the potential association between SULT1A1 copy number and survival in women enrolled in North Central Cancer Treatment Group (NCCTG) 89-30-52 who were treated with tamoxifen monotherapy [21,26,28]. In addition, we sought to determine the association between the CYP2C19*17 allele with clinical outcome – with our entire cohort, as well as the subset of patients with impaired CYP2D6 activity.
Materials & methods
Patient samples (NCCTG 89-30-52)
The North Central Cancer Treatment Group conducted a randomized Phase III clinical trial in postmenopausal women with resected ER-positive breast cancer to assess the addition of fluoxymesterone to tamoxifen in year 1 of the first 5 years of adjuvant therapy (NCCTG 89-30-52). Details and results from this trial have been reported elsewhere [29]. Briefly, this study enrolled postmenopausal women of any age with T1cN0M0 or T2N0M0 and women 65 years of age or older with a tumor stage T1N1M0 or T2N1M0. The invasive breast tumor must have been positive for ER by a standard biochemical assay (≥ 10 fmol/mg cytosol protein) or by immunohistochemistry. Patients were surgically treated with either a modified radical mastectomy or breast conservative therapy, including lumpectomy, axillary nodal dissection, and radiation therapy. A paraffin-embedded tumor block was submitted to the NCCTG operations office. From January 1991 to April 1995, 541 women were enrolled. Our current study utilized the 256 women randomized to the tamoxifen-only arm. The study was approved by the institutional review board of the Mayo Clinic (Rochester, MN, USA) and the individual NCCTG sites that enrolled patients onto the clinical trial.
Genotyping
For each patient, three 10-μm thick tissue sections were prepared from a block of their formalin-fixed paraffin-embedded breast tumor and the entire paraffin section was scraped from the slides, and DNA extracted using a modification of the method utilized by Schroth et al. [17].
SULT1A1 copy number genotype was determined by quantitative multiplex PCR as described previously [27]. In brief, PCR was performed with primers that co-amplified a 212-bp fragment spanning exons 3 and 4 of SULT1A1 and a 208-bp fragment within SULT1A2. The amplicons were separated by size and quantitated on an Applied Biosystems 3730 (Carlsbad, CA, USA). The peak height, corresponding to quantity, for both the 208-bp fragment (SULT1A1) and the 212-bp fragment (SULT1A2) were measured using GeneMarker® version 1.51 (SoftGenetics, PA, USA), and the ratio between the peak heights determined copy number.
CYP2C19*17 (−806C>T) was genotyped by first amplifying an approximately 360-bp segment in the promoter region of CYP2C19 and then sequencing in both the forward and reverse directions. Sequence chromatograms for each sample were analyzed using Mutation Surveyor version 3.12 (SoftGenetics) and comparing to a reference sequence (NT_030059). Samples were characterized as homozygous wild-type, heterozygous or *17/*17 at the −806 locus.
Statistical analysis
The primary aim of this study was to assess whether disease-free survival (DFS) differed with respect to SULT1A1 copy number or CYP2C19*17 genotype. DFS was defined as the time from randomization to documentation of the first of the following events: local, regional or distant recurrence, contralateral ductal carcinoma in situ or breast cancer, other nonbreast invasive second primary cancers, or death from any cause. Patients who were alive without any of these events were censored at the date of their last disease evaluation. The log-rank test and the generalized Wilcoxon tests were utilized to assess whether DFS differed with respect to genotype. Multivariate cox modeling was performed to examine the strength of association between genotype and DFS after adjusting for traditional prognostic factors. As SULT1A1 activity has been shown to be similar among patients with more than two gene copies [27] and less than 5% of the study cohort had one gene copy, we chose to examine whether clinical outcome differed between those with two or fewer gene copies and those with more than two gene copies.
In addition, the impact of SULT1A1 copy number and CYP2C19*17 genotype on DFS was assessed in the subset of patients with impaired CYP2D6 metabolism, defined as those with a reduced (*10, *17 and *41) or null (*3, *4 and *6) allele or any patient taking a weak or potent CYP2D6 inhibitor, as described previously [30].
Results
Paraffin-embedded tumor blocks from primary surgical specimens were available from 223 out of the 256 women randomized to the tamoxifen-only arm of NCCTG 89-30-52. The study cohort consists of the 190 women for whom DNA extraction and genotyping was successful for either SULT1A1 copy number, CYP2C19*17 or both. The pretrial characteristics of the study cohort and the 66 women enrolled onto NCCTG 89-30-52 who lacked genotype data are listed in Table 1. Tumor size in the 66 women who lacked genotype data tended to be smaller (<3 cm in size) compared with those with genotype data (Fisher’s exact p = 0.053).
Table 1.
Patient and tumor characteristics prior to tamoxifen treatment for women enrolled in North Central Cancer Treatment Group 89-30-52 with genotyping results (the present study cohort) and those without genotyping results.
| Patient/tumor characteristics | Those with genotyping data available
|
Those without genotyping data available
|
|---|---|---|
| Study cohort (n = 190) (%) | Excluded (n = 66) (%) | |
|
Age
| ||
| <65 years | 26.8 | 36.4 |
|
| ||
| ≥65 years | 73.2 | 63.6 |
|
| ||
|
Race
| ||
| Caucasian | 94.7 | 84.9 |
|
| ||
| African–American | 1.6 | 3.0 |
|
| ||
| American Indian or Alaska Native | 0.5 | 0 |
|
| ||
| Not reported | 3.2 | 12.1 |
|
| ||
|
Extent of surgery
| ||
| Mastectomy | 83.7 | 75.8 |
|
| ||
| Breast conservation | 16.3 | 24.2 |
|
| ||
|
Tumor size
| ||
| <3 cm | 75.8 | 87.9 |
|
| ||
| ≥3 cm | 24.2 | 12.1 |
|
| ||
|
Estrogen receptors
| ||
| 10–49 fmols | 19.5 | 21.2 |
|
| ||
| ≥50 fmols | 68.4 | 60.6 |
|
| ||
| Positive | 12.1 | 18.2 |
|
| ||
|
Number of positive nodes
| ||
| 0 | 60.5 | 68.2 |
|
| ||
| 1–3 | 27.9 | 16.6 |
|
| ||
| 4–9 | 7.9 | 9.1 |
|
| ||
| 10+ | 3.7 | 6.1 |
|
| ||
|
CYP2D6 genotype
| ||
| EM/EM | 43.7 | 18.2 |
|
| ||
| EM/IM, EM/PM, IM/IM or IM/PM | 50.0 | 13.6 |
|
| ||
| PM/PM | 4.2 | 3.0 |
|
| ||
| Unknown | 2.1 | 65.2 |
|
| ||
|
CYP2D6 inhibitor use while receiving tamoxifen
| ||
| Potent inhibitor† | 1.6 | 0 |
|
| ||
| Weak inhibitor‡ | 5.3 | 1.5 |
|
| ||
| None | 80.5 | 89.4 |
|
| ||
| Unknown | 12.6 | 9.1 |
|
| ||
|
CYP2C19*17 genotype
| ||
| wt/wt | 57.4 | – |
|
| ||
| wt/*17 | 25.3 | – |
|
| ||
| *17/*17 | 6.8 | – |
|
| ||
| Unknown | 10.5 | – |
|
| ||
|
SULT1A1 copy number
| ||
| 1 | 3.7 | – |
|
| ||
| 2 | 60.0 | – |
|
| ||
| 3 | 17.4 | – |
|
| ||
| 4+ | 7.9 | – |
|
| ||
| Unknown | 11.1 | – |
Potent inhibitors are fluoxetine or paroxetine.
Weak inhibitors are sertraline, cimetidine, amiodarone, doxepin, ticlopidine or haloperidol.
EM: *1 or *2 allele; IM: *10, *17 or *41 allele; PM: *3, *4 or *6 allele; wt: Wild-type.
The median length of follow-up among the 86 women still living was 14.3 years (range: 5.7–18.7 years). At the time of writing, 70 women were alive without a disease event, four were alive with disease progression, 12 were alive with second primary disease, 38 had died following disease progression, 15 died following development of a second primary disease, two committed suicide, 32 died of other causes without disease progression and 17 died of unknown causes.
SULT1A1 and CYP2C19*17 genotyping was successful in 169 and 170 cases, respectively. The observed SULT1A1 copy numbers were one copy (4.1%), two copies (67.5%), three copies (19.5%) and four or more copies (8.9%). The CYP2C19*17 minor allele frequency was 22%, and the observed phenotypes were wild-type (wt)/wt (64.1%), wt/*17 (28.2%) and *17/*17 (7.6%). The genotype frequencies for CYP2C19*17 were in Hardy–Weinberg equlibrium.
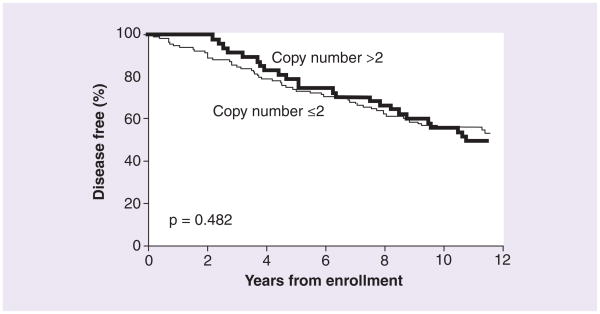
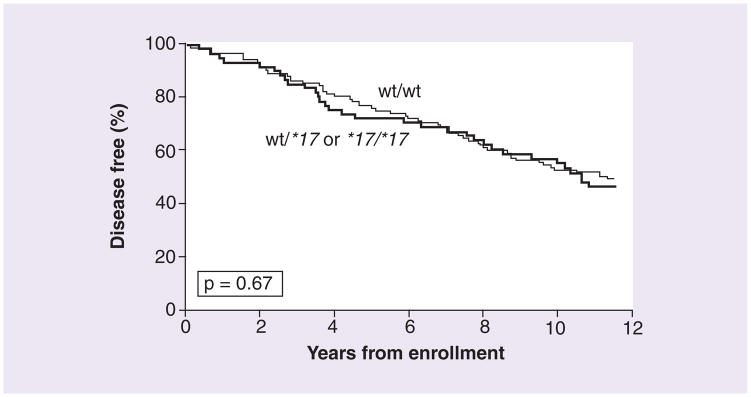
In a univariate analysis, DFS was not found to differ with respect to SULT1A1 copy number (>2 vs ≤2), p = 0.482 (Figure 2), or with respect to CYP2C19*17 genotype (wt/wt vs wt/*17 and *17/*17), p = 0.667 (Figure 3). In addition, after adjusting for tumor size and nodal status, neither SULT1A1 copy number (adjusted hazard ratio [HR]: 1.18; 95% CI: 0.76–1.83), nor CYP2C19*17 genotype was found to be associated with DFS (HR: 0.93; 95% CI: 0.64–1.37). In the subset of patients with impaired CYP2D6 activity, DFS was not found to differ with respect to SULT1A1 copy number (>2 vs ≤2, p = 0.198, n = 98) or CYP2C19*17 genotype (p = 0.871, n = 100). This finding held after adjusting for tumor size and nodal status (SULT1A1, HR: 1.2, 95% CI: 0.66–2.11; CYP2C19*17, HR: 0.97; 95% CI: 0.59–1.59).
Figure 2.
Percentage of patients remaining disease-free over time, analyzed by SULT1A1 copy number.
Figure 3.
Percentage of patients remaining disease-free over time, analyzed by CYP2C19*17 genotype
wt: Wild-type.
Discussion & conclusion
Using samples from patients randomized to tamoxifen monotherapy in NCCTG 89-30-52, in which the median follow-up for living patients is 14 years, we performed SULT1A1 copy number and CYP2C19*17 genotyping to determine whether these genetic variants were associated with the clinical outcome of disease-free survival. This study expands our previous CYP2D6 work to a more comprehensive study of the role of genetic variation in tamoxifen metabolism. In our earlier reports [30,31], which were recently updated in the context of a much larger cohort [32] of women with ER positive breast cancer treated with tamoxifen who underwent comprehensive CYP2D6 genotyping [17], there was a significant association between CYP2D6 genotype/phenotype and DFS. However, we found no association between CYP2C19*17 or SULT1A1 copy number and DFS in this study, either in the entire cohort or in those with impaired CYP2D6 metabolism.
SULT1A1 copy number, which we have found to be highly associated with SULT1A1 activity, was predicted to potentially be a biomarker for tamoxifen response because it metabolizes 4-OH tamoxifen and endoxifen, resulting in inactivation of the 4-OH metabolites. Therefore, in principle, high SULT1A1 activity could result in decreased activity and concentrations of the active metabolites of tamoxifen. While previous studies have not demonstrated a consistent association between SULT1A1 SNPs and tamoxifen clinical outcome [18,21,23], no studies have evaluated the association between SULT1A1 copy number with DFS in tamoxifen-treated patients [18,19,21,23,26,28]. In this study, we did not identify an association between SULT1A1 copy number and DFS, either in the entire group or the subset with impaired CYP2D6 activity. While our data suggest that we can rule out a large difference in DFS based on SULT1A1 copy number (i.e., HR > 1.8), further studies with larger sample size and ability to subset patients by CYP2D6 activity would be required to reveal small differences in outcome based on SULT1A1, if there is an effect.
CYP2C19 was predicted to be an important biomarker for response to tamoxifen because it has similar in vitro activities to CYP2D6 and can also catalyze the conversion of tamoxifen to endoxifen [33]. CYP2C19*17 has a relatively high minor allele frequency (24%) in individuals of European descent and results in ultra-rapid metabolism. Findings from the subset of women in our cohort with impaired CYP2D6 metabolism did not provide evidence to suggest that the CYP2C19*17 polymorphism is associated with DFS in this subgroup. However, similar to our findings with SULT1A1 copy number, a much larger patient cohort would be necessary to identify a small effect size of CYP2C19 genotype and to fully elucidate the interplay of CYP2D6 and CYP2C19 genetic variation. While proton pump inhibitors are known to influence CYP2C19 enzyme activity, and therefore may affect tamoxifen metabolism, patients in this study were enrolled in the pre-proton pump inhibitor era, so it is unlikely to impact our findings. Finally, a recent study by Gjerde et al. demonstrated an association between CYP2C19 genotype and not only tamoxifen metabolite levels, but also estrogen levels [26]. Another recent report suggested that increased catabolism of estrogens by CYP2C19 may lead to decreased estrogen level and reduced risk of breast cancer [34]. Additional studies are necessary to elucidate the role of estrogen levels during tamoxifen therapy to fully understand the role of variation in enzymes that affect both tamoxifen and estrogen levels in breast cancer hormonal therapy.
There are several limitations to our study. First, this is a secondary analysis of a prospective study involving primarily Caucasian (95%) women, so it is unclear how these results apply to other populations. Notably, the frequency of the SULT1A1 copy number variation is much higher in subjects of African descent, where up to 62% may have increased copy number. Second, given the complexity of the tamoxifen pathway and the many enzymes involved in activation, inactivation and regulation of tamoxifen, the impact of a single variation can be disguised by other compensating variables, yet potentially important in some clinical contexts. Our sample size (n = 190 patients) was relatively small, thereby limiting our ability to detect small but potentially clinically important differences in DFS. Therefore, if SULT1A1 copy number or CYP2C19*17 resulted in a small difference in the steady state concentrations of endoxifen, a much larger patient cohort may be required to detect their importance. Third, we were unable to assess for the common null CYP2C19 alleles (e.g., *2 and *3), which significantly affect CYP2C19 enzyme activity [35]. Although other studies have not shown an association between these variants with steady concentrations of endoxifen [35] or with tamoxifen clinical outcomes [16], further studies are necessary to fully evaluate these low-activity alleles. Finally, our study did not evaluate the important UGT isoforms that have previously been implicated in the glucuronidation of tamoxifen and the active metabolites. While recent reports have not demonstrated an association between UGT genetic variants and endoxifen concentrations [35], additional studies will be necessary to determine whether genetic and environmental factors that affect glucuronidation are associated with the DFS in tamoxifen treated patients.
Genetic variation in CYP2D6 is the strongest predictor of endoxifen concentrations, accounting for 38% of the variation in concentration of z-endoxifen; however, a substantial amount of the variation in endoxifen steady state concentrations are unexplained by CYP2D6 genetic variation [35]. Furthermore, the association between CYP2D6 genotype and tamoxifen clinical outcome continues to be controversial. Therefore, additional studies are needed to identify additional environmental and genetic markers that would allow caregivers to individualize treatment for women with ER-positive breast cancer.
Future perspective
Tamoxifen metabolism is complex, with multiple different enzymes generating metabolites with varying degrees of affinity for the ER. While our study did not demonstrate evidence of an association between either SULT1A1 copy number or CYP2C19*17 genotype and disease-free survival in tamoxifen treated breast cancer, future studies in larger patient cohorts evaluating these and additional genetic and environmental factors that are associated with variation in tamoxifen metabolism are needed, given the possibility of a small effect size that is overshadowed by other variables including drug interactions, hormonal status and combinations of genetic variation.
Our study demonstrates the complexity of developing personalized therapy for the treatment of breast cancer. Ideally, as additional drugs are developed to effectively treat breast cancer, biomarkers (both host and tumor) predictive of drug effect will be established that will allow physicians to individualize therapy. Achievement of that goal will require that these biomarkers not only be reproducible and validated in well annotated cohorts, but these biomarkers will require prospective validation as well.
Executive summary.
Background
In addition to CYP2D6, SULT1A1 and CYP2C19 have been suggested to play a role in tamoxifen metabolism.
The number of copies of SULT1A1 varies among individuals and the activity of the enzyme is correlated with the number of copies of the gene.
CYP2C19*17 has recently been identified as a new allele, characterized by two SNPs in the 5′-flanking region, and is associated with higher enzyme activity.
Methods & materials
A total of 190, primarily Caucasian, tamoxifen-treated women enrolled in North Central Cancer Treatment Group 89-30-52 with estrogen receptor-positive early breast cancer, were genotyped for SULT1A1 copy number and CYP2C19*17, and evaluated for an association between genotype and disease-free survival.
Results
No association was identified for either marker, either adjusting for traditional prognostic factors or not.
No association was identified for either marker in the subset of women with impaired CYP2D6 activity.
Conclusion
Further studies in a larger cohort will be important to understand if these genetic variants have a small, but important effect on outcome in tamoxifen treatment.
Further pharmacogenomic studies involving additional genes will be necessary to fully understand the complex biochemical pathways regulating tamoxifen therapy and to individualize hormonal therapy in breast cancer.
Acknowledgments
The authors would like to thank Raquel Ostby for her assistance in the preparation of this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported in part by NIH grants CA90628 (Matthew P Goetz and Vera J Suman), Mayo Clinic Breast Cancer Specialized Program of Research Excellence CA116201 (Matthew P Goetz, Vera J Suman and James N Ingle), and U01 GM61388 (Matthew P Goetz, James N Ingle and Richard M Weinshilboum). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 3.Lien EA, Anker G, Lonning PE, Solheim E, Ueland PM. Decreased serum concentrations of tamoxifen and its metabolites induced by aminoglutethimide. Cancer Res. 1990;50:5851–5857. [PubMed] [Google Scholar]
- 4.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- 5.Lien EA, Solheim E, Ueland PM. Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment. Cancer Res. 1991;51:4837–4844. [PubMed] [Google Scholar]
- 6.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 7.Nishiyama T, Ogura K, Nakano H, et al. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol. 2002;63:1817–1830. doi: 10.1016/s0006-2952(02)00994-2. [DOI] [PubMed] [Google Scholar]
- 8.Ogura K, Ishikawa Y, Kaku T, et al. Quaternary ammonium-linked glucuronidation of trans-4-hydroxytamoxifen, an active metabolite of tamoxifen, by human liver microsomes and UDP-glucuronosyltransferase 1a4. Biochem Pharmacol. 2006;71:1358–1369. doi: 10.1016/j.bcp.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome p450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 10.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC. The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor α for degradation in breast cancer cells. Cancer Res. 2009;69:1722–1727. doi: 10.1158/0008-5472.CAN-08-3933. [DOI] [PubMed] [Google Scholar]
- 12.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10:825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leyland-Jones B, Regan MM, Bouzyk M, et al. Outcome according to CYP2D6 genotype among postmenopausal women with endocrine-responsive early invasive breast cancer randomized in the BIG 1–98 trial. Cancer Res. 2011;70:Abstracts S1–S8. [Google Scholar]
- 15.Rae JM, Drury S, Hayes DF, et al. Lack of correlation between gene variants in tamoxifen metabolizing enzymes with primary endpoints in the ATAC trial. Cancer Res. 2010;70:Abstracts S1–S7. [Google Scholar]
- 16.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 17▪▪.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. CYP2D6 variation was found to be associated with clinical outcomes. Specifically, the presence of two functional CYP2D6 alleles was associated with better clinical outcomes than the presence of nonfunctional or reduced-function alleles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano D, Lazzeroni M, Zambon CF, et al. Efficacy of tamoxifen based on cytochrome p450 2D6, CYP2C19 and SULT1A1 genotype in the italian tamoxifen prevention trial. Pharmacogenomics J. 2011;11:100–107. doi: 10.1038/tpj.2010.17. [DOI] [PubMed] [Google Scholar]
- 19.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 21.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnant M, Mlineritsch B, Stoeger H, et al. Mature results from ABCSG-12: adjuvant ovarian suppression combined with tamoxifen or anastrozole, alone or in combination with zolendronic acid, in premenopausal women with endocrine-responsive early breast cancer. J Clin Oncol. 2010;28:Abstract 533. [Google Scholar]
- 23.Nowell S, Sweeney C, Winters M, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94:1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 24▪.Sim SC, Risinger C, Dahl ML, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. Characterizes a novel CYP2C19*17 allele that has increased transcriptional activity resulting in increased drug metabolism. [DOI] [PubMed] [Google Scholar]
- 25.Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant. CYP2C19*17. Br J Clin Pharmacol. 2010;69:222–230. doi: 10.1111/j.1365-2125.2009.03578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjerde J, Geisler J, Lundgren S, et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer. 2010;10:313. doi: 10.1186/1471-2407-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27▪.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16:463–470. doi: 10.1093/hmg/ddl468. Characterizes a copy number polymorphism of the SULT1A1 gene as well as several promoter SNPs. The authors demonstrate that although promoter haplotypes influence enzyme activity, the number of copies of SULT1A1 present best explains the level of enzyme activity, which may have pharmacogenetic implications. [DOI] [PubMed] [Google Scholar]
- 28.Grabinski JL, Smith LS, Chisholm GB, et al. Genotypic and allelic frequencies of SULT1A1 polymorphisms in women receiving adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2006;95:13–16. doi: 10.1007/s10549-005-9019-5. [DOI] [PubMed] [Google Scholar]
- 29.Ingle JN, Suman VJ, Mailliard JA, et al. Randomized trial of tamoxifen alone or combined with fluoxymesterone as adjuvant therapy in postmenopausal women with resected estrogen receptor positive breast cancer. North Central Cancer Treatment Group trial 89-30-52. Breast Cancer Res Treat. 2006;98:217–222. doi: 10.1007/s10549-005-9152-1. [DOI] [PubMed] [Google Scholar]
- 30.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome p450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 31.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 32.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EM. Metabolism of tamoxifen by recombinant human cytochrome p450 enzymes: formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:869–874. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- 34.Justenhoven C, Hamann U, Pierl CB, et al. CYP2C19*17 is associated with decreased breast cancer risk. Breast Cancer Res Treat. 2009;115:391–396. doi: 10.1007/s10549-008-0076-4. [DOI] [PubMed] [Google Scholar]
- 35.Murdter TE, Schroth W, Bacchus-Gerybadze L, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of Phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther. 2011;89:708–717. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]