Abstract
Recent years have witnessed an explosion of research on the role of epigenetic modifications, such as DNA methylation and histone protein acetylation and phosphorylation, in neuroscience. These changes exert control over gene expression and have been shown to play important roles in a variety of neural processes, including learning and memory. We and others have also recently shown that epigenetic changes may contribute to neurodegenerative disorders, such as Alzheimer’s disease. Western blot analysis with antibodies raised against specific histone modifications is a relatively simple technique able to reveal the type, location, and degree of histone posttranslational modifications produced by an experimental manipulation. Here we provide a step-by-step protocol for isolating histone proteins from tissue and measuring these posttranslational modifications.
Keywords: Epigenetics, Histone, Chromatin, Acetylation, Phosphorylation, Methylation, Western blot
1. Introduction
The term epigenetics, coined by Conrad Waddington (1), refers to modifications that occur above (“epi-”) the genetic level. These changes do not alter the underlying DNA nucleotide sequence, but are capable of persisting through cell division and influencing gene transcription. The most commonly studied epigenetic modifications are DNA methylation and chromatin modification. This chapter will focus on a method used to assay the latter, chromatin modifications.
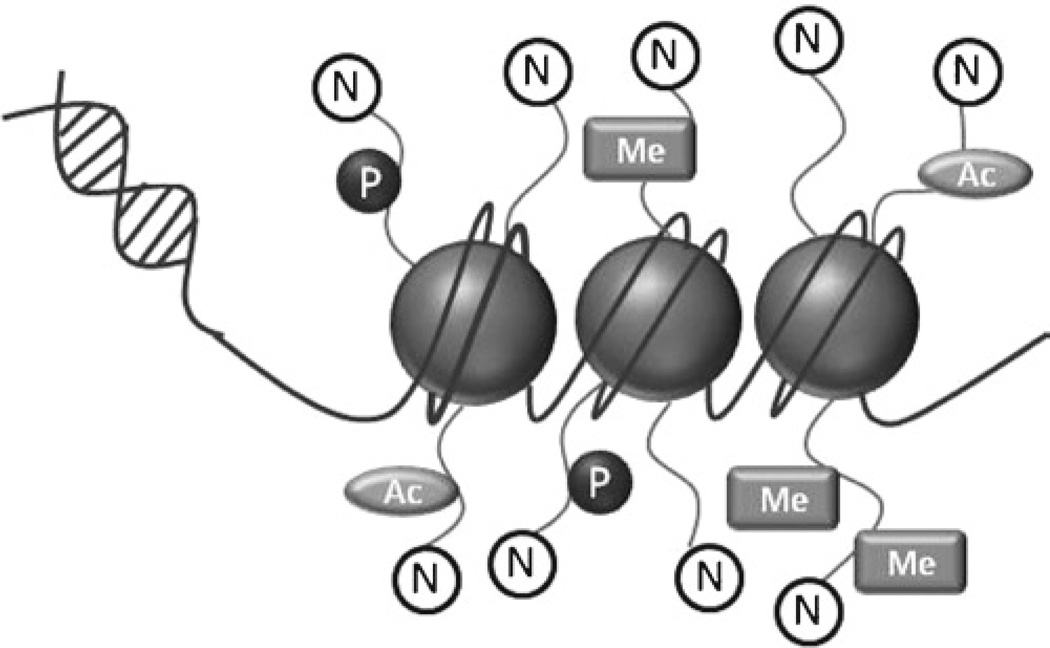
Chromatin consists of a complex of DNA and core histone proteins, which act as a spool to package DNA to fit inside the nucleus. These core histone proteins (H2A, H2B, H3, and H4) are subject to posttranslational modification on their N-terminal tails. The most common modifications are acetylation, phosphorylation, and methylation, all of which are capable of influencing the rate of gene transcription by controlling physical access to the gene (Fig. 1). Both phosphorylation and acetylation are associated with transcriptional activation, while histone methylation’s transcriptional consequences are dependent on the number and location of the methyl groups (2).
Fig. 1.
Schematic representation of DNA spooled around core histone proteins to form chromatin, along with potential posttranslational modifications (P phosphorylation, Ac acetylation, Me methylation) of the N-terminal tails.
In recent years, the field of neuroscience has begun to realize the importance of epigenetic mechanisms in both development and cognition. A critical role for chromatin modifications in learning has now been demonstrated in several rodent behavioral models, including tasks dependent on the hippocampus and cortex (3–7). Moreover, chromatin modifications play an important role in psychiatric disorders such as schizophrenia, depression, and drug addiction (8).
Histone deacetylases (HDACs) are the enzymes responsible for the removal of acetyl groups from the N-terminal of histones. Several HDAC inhibitors have been developed, including SAHA, a compound approved by the FDA for the treatment of T-cell lymphoma. With the discovery that HDAC inhibitors not only increase acetylation levels, but also enhance cognition (9, 10), a number of laboratories have turned a therapeutic eye to the role of histone modifications in Alzheimer’s disease. In accordance with this, a recent publication from our group reported that memory deficits in a mouse model of Alzheimer’s disease can be rescued by treatment with HDAC inhibitors that specifically target class I HDACs (11).
The following protocol will provide investigators with an excellent technique to begin their own research in the field of epigenetics.
2. Materials
2.1. Extracting Histone Proteins
Tissue to be assayed.
1 M Tris-HCl stock, pH 7.4.
1 M KCl stock. Dissolve 3.73 g of KCl in 50 mL of ddH2O.
100 mg/mL (900 mM) sodium butyrate. Dissolve 100 mg in 1 mL of ddH2O. Store aliquots at −20°C. Sodium butyrate is used as a HDAC inhibitor.
100 mM sodium orthovanadate. Dissolve 18.39 mg of sodium orthovanadate in 1 mL of ddH2O. Store aliquots at −20°C. Sodium orthovanadate is used as a phosphotyrosine phosphatase inhibitor.
0.2 M phenylmethylsulphonyl fluoride (PMSF). Dissolve 34.8 mg of PMSF in 1 mL of 100% EtOH. Store at 4°C. PMSF is used as a serine protease inhibitor.
100× protease inhibitor cocktail (Sigma).
Homogenization buffer: 50 mM Tris-HCl, pH 7.5, 25 mM KCl, 250 mM sucrose, 2 mM sodium butyrate, 1 mM sodium orthovanadate, 0.5 mM PMSF, 1× protease inhibitor cocktail. To make 10 mL, mix 855.8 mg of sucrose, 500 µl of 1 M Tris-HCl stock, and 250 of µl 1 M KCl stock in 9.13 mL of ddH2O. Then add 22 µl of 100 mg/mL sodium butyrate, 100 µl of 100 mM sodium orthovanadate, 25 µl of 0.2 M PMSF, and 100 µl of 100× protease inhibitor cocktail immediately before use. Large volume stocks containing the first three ingredients can be prepared and stored at 4°C, but the inhibitors should always be added immediately before use.
Dounce homogenizers or RNase-free tubes with tissue grinding pestles.
0.4 N H2SO4 (sulfuric acid): Mix 111.12 µl of 18 M H2SO4 in 9.88 mL of ddH2O. Store at 4°C.
Trichloroacetic acid (TCA) with 10 mM sodium deoxycholate. Mix 160 mg of sodium deoxycholate in 40 mL of 6.1 N TCA. Store at 4°C.
Acidified acetone: Mix 5 mL of acetone (HPLC grade) and 5 mL of hydrochloric acid (32–38% trace metal grade). Store at 4°C.
100% acetone. Store at 4°C so it is cold for use.
10 mM Tris-HCl, pH 8.0.
Bradford reagent for protein assay.
2.2. Western Blotting for Posttranslational Histone Modifications
Gel casting and electrophoresis apparatus, including power supply.
30% Acrylamide/Bis (37.5:1) solution.
1.5 M Tris-HCl, stock, pH 8.8.
10% sodium dodecyl sulfate (SDS) solution (w/v). Dissolve 1 g of SDS in 10 mL of ddH2O.
50% glycerol solution (v/v). Disolve 50 mL of glycerol in 50 mL of ddH2O.
10% ammonium persulfate (APS) solution (w/v). Dissolve 1 g of APS in 10 mL of ddH2O, and allow it to settle.
TEMED.
15% Resolving gel solution: 375 mM Tris-HCl, pH 8.8, 15% acrylamide/bis (37.5:1), 0.1% SDS, 2.5% glycerol, 0.1% APS, 0.1% TEMED. To make 40 mL, mix 20 mL of 30% acrylamide, 10 mL of 1.5 M Tris-HCl, 400 µl of 10% SDS, 2 mL of 50% glycerol, and 400 µl of 10% APS with 7.2 mL of ddH2O. Add 40 µl of TEMED immediately prior to pouring gel.
Stacking gel solution: 360 mM Tris-HCl, pH 8.8, 4% acrylamide/bis (37.5:1), 0.1% SDS, 15% glycerol, 0.1% APS, 0.1% TEMED. To make 20 mL, mix 2.6 mL of 30% acrylamide, 4.8 mL of 1.5 M Tris-HCl, 200 µl of 10% SDS, 6 mL of 50% glycerol, and 200 µl of 10% APS with 6.2 mL of ddH2O. Add 20 µl of TEMED immediately prior to pouring gel.
Running buffer: 250 mM glycine, 25 mM Tris-HCl base, 0.1% SDS. To prepare 2 L, mix 37.54 g of glycine, 6 g of Tris-HCl base, and 2 g of SDS in ddH2O.
Protein molecular weight standard.
Transfer buffer: 250 mM glycine, 25 mM Tris-HCl base, 10% methanol. To prepare 2 L, mix 37.54 g of glycine, 6 g of Tris-HCl base, and 200 mL of methanol in ddH2O.
Gel transfer apparatus.
PVDF membrane and filter paper.
Methanol.
Tris-Buffered Saline (TBS): 50 mM Tris-HCl, pH 7.5, 150 mM NaCl. To prepare 2 L, mix 17.54 g of NaCl and 100 mL of 1 M Tris-HCl, pH 7.5, in ddH2O.
TBS with Tween-20 (TBST): 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20. To prepare 1 L, mix 999 mL of TBS with 1 mL of Tween-20.
TBST with 5% bovine serum albumin (BSA): 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20, 5% BSA. To prepare 200 mL, dissolve 10 g of BSA in TBST.
Sealable plastic pouches for antibody incubation.
Primary Antibodies. See Table 1. We use each of these antibodies at 1:1,000, but conditions may need to be reoptimized in each laboratory.
Secondary Antibodies: HRP-conjugated anti-rabbit and anti-mouse IgG (depending on the source of primary antibody).
ECL Western blotting substrate.
Kodak bioMax light film.
0.2 N sodium hydroxide (NaOH): Dilute 10 N NaOH 1:50.
Table 1.
Primary antibodies for Western blotting to detect changes in histone modifications
| To Detect | Antibody | Source |
|---|---|---|
| Acetylated H3 | Rabbit anti-acetyl-histone H3 (Lys9/Lys14) | Cell Signaling Technology 9677 |
| Phosphorylated H3 | Rabbit anti-phospho-histone H3 (Ser28) | Millipore 07-145 |
| Trimethylated H3 | Rabbit anti-trimethyl-histone H3 (Lys4) | Millipore 07-473 |
| Total H3 | Mouse anti-histone H3 (clone 96C10) | Cell Signaling Technology 3638 |
| Acetylated H4 | Rabbit anti-acetyl-histone H4 (Lys5/Lys8/Lys12/Lys16) | Millipore 06-598 |
| Total H4 | Rabbit anti-histone H4 | Millipore 07-108 |
2.3. Analysis of Western Blots
Flat bed scanner.
ImageJ analysis software. You can download this freeware program at http://rsbweb.nih.gov/ij/index.html.
3. Methods
The protocol described below outlines procedures for extraction of histone proteins from a tissue sample, performing the Western blot analysis of histone posttranslational modifications, and quantifying and interpreting the results.
3.1. Extracting Histone Proteins
Prepare two sets of 1.5-mL sterile microfuge tubes per treatment condition.
Prepare fresh homogenization buffer (1.5 mL per sample) according to Subheading 2.1, step 8.
Fill Dounce homogenizers or RNase-free tubes with tissue grinders with 1 mL of homogenization buffer and place on ice (see Note 1).
Transfer tissue from dry ice or −80°C storage into tube and homogenize with approximately six strokes.
Transfer homogenate to microfuge tube.
Centrifuge at 7,700 × g for 1 min at 4°C to pellet nuclei (see Note 2).
Transfer supernatant to a fresh 1.5-mL microfuge tube and store at −20°C if desired (see Note 3).
Resuspend pellets containing nuclear fraction in 500 µl of 0.4 N H2SO4 by thorough trituration. Vortex samples if necessary.
Incubate on ice for 30 min, with brief vortexing every 10 min.
Centrifuge extracts at maximum speed for 10 min at 4°C.
Transfer supernatant to fresh 1.5-mL tube and discard pellet (see Note 4).
Add 250 µl of trichloroacetic acid and sodium deoxycholate to supernatant to precipitate histones.
Slowly invert the tubes to mix by hand; the solution should immediately turn cloudy.
Incubate on ice for 30 min.
Centrifuge solution at maximum speed for 30 min at 4°C.
Discard supernatant (see Note 5), rinse tube with 1 mL of cold, acidified acetone (see Note 6), and invert to mix.
Incubate on ice for 5 min.
Centrifuge at maximum speed for 5 min at 4°C.
Discard supernatant and rinse tube with 1 mL cold (4°C) 100% acetone as in Note 6. This provides a safe method of removing acid from the solution without affecting the histone (protein) pellet. Carefully invert tube to mix and incubate on ice for 5 min.
Centrifuge at maximum speed for 5 min at 4°C.
Decant supernatant and allow the pellet to dry for approximately 5 min (see Note 7).
Resuspend protein extract in 50 µl of 10 mM Tris-HCl, pH 8.0 (see Note 8).
Quantify the protein concentration of each sample using a Bradford reagent-based commercial protein assay kit.
Store sample at −80°C. We prefer to add SDS-PAGE sample buffer before storage, to further protect the samples.
3.2. Western Blotting for Posttranslational Histone Modifications
Determine the number of gels needed and the number of lanes per gel.
Prepare gel casting apparatus.
Make Resolving and Stacking Gel solutions, but do not add TEMED until immediately before pouring each of the solutions, in steps 4 and 5.
Pour Resolving Gel solution between glass plates, taking care not to produce any bubbles, and wait for 10 min.
Pour Stacking Gel solution on top of resolving gel and insert gel combs.
Wait for 15 min for the gel to set.
While the gels are setting up, make Running Buffer.
Load gel in electrophoresis apparatus and fill with Running Buffer.
Remove gel combs and load 1 µg of histone extract per lane. Load molecular weight standards in desired lane(s).
Run the gel at 100 V for 1 h and 45 min, or until protein marker has run down the gel.
While the gel is running, prepare Transfer Buffer and make distinct marks on each PVDF membrane (e.g., cut a corner off with scissors) for later identification.
Prepare PVDF membranes for transfer by pre-incubation in methanol for 1 min, followed by a dip in ddH2O. Leave membrane in Transfer Buffer until the gel finishes running.
Prepare the gel for transfer by setting up the transfer cassette in the following order: (a) pad, (b) filter paper, (c) gel, (d) PVDF membrane, (e) filter paper and (f) pad (see Note 9).
Set up transfer chamber with stir bar and place in an ice bath on stir plate.
Run at 75 V for 2 h.
Make TBS, TBST, and TBST with 5% BSA during transfer.
Remove blots from cassettes and place in methanol for 1 min.
Dry at room temperature for 15 min (see Note 10) and prepare primary antibody solutions.
Cut plastic sealing bags to size for blots and incubate blots with primary antibody in TBST + 5% BSA on rotator overnight at 4°C (see Note 11).
Remove the blot from the primary antibody and perform 3 × 5 min washes in TBST.
During these washes, prepare the secondary antibody solution.
Incubate blots with secondary antibody, 1:5,000 in TBST + 5% BSA, for 60 min at room temperature (see Note 12).
Perform 3 × 5 min washes in TBST, followed by 3 × 5 min washes in TBS.
Set up for development by chemiluminescence by preparing ECL substrate (1 mL per blot) and sections of plastic wrap for each blot.
At the end of washing, place each blot on square of plastic wrap.
Add 1 mL of ECL reagent to each blot and wait for 1 min.
Wrap blots in plastic wrap, being careful not to create any uneven surfaces.
Expose to film in dark room, developing after 30 s, 2 min, and 10 min (see Note 13).
After exposing to film, the antibody must be stripped and reprobed with a second antibody (see Note 14). Place blot in a solution of 0.2 N NaOH for 15 min, rotating on an orbital shaker at maximum speed.
Perform 1 × 5 min wash in TBS.
Block the membrane in TBST with 5% BSA.
Continue Western blotting with control antibody, as described above in steps 19–28.
3.3. Analysis of Western Blots
To quantify the relative change in a histone modification, it is necessary to measure bands from both the blot exposed to the antibody of interest, and the blot exposed to a control protein (such as total histone H4). This can be done using an image analysis program. ImageJ is a freely available program capable of providing a quantitative readout of band densities.
Take exposed films of both blots (let us use the example of acetylated H3 [AcH3] and total H4 [TH4]) and scan each (using a consumer-grade scanner) to a separate TIFF file at a resolution of 300 dpi. Turn off all image processing filters and acquire the image in 8-bit grayscale.
Open the AcH3 scan (blot) with Image J.
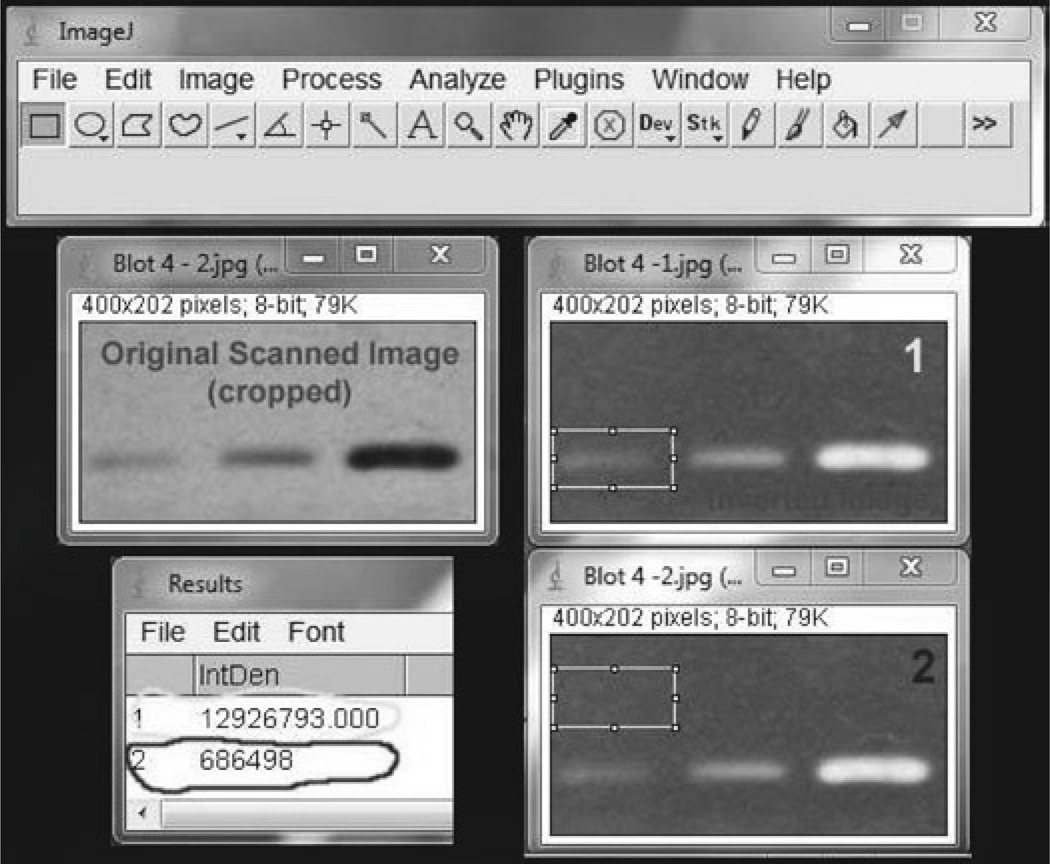
Crop the image close to the bands to remove unnecessary area.
Invert the image (CTRL + SHFT + I). This command results in the band having higher gray values relative to the film (see Fig. 2).
Open the “Set Measurements” dialog box located in the “Analyze” menu.
Check the “Integrated Density” box.
Click on the image and draw a rectangle around the first band – Inverted Image 1 (see Fig. 2).
To measure the density of this area, use the “Measure” command in the “Analyze” menu. You can also use the shortcut (CTRL + M). This command will sum the value of all pixels (see Fig. 2).
To uncover the “true integrated density” of the band, you must now measure and subsequently subtract the value of the film in this region. This is equivalent to a background subtraction. To do this, move the rectangle to a blank area above the band and repeat the “measure” command (see Fig. 2). The integrated density for this region is smaller than the first measurement. Subtract this value from the first to yield the true integrated density of your band’s signal. It is essential to use the same size rectangle for all measurements on a given scan (blot). Different sized rectangles can be used for different scans (blots).
Repeat this procedure for all bands.
Open the scan from your “TH4” blot.
Repeat steps 3–10 for the “TH4” blot.
For each sample/lane, divide the “true integrated density” of the AcH3 band by the “true integrated density” of the TH4 band. This normalization yields a relative acetylated histone value for each sample.
Fig. 2.
Using ImageJ software to quantify the intensity of bands after Western blotting.
Footnotes
We typically work with 40 mg or more of starting brain tissue. However, the protocol can easily be adapted for smaller tissue amounts, as we have successfully isolated sufficient histone protein concentrations from just a few 1 mm2 tissue punches. Simply reduce the volume of the homogenization buffer (e.g., 500 µl).
It is particularly important to develop a standard practice of orienting the tubes in the same direction for each spin in the centrifuge. In many cases, the small histone pellet’s location will not be obvious and will be inferred from how the tube was oriented during the spin.
This is the cytoplasmic fraction and can, if desired, be used for other assays.
Acid extraction by H2SO4 is a common method for separating histones. Following incubation on ice and centrifugation, nuclear proteins and nucleic acids will precipitate and pellet. Investigators specifically interested in examining histone phosphorylation may consider a high salt-based extraction method instead. Phosphorylation is particularly acid labile.
Do not expect to see a pellet at this point.
When rinsing the pellet with acidified acetone, run the acetone down the side of the tube that is expected to have product based on the way the tube was oriented in the centrifuge. Carefully run your pipette tip along the tube’s side to help “peel” the sample off the side of the tube and into the acetone.
Use care to not over-dry the pellet, as this will make resuspension extremely difficult.
Resuspension requires patience. Carefully pipette the Tris-HCl down the side of the tube where the product is from centrifugation. Repeat by pulling up Tris-HCl at the bottom of the tube and running it down the side again. Then let Tris-HCl sit for several minutes while you prepare protein assay or add Tris-HCl to additional samples. Finally, gently triturate to complete resuspension. Some of the pellet will likely remain at the bottom of the tube, but histones have resuspended.
To ensure complete transfer, be careful to remove any bubbles between layers in transfer cassette using a 15-mL conical vial or similar.
This drying step precludes the need to include a blocking step.
We typically incubate blots with the primary antibody overnight at 4°C at a concentration of 1:1,000. However, a rapid version of this protocol can be performed in which blots are incubated with the primary antibody for 90 min at room temperature. These times and concentrations may need to be optimized in your laboratory. Be sure all bubbles are removed from plastic bags before sealing to ensure proper contact between all parts of blot and antibody during incubation.
As with the primary antibody, the incubation time and secondary antibody concentration will need to be optimized in your laboratory. We have found that the time can range from 45 min to 2 h, with a concentration range of 1:5,000–10,000.
In order to use this analysis for publication, one must first confirm that their detection system is in the “linear range.” To test if your system is indeed linear, perform a serial dilution (over a tenfold concentration) of your sample and then blot with one of your antibodies. Then perform the analysis from Subheading 3.3 to determine the linear range. Using an HRP-conjugated secondary antibody, ECL, and high quality film, one should have at least a fivefold range of linearity using this system. If you do not, then the Western blotting protocol and exposure time must be optimized. The two most common causes of reduced linearity using this system are film over-exposure and suboptimal primary/secondary antibody ratios/incubation times. One can achieve >100-fold linearity using more sophisticated detection systems based on direct fluorescence (e.g. LiCOR or Typhoon).
A second (control) antibody for a protein that is not expected to change with the experimental manipulation should be used to normalize the signal from the first antibody. This serves as an excellent loading control in addition to normalizing based on the results of the protein assay. We recommend that the control antibody recognize an antigen with a different molecular weight than the first. This ensures that the first antibody will not influence the results of the second, in case the stripping did not fully remove all of the first antibody. For example, if probing for acetylation of histone H3 on a blot is performed first, we recommend normalizing to total H4 protein to avoid the confounding of both antibodies having a molecular weight of 17 kDa. Also, we recommend stripping a blot no more than once to avoid stripping protein from the blot.
References
- 1.Waddington CH. The Strategy of Genes. New York: Macmillan; 1957. [Google Scholar]
- 2.Spencer VA, Davie JR. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 3.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 4.Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J. Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koshibu K, Graff J, Beullens M, Heitz FD, Berchtold D, Russig H, Farinelli M, Bollen M, Mansuy IM. Protein phosphatase 1 regulates the histone code for long-term memory. J. Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav. Brain. Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007;27:128–140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction and reconsolidation of conditioned fear. Learn. Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class I histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropharm. 2009;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]