Abstract
With an estimated 170 million infected individuals, hepatitis C virus (HCV) has a major impact on public health. The liver is the primary target organ of HCV, and the hepatocyte is its primary target cell. Attachment of the virus to the cell surface followed by viral entry is the first step in a cascade of interactions between the virus and the target cell that is required for successful entry into the cell and initiation of infection. Using recombinant HCV envelope glycoproteins and HCV pseudotype particles, several cell surface molecules have been identified interacting with HCV during viral binding and entry. These include CD81, highly sulfated heparan sulphate, the low-density lipoprotein receptor, scavenger receptor class B type I and claudin-1. Treatment options for chronic HCV infection are limited and a vaccine to prevent HCV infection is not available. Interfering with HCV entry holds promise for drug design and discovery as the understanding of molecular mechanisms underlying HCV interaction with the host cell is advancing. The complexity of the virus entry process offers several therapeutic targets.
Keywords: Antiviral Agents, therapeutic use, Hepacivirus, metabolism, physiology, Hepatitis C, drug therapy, Humans, Membrane Fusion
Keywords: hepatitis C virus, viral entry, entry inhibitor, neutralizing antibodies
2. INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic hepatitis worldwide (1). Treatment options for chronic HCV infection are limited and a vaccine to prevent HCV infection is not available (2). Moreover, chronic hepatitis C may progress to liver cirrhosis and ultimatively lead to hepatocellular carcinoma. HCV is a small enveloped positive-strand RNA virus that has been classified in the genus Hepacivirus of the Flaviviridae family. In vivo, HCV infects only humans and chimpanzees (3). The liver is the primary target organ of HCV, and the hepatocyte is its primary target cell. Replication of the HCV genome has been demonstrated in vivo and in vitro in liver hepatocytes, and hematopoietic cells including dendritic cells and B lymphocytes (4, 5). Attachment of the virus to the cell surface followed by viral entry is the first step in a cascade of interactions between the virus and the target cell that is required for successful entry into the cell and initiation of infection (6). This step is an important determinant of tissue tropism and pathogenesis, it thus represents a major target for antiviral host cell responses, such as antibody-mediated virus neutralization, and antiviral therapy.
3. MOLECULAR MECHANISMS OF HCV ENTRY INTO TARGET CELLS
3.1. Viral determinants: envelope glycoproteins E1 and E2
The HCV genome encodes a single precursor polyprotein of about 3,000 amino acids that is cleaved co- and post-translationally into functional structural and non-structural proteins by host and viral proteases including three structural proteins: the core protein forming the viral nucleocapsid and two envelope glycoproteins, E1 and E2. HCV particles have a size of about 55–60 nm in diameter (7–9). In analogy to other members of the Flaviviridae family, HCV is thought to adopt a classical icosahedral scaffold in which the two envelope glycoproteins E1 and E2 are anchored to the host cell-derived double-layer lipid envelope (10). E1 and E2 are type I transmembrane glycoproteins containing up to 6 and 11 potential glycosylation sites, respectively (11) and forming noncovalent heterodimers. The nucleocapsid is probably composed of multiple copies of the core protein in complex with the viral genome and lies underneath the envelope (10).
Studies of the HCV life cycle have long been hampered by the lack of an efficient cell culture system to generate infectious virus in vitro. Several model systems have thus been developed for the study of defined aspects of the HCV life cycle such as viral entry, replication, assembly and release (for review see (12)). Recombinant HCV envelope glycoproteins (13), HCV-like particles (HCV-LPs) (14–16) and retroviral HCV pseudotypes (HCVpp) (17, 18) have been successfully used to analyze virus attachment and entry. Most recently, efficient in vitro model systems for the production of infectious recombinant virions (HCVcc) have been described (9, 19, 20). Using these model systems, it could be demonstrated that envelope glycoproteins E1 and E2 are crititcal for host cell entry. In fact, HCVpp assembled with either E1 or E2 glycoproteins were significantly less infective than HCVpp containing both envelope glycoproteins (17). Furthermore, HCVcc generated from a construct with an in-frame-deletion of the HCV envelope protein coding sequence are not infectious (9).
Monoclonal or polyclonal antibodies targeting both linear and conformational epitopes of envelope glycoprotein E2 have been shown to inhibit cellular binding of HCV-LP binding, entry of HCVpp and infection of HCVcc (9, 14–20) suggesting that envelope glycoprotein E2 plays a key role for host cell surface interaction. Within the E2 envelope glycoprotein sequence hypervariable regions have been identified. These amino acids differ by up to 80% among HCV genotypes, and even among different subtypes of the same genotype. The N-terminal 27 residues of E2 (aa 384-410) show a very high degree of variation and this portion of the sequence has been termed hypervariable region 1 (HVR-1) (21). This region plays a critical role in HCV interaction with host cells as HVR-1-delted HCVpp demonstrate reduced infectivity (22, 23). The important role of HVR-1 in HCV infectivity is further supported by studies demonstrating that antibodies targeting regions within HVR-1 inhibit cellular recombinant E2 (24, 25) and HCV-LP binding (15, 26) as well as HCVpp entry into target cells (18, 27). The exact role of E1 still remains unknown. E1 may directly interact with cell surface molecules and/or contribute to proper folding and processing of E2. Interestingly, antibodies targeting the N-terminal region of E1 have been shown to inhibit HCV-LP binding (15, 28) as well as HCV infection of a B-cell-derived cell line (29) suggesting that E1-cell surface interaction may contribute to viral binding and entry. In addition, HCV envelope proteins E1 and E2 are thought to induce fusion between the viral envelope and host cell membranes (30).
3.2. Cellular determinants
Using recombinant HCV envelope glycoproteins and HCV-LPs, several cell surface molecules have been identified interacting with HCV during viral binding and entry. These include CD81 (13), the low-density lipoprotein (LDL) receptor (31), highly sulfated heparan sulfate (32), scavenger receptor class B type I (SR-BI) (24) and DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3 grabbing non integrin )/L-SIGN (DC-SIGNr, liver and lymph node specific) (33, 34). Experimental data using HCVpp and HCVcc have confirmed functional roles for heparan sulfate (35) as well as CD81 (9, 19, 35) and SR-BI (22, 36–38) in HCV binding and entry, respectively. Most recently, an additional entry factor, claudin-1, has been identified (39).
3.2.1. Capture molecules
3.2.1.1. Glycosaminoglycans: heparan sulfate
Glycosaminoglycan (GAG) chains on cell surface proteoglycans provide primary docking sites for the binding of various viruses and other microorganisms to eukaryotic cells. The GAG heparan sulfate is an important cellular binding molecule for several viruses and may serve as the initial docking site for HCV attachment before the virus is transfered to high-affinity entry receptors. In fact, incubation of HCV-LPs with heparin - a structural homolog of highly sulfated heparan sulfate – reduced HCV-LP binding to human hepatoma cells resulting in a decreased internalization of theses particles (32). In addition, incubation of serum-derived HCV or VSV/HCV pseudotypes with heparin markedly inhibited virus binding to target cells (40, 41). Most recently, using the HCVcc system, it could be confirmed that heparan sulfate plays an important role for HCV binding as heparin and pre-treatment of cells with heparinases reduced HCVcc infectivity (35).
3.2.1.2. Lectins: DC-SIGN/L-SIGN
C-type lectins may act both as adhesion molecules and as pathogen recognition receptors. The mannose binding C-type lectins DC-SIGN and L-SIGN serve as adhesion receptors to mediate contact between dendritic cells, T lymphocytes and endothelial cells. Both lectins are not expressed in hepatocytes. DC-SIGN is expressed in Kupffer cells, which are immobile liver macrophages localized close to LSEC and hepatocytes (42). L-SIGN is highly expressed in liver sinusoidal endothelial cells. DC-SIGN and L-SIGN recognize carbohydrate structures on pathogens (43) and have been shown to bind envelope glycoprotein E2 with high affinity (33, 44). L-SIGN and DC-SIGN expressed on human Radji B cells or HeLa cells have also been shown to capture and transmit HCVpp to human hepatoma Huh7 cells in coculture model systems (34, 45). Capture of circulating HCV particles by liver sinusoidal cells may thus facilitate viral infection of neighbouring hepatocytes which are not in direct contact with circulating blood and may, therefore, contribute to the pathogenesis of viral infection.
3.2.2. Entry receptors
3.2.2.1. Tetraspanins: CD81 and claudin-1
Tetraspanins are widely expressed proteins that regulate cell morphology, motility, invasion, fusion and signalling (46). These proteins contain four transmembrane domains, short intracellular domains and two extracellular loops, namely the small extracellular loop and the large extracelullar loop (LEL). CD81 has been identified as an HCV E2 binding molecule by expression cloning (13) using a cDNA library derived from a subclone of the human T cell lymphoma cell line Molt-4, which exhibits a high E2-binding capacity (25). Anti-CD81 antibodies as well as a soluble form of CD81 LEL have been shown to inhibit HCVpp and HCVcc entry into Huh-7 hepatoma cells and human heptocytes (9, 17–20, 45, 47, 48). Furthermore, silencing of CD81 expression in hepatoma cells by small interfering RNAs inhibited HCVpp entry as well as HCVcc infectivity and expression of CD81 in hepatoma cell lines that are resistant to HCVpp and HCVcc infection conferred susceptibility to HCV infection (22, 45, 47, 49). In addition, it has been demonstrated that CD81 expression levels on hepatoma cells correlate with HCV infectivity (49, 50). These results suggest that susceptibility to HCV infection may be linked to CD81 density on the cell surface and thus provide an explanation for HCV tissue tropism in vivo. Interestingly, CD81 from HCV refractory species are able to bind HCV E2 (51) and CD81 of various species may confer susceptibility to HCV infection (52) suggesting that CD81 is not the determinant for the species specificity of HCV. Recent studies using the HCVpp and HCVcc model system demonstrated the ability of anti-CD81 antibodies to inhibit HCV entry at a step post binding (35, 53) suggesting that CD81 functions as an HCV entry co-receptor after docking of the virus to attachment molecules. Most recently, another member of the tetraspanin family claudin-1 (CLDN1), has been identified as an HCV co- entry factor by expression cloning (39). CLDN1 is highly expressed in the liver but also in other tissues (54). However, CLDN1 expression correlates with HCV permissiveness and expression of CLDN1 in non-hepatic 293T cells renders them susceptible to HCVpp entry (39). In addition, overexpression of this molecule in CD81-deficient HepG2 hepatoma cells did increase their HCV permissivity, suggesting that CLDN1 is not an alternative entry pathway to CD81 (39). In addition, kinetic studies showed that CLDN1 acts at a post binding step after HCV interaction with CD81 (39). As tetraspanins are able to form associations with a wide variety of proteins as well as cholesterol and gangliosides (46), this suggests that several HCV co-receptors may be recruited to discrete membrane domains allowing the formation of an HCV entry receptor complex. Interestingly, murine CLDN1 also supports HCVpp entry, demonstrating that CLDN1, as CD81, is not a determinant for species host range (39).
3.2.2.2. Scavenger receptor SR-BI
SR-BI or CLA-1 (CD36 and LIMPII Analogous-1) is a 509 amino acid glycoprotein with a large extracellular loop anchored to the plasma membrane at both the N- and C- termini by transmembrane domains with short extensions into the cytoplasm (55). SR-BI is involved in bidirectional cholesterol transport at the cell membrane and is a multiligand receptor as it can bind both native high-density lipoprotein (HDL) and low density lipoproteins (LDL) as well as modified lipoproteins such as oxidized LDL (oxLDL). SR-BI is highly expressed in liver and steroidogenic tissues (55) as well as human monocyte-derived dendritic cells but not on any other peripheral blood mononuclear cells (56). Furthermore, SR-BI and its splicing variant SR-BII, have been found to mediate binding and uptake of a broad range of bacteria into nonphagocytic human epithelial cells overexpressing SR-BI and SR-BII (57, 58) suggesting that class B scavenger receptors may serve as pattern recognition receptors for bacteria. Cross-linking studies using recombinant C-terminally truncated HCV envelope glycoprotein E2 isolated SR-BI on HepG2 cells as a cell surface protein binding HCV envelope glycoprotein E2 (24). SR-BI recognition by HCV E2 required the hypervariable region HVR-1 (24). Moreover, antibodies directed against cell surface expressed SR-BI inhibited binding of recombinant envelope glycoproteins and HCV-like particles to primary hepatocytes (15) as well as HCVpp entry (22, 59, 60). In addition, it has been shown that physiological SR-BI ligands, such as HDL or oxLDL, can modulate HCV infection: HDL and oxLDL have been shown to enhance and inhibit HCVpp entry, respectively (61–63), whereas both HDL and LDL inhibited HCV replication in human hepatocytes infected with serum-derived HCV (64). These results suggest the existence of a complex interplay between lipoproteins, SR-BI and HCV envelope glycoproteins for HCV entry. Most recently, the important role of SR-BI in productive HCV infection has been confirmed using the HCVcc system. Overexpression of SR-BI and SR-BII was able to increase HCVcc infectivity (36) while down-regulation of this receptor by SR-BI specific siRNA markedly reduced the susceptibility of human hepatoma cells to HCVcc infection (Zeisel MB, Schnober EK, Haberstroh A, Lavillette D, Cosset FL, Wakita T, Jaeck D, Royer C, Schuster C, Stoll-Keller F, Blum H, Barth H, Baumert TF. 13th International Meeting on Hepatitis C Virus & Related Viruses 2006, Cairns, Australia). Moreover, anti-SR-BI antibodies directed against epitopes of the SR-BI extracellular loop specifically inhibited HCVcc infection and kinetic studies demonstrated that SR-BI acts predominately following binding of HCV at an entry step occurring at a similar time point as CD81-HCV interaction (38).
3.2.2.3. LDL receptor
The LDL receptor (LDLR) transports cholesterol-containing lipoproteins into the cell by endocytosis via clathrin-coated pits. Receptor-ligand complexes are delivered into endosomes where low pH induces the release of lipoproteins which then proceed to lysosomes where free cholesterol is generated by cholesterol ester hydrolysis (65). The apolipoprotein B (apoB)-containing LDL and apolipoprotein E (apoE)-containing very low-density lipoproteins (VLDL) are the major LDLR ligands. As HCV is able to associate with LDL and VLDL in serum (66, 67), the LDLR was suggested to be a putative HCV receptor candidate. The LDLR has been shown to mediate internalize serum-derived HCV by binding virus-LDL particles (31). Anti-LDLR antibodies as well as anti-apoB and apoE antibodies were able to inhibit HCV endocytosis (17, 31, 40, 68). It could also be demonstrated that LDLR plays a role in an early step of serum-derived HCV infection of primary human hepatocytes (64). However, studies using the HCVpp system where HCV is not associated with lipoproteins suggest that LDLR does not appear to play a role for infection of Huh7 cells with HCVpp (17). Further studies using HCVcc and human hepatocytes will allow gaining more insight into the role of LDLR in HCV infection.
Despite the numerous experimental data demonstrating the importance of the above described receptors in HCV infection, none of these molecules has a liver-specific expression profile as it would be expected for receptors of a hepatotropic virus. Moreover, all HCV permissive cell lines identified so far express CD81, SR-BI and CLDN1 and are of hepatic origin but various cell lines of non-hepatic origin expressing these receptors are non permissive for HCV (17, 22, 27, 47), suggesting that additional liver specific factor(s) which are still to be discovered are required for HCV infection.
3.3. Mechanisms of HCV internalization and fusion
To multiply, viruses must deliver their genome into host cells. The critical step subsequently leading to viral replication is the penetration of the viral genome through a host cell membrane. Cell attachment of other members of the Flaviviridae family such as flaviviruses leads to endocytosis of bound virions (69). Clathrin-mediated endocytosis is the most commonly route of endocytosis for viruses that require internalization. It transports incoming viruses together with their receptors into early and late endosomes (6). It has been shown that early and late endosomes constitute distinct entry sites depending on the pH threshold of viral proteins. The acidic pH in endosomes provides an essential cue that triggers penetration and uncoating. Penetration of enveloped virus occurs by membrane fusion catalyzed by fusion peptides embedded in the viral envelope glycoproteins (70). In some cases, acidic pH is not sufficient to induce fusion and viral proteins need to be cleaved by endosomal proteases to become fusion competent. Two classes of viral fusion proteins (class I and II) mediating entry of enveloped viruses have been defined. Type II fusion proteins, occuring in flaviviruses and alpha viruses, are synthezised as heterodimers with other proteins that dissociate at the acidic pH in the endosome and assemble into more stable homotrimers that destabilizes the target cell membrane and then leads to the formation of a fusion pore (6, 71). Recent studies using HCVpp and HCVcc have demonstrated that HCV entry into both hepatoma cells and primary human hepatocytes depends on clathrin-mediated endocytosis (72–74). Structural homology with fusion proteins from flaviviruses suggests that HCV envelope glycoproteins may belong to class II fusions proteins (10, 75–77). Although – in contrast to flaviviruses-HCV glycoproteins are not matured by a cellular endoprotease during their transport through the secretory pathway (77), similar membrane fusion mechanisms may operate in HCV. This hypothesis is supported by the observation that HCVpp entry into Huh-7 cells is pH-dependent (17, 18, 78). In addition, HCVcc infection was markedly inhibited by agents preventing the acidification of endosomal compartments, suggesting that a pH-dependent membrane fusion process may be required for delivery of the HCV genome into the host cell cytosol (72, 79). Finally, it has been shown that HCVpp are delivered to early but not late endosomes (73). As HCV fusion kinetics are delayed as compared to other viruses, it has been suggested that after internalization, HCVpp entry necessitates additional, low-pH-dependent interactions, modifications, or trafficking (73). However, neither HCVpp nor HCVcc require cleavage by endosomal proteases for fusion (73, 79).
4. VIRAL ENTRY: TARGET FOR ANTIVIRAL THERAPY
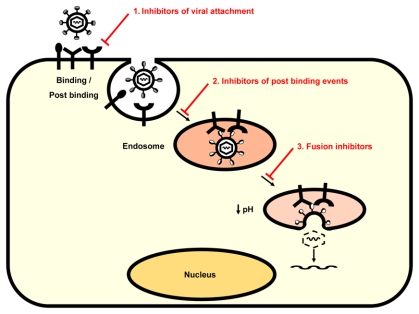
Current approaches for HCV antiviral drug development targeting viral enzymes comprise protease inhibitors, inhibitors of protein translation and polymerase inhibitors (for review see (80)). Interfering with HCV entry also holds promise for drug design and discovery as the understanding of molecular mechanisms underlying HCV interaction with the host cell is growing. Viral entry may be inhibited (i) by blocking interaction between the virus and the target cell, (ii) by interfering with post binding events, and (iii) by interfering with viral fusion (Fig. 1).
Fig. 1. HCV entry: targets for antiviral therapy.
Both virus and host cell components involved in virus entry may serve as targets for the development of HCV entry inhibitors. Viral entry may be inhibited (1) by blocking interaction between the virus and the target cell, (2) by interfering with post binding events, and (3) by interfering with viral fusion. Future anti-HCV therapies might be combinations of drugs targeting distinct steps of HCV infection, i. e. entry inhibitors, protease inhibitors and polymerase inhibitors that might have complementary effects and delay the emergence of drug resistance.
4.1. Neutralizing antibodies
Viral attachment and entry is a major target of adaptive host cell defences. Viral proteins are recognized as non-self by the host’s immune system and induce the production of antibodies. A small proportion of these antibodies exhibit antiviral activity in vitro and are defined as virus-neutralizing antibodies. These antibodies render virions non-infectious by interfering with receptor binding and cell entry. Many successful antiviral vaccines are based on the induction of neutralizing antibodies. Isolation and characterization of antibodies targeting distinct steps of HCV entry is an important strategy for protection against this virus and provide a rational basis for HCV vaccine development. Antibody-mediated neutralization occurs during HCV infection in vivo but the role of antibodies for the control of HCV infection is still unclear. Antibodies with HCV neutralizing properties have been first described in experimental infection of chimpanzees (81, 82). These antibodies were directed against epitopes in the hypervariable region (HVR-1) of HCV envelope glycoprotein E2 and appeared to be isolate-specific (81, 82). The presence of antibodies directed against HVR-1 has also been associated with viral clearance in HCV-infected humans (83) and HCV-infected patients with primary antibody deficiencies have been reported to have accelerated rates of disease progression (84, 85). However, HCV infection is established despite the induction of an humoral immune response that targets various epitopes of the HCV envelope glycoproteins (26, 27, 86–88). Until recently, functional studies analyzing the neutralizing antibody response during acute and chronic HCV infection using HCV model systems demonstrated a lack of neutralizing antibodies in the majority of patients with acute HCV infection (26, 27, 86, 89). These studies were limited by the fact that the viral surrogate ligand was derived from a different isolate than the virus present in the infected patient thus precluding the detection of isolate-specific antibodies. Most recently, studies using well defined nosocomial or single-source HCV outbreaks with a defined inoculum enabled for the first time to study the role of isolate-specific neutralizing antibodies for HCV clearance in humans. Using the HCVpp model system, two studies demonstrated that neutralizing antibodies are induced in the early phase of infection by patients who subsequently clear the virus (90) or control viral infection (60). These results suggest that a strong early neutralizing antibody response may play a role in the outcome of HCV infection. Patients who do not clear the virus develop high-titer and even cross-neutralizing antibodies during the chronic phase of infection (9, 26, 27, 86, 90). Viral escape from antibody-mediated neutralization in these individuals may occur on several levels: (i) HCV exists as a quasispecies with distinct viral variants in infected individuals changing constantly over time and his variability has been shown to represent a mechanism of escape from antibody-mediated neutralization in the chimpanzee model (27); (ii) the interplay of HCV glycoproteins with high-density lipoprotein and the scavenger receptor BI has been shown to mediate protection from neutralizing antibodies present in sera of acute and chronic HCV-infected patients (61, 91); and (iii) as shown for other viruses such as HIV, escape from neutralizing antibodies may occur through a combination of different mechanisms, for instance point mutations, insertions/deletions or changes in glycosylation patterns of the viral envelope (92) or conformational masking of receptor binding sites following envelope-antibody interaction (93) preventing neutralizing antibody binding. The HCV tissue culture model systems and patient cohorts with well defined viral isolates will now allow to adress these questions and define therapeutic strategies based on neutralizing antibodies. Several viral epitopes targeted by neutralizing antibodies have already been identified: epitopes of the E2 HVR-1 region (aa 384-410) (18, 25, 61), an epitope adjacent to the N-terminal region HVR-1 region (aa 408-422) (26, 94), the E2 CD81 binding region (aa 474-494 and aa 522-551) (18, 47, 94) and conformational epitopes within glycoprotein E2 (95, 96). These epitopes may represent potential candidate targets for antibodies in passive immunoprophylaxis. Indeed, two studies have demonstrated that monoclonal antibodies directed against conformational epitopes (95) or epitope aa 412-423 exhibited broad cross-neutralizing activity among all major genotypes of HCVpp entry (94) as well as HCVcc infectivity (97). Future in vivo studies are required to study the relevance of these findings for antibody-mediated prevention of HCV infection. This may have important implications for the development of novel preventive and curative antiviral strategies, e. g. passive immunoprophylaxis after accidental exposure to HCV and prevention of reinfection of liver grafts after liver transplantation.
4.2. Entry inhibitors
4.2.1. Inhibitors of viral attachment
The lectin cyanovirin-N (CV-N) has initially been discovered as an active compound against HIV and was then shown to present antiviral activity against other enveloped viruses (98, 99). This antiviral activity results from interactions between CV-N and high-mannose oligosaccharides on viral envelope glycoproteins (100). HCV envelope glycoproteins are highly glycosylated and contain oligomannose glycans. It has been shown that these oligomannose glycans interact with CV-N resulting in HCV antiviral activity by blocking HCV entry into target cell (101). As most of the HCV glycosylation sites are highly conserved, drugs that target glycans on HCV glycoproteins may not lead so rapidly viral escape/resistance as it is the case for HIV (92). Other carbohydrate-binding agents, such as plant lectins, monoclonal antibodies and the mannose-specific non-peptidic antibiotic Pradimicin A have been shown to prevent HIV infectivity (102). Such substances might also be efficient against other viruses that require a glycosylated envelope for entry into target cells.
Another approach to target HCV attachment might be the development of heparin-derived molecules, as heparin has been shown to potently inhibit HCV E2, HCVpp, HCV-LP as well as HCVcc binding to hepatoma cells (32, 35, 53, 103). The systematic generation and screening of heparan sulfate-like molecules and semisynthetic derivatives is already explored as an antiviral approach against dengue virus infection (104).
4.2.2. Inhibitors of post binding steps
Antiviral compounds targeting viral entry may either act on conserved mechanisms or target specific cell surface molecules. Recent studies have shown that long phosphorothioate oligonucleotides (PS-ON), that are amphipathic DNA polymers, displays a sequence independent antiviral activity against HIV by blocking virus-cell fusion (105). Most recently, it could be demonstrated that PS-ON inhibit HCV fusion and entry (Matsumara T, Kato K, Hu Z, Juteau JM, Vaillant A, Liang JT. The 57th Annual Meeting of the American Association for the Study of Liver Diseases, 2006, Boston, USA). The PS-ON are a promising new class of antiviral compounds that may have a broad spectrum in all families of enveloped viruses.
Structural information of HCV envelope glycoprotein E2 and CD81 was used to identify imidazole based compounds that mimic an alpha helix in the LEL of CD81 and compete for the binding of HCV E2 to CD81 expressed on target cells. These drugs bind HCV E2 in a reversible manner and block HCV E2 interaction with CD81 while having no effect on CD81 expression nor on CD81-interaction with physiological partner molecules (106). However, data of the effect of these drugs on HCVcc infection are not yet available. Recently, SR-BI has been demonstrated to bind and internalize serum amyloid A (SAA), an acute phase protein mainly produced in the liver and known to mediate pro-inflammatory cellular responses (107, 108). SAA was shown to inhibit HCV entry by interacting with the virus thereby reducing its infectivity (109). Thus, SAA analogues might present potent anti-HCV activity. In addition to small molecule inhibitors, peptides that mimic conserved regions of HCV E2 interacting with cell entry receptors may also provide an interesting approach to prevent HCV infection but have weaknesses as drugs because they are not orally available and expensive to produce.
4.2.3. Fusion inhibitors
Insights into the molecular mechanisms of HCV fusion are just about to arise and molecules likely to interfere with HCV penetration have not yet been described. As HCV enters the host cell through endocytosis and requires low pH for delivery of HCV genome, agents preventing the acidification of endosomal compartments, such as chloroquine, are able to prevent infection (72, 79). Potential targets for anti-fusion drugs might arise in the next few years while insights into HCV fusion process are growing. Peptide-based fusion inhibitors have already been established for the treatment of other viral infections such as HIV infection. Enfuvirtide which blocks HIV fusion to host cells is the first compound of this family approved for clinical use (110).
5. PERSPECTIVES
The development of novel HCV model systems allowed to increase the understanding of the complex viral entry process thereby offering new therapeutic targets to prevent the virus to reach its site of replication. Major progress has been made over the past few years in the characterization of host cell molecules involved in virus entry and the sequence of events ultimatively leading to viral replication. Both virus and host cell components involved in virus entry may serve as targets for the development of HCV entry inhibitors (Table 1). As for other infectious diseases, it might be preferable to target viral proteins than host cell components because of potential adverse effects resulting form interference with normal cell functions. The optimal entry inhibitor would block viral binding sites on receptors without affecting functional physiological ligand binding. Future anti-HCV therapies might be combinations of drugs targeting distinct steps of HCV infection, i. e. entry inhibitors, protease inhibitors and polymerase inhibitors that might have complementary effects and delay the emergence of drug resistance.
Table 1.
HCV entry: viral and cellular targets for antiviral therapy
| Antiviral target | Antiviral compounds |
|---|---|
| Viral targets | |
| HCV envelope glycoproteins | Neutralizing antibodies |
| Carbohydrate binding agents, e. g. cyanovirin | |
| Heparin-derived molecules | |
| Imidazole-based compounds | |
| Serum amyloid A | |
| Cellular targets | |
| HCV entry molecules | HCV E2 peptides |
| Phosphorothioate oligonucleotides | |
| Viral fusion mechanisms | Endososomal acidification inhibitors, e. g. chloroquine |
| Peptide-based fusion inhibitors | |
Acknowledgments
Authors are supported by grants of the European Union (VIRGIL Network of Excellence), Belgium, the Deutsche Forschungsgemeinschaft (Ba1417/11-2), Germany, Inserm, France, University Louis Pasteur, France, the chair of excellence program of the Agence Nationale de la Recherche (ANR), France, and the Agence Nationale de la Recherche sur le SIDA et les Hépatites Virales (ANRS), France. MBZ is supported by the Inserm Poste Vert Program in the frame work of Inserm European Associated Laboratory Freiburg-Strasbourg.
Abbreviations
- aa
amino acid
- apo
apolipoprotein
- CLDN1
claudin-1
- DC-SIGN
dendritic cell-specific intercellular adhesion molecule 3 grabbing non integrin
- HCV
hepatitis C virus
- HCVcc
cell culture-derived HCV
- HCV-LP
HCV-like particles
- HCVpp
HCV pseudoparticles
- HDL
high-density lipoprotein
- HIV
human immunodeficiency virus
- HVR-11
hypervariable region-1
- LDL
low-density lipoprotein
- LDLR
LDL receptor
- L-SIGN
DC-SIGNr, liver and lymph node specific
- siRNA
small interfering RNA
- SR-BI
scavenger receptor class B type I
- VLDL
very low-density lipoprotein
References
- 1.Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–2. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 2.De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–60. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach BDCM. Rice: Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 4. Lippincott Williams & Wilkins; 2001. pp. 991–1041. [Google Scholar]
- 4.Sung VM, Shimodaira S, Doughty AL, Picchio GR, Can H, Yen TS, Lindsay KL, Levine AM, Lai MM. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J Virol. 2003;77:2134–46. doi: 10.1128/JVI.77.3.2134-2146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goutagny N, Fatmi A, De Ledinghen V, Penin F, Couzigou P, Inchauspe G, Bain C. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J Infect Dis. 2003;187:1951–8. doi: 10.1086/375350. [DOI] [PubMed] [Google Scholar]
- 6.Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–40. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, Konishi M, Yokoi M, Ishida S, Suzuki S, Kohara M. Hepatitis C virus particle detected by immunelectron microscopic study. J Gen Virol. 1994;75:1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu YK, Feinstone SM, Kohara M, Purcell R, Yoshikura H. Hepatitis C virus: detection of intracellular virus particles by electron microscopy. Hepatology. 1996;23:205–209. doi: 10.1002/hep.510230202. [DOI] [PubMed] [Google Scholar]
- 9.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–6. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 11.Op De Beeck A, Montserret R, Duvet S, Cocquerel L, Cacan R, Barberot B, Le Maire M, Penin F, Dubuisson J. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J Biol Chem. 2000;275:31428–37. doi: 10.1074/jbc.M003003200. [DOI] [PubMed] [Google Scholar]
- 12.Barth H, Liang TJ, Baumert TF. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology. 2006;44:527–35. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- 13.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–41. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 14.Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827–36. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth H, Cerino R, Arcuri M, Hoffmann M, Schurmann P, Adah MI, Gissler B, Zhao X, Ghisetti V, Lavezzo B, Blum HE, von Weizsacker F, Vitelli A, Scarselli E, Baumert TF. Scavenger receptor class B type I and hepatitis C virus infection of primary tupaia hepatocytes. J Virol. 2005;79:5774–85. doi: 10.1128/JVI.79.9.5774-5785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellnitz S, Klumpp B, Barth H, Ito S, Depla E, Dubuisson J, Blum HE, Baumert TF. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J Virol. 2002;76:1181–93. doi: 10.1128/JVI.76.3.1181-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003;197:633–42. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 20.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–8. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 22.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003 doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 23.Callens N, Ciczora Y, Bartosch B, Vu-Dac N, Cosset FL, Pawlotsky JM, Penin F, Dubuisson J. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry. J Virol. 2005;79:15331–41. doi: 10.1128/JVI.79.24.15331-15341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A, Lau JYN, Choo QL, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmann D, Barth H, Gissler B, Schürmann P, Adah MI, Gerlach JT, Pape GR, Depla E, Jacobs D, Maertens G, Patel AH, Inchauspé G, Liang TJ, Blum HE, Baumert TF. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J Virol. 2004;78:9030–9040. doi: 10.1128/JVI.78.17.9030-9040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A. 2003;100:14199–204. doi: 10.1073/pnas.2335981100. Epub 2003 Nov 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triyatni M, Saunier B, Maruvada P, Davis AR, Ulianich L, Heller T, Patel A, Kohn LD, Liang TJ. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J Virol. 2002;76:9335–9344. doi: 10.1128/JVI.76.18.9335-9344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keck ZY, V, Sung M, Perkins S, Rowe J, Paul S, Liang TJ, Lai MM, Foung SK. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J Virol. 2004;78:7257–63. doi: 10.1128/JVI.78.13.7257-7263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takikawa S, Ishii K, Aizaki H, Suzuki T, Asakura H, Matsuura Y, Miyamura T. Cell fusion activity of hepatitis C virus envelope proteins. J Virol. 2000;74:5066–74. doi: 10.1128/jvi.74.11.5066-5074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–71. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barth H, Schäfer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Weizsäcker Fv, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–12. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 33.Pohlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C Virus Glycoproteins Interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozach PY, Amara A, Bartosch B, Virelizier JL, Arenzana-Seisdedos F, Cosset FL, Altmeyer R. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J Biol Chem. 2004;279:32035–45. doi: 10.1074/jbc.M402296200. [DOI] [PubMed] [Google Scholar]
- 35.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–20. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, Moreau M, Owen JS, Leroux-Roels G, Balfe P, McKeating JA. Scavenger receptor BI and BII expression levels modulate Hepatitis C virus infectivity. J Virol. 2007 doi: 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of Hepatitis C Virus Infection Is Dependent on Cholesterol and Cooperativity between CD81 and Scavenger Receptor B Type I. J Virol. 2007;81:374–83. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset F-L, Wakita T, Jaeck D, Doffoel M, Royer C, Soulier E, Schvoerer E, Schuster C, Stoll-Keller F, Bartenschlager R, Pietschmann T, Barth H, Baumert TF. Scavenger receptor BI is a key host factor for Hepatitis C virus infection required for an entry step closely linked to CD81. doi: 10.1002/hep.21994. Submitted. [DOI] [PubMed] [Google Scholar]
- 39.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007 doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 40.Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J Med Virol. 2002;68:206–15. doi: 10.1002/jmv.10196. [DOI] [PubMed] [Google Scholar]
- 41.Basu A, Beyene A, Meyer K, Ray R. The hypervariable region 1 of the E2 glycoprotein of hepatitis C virus binds to glycosaminoglycans, but this binding does not lead to infection in a pseudotype system. J Virol. 2004;78:4478–86. doi: 10.1128/JVI.78.9.4478-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kooyk Y, Geijtenbeek TB. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 43.Koppel EA, van Gisbergen KP, Geijtenbeek TB, van Kooyk Y. Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell Microbiol. 2005;7:157–65. doi: 10.1111/j.1462-5822.2004.00480.x. [DOI] [PubMed] [Google Scholar]
- 44.Gardner JP, Durso RJ, Arrigale RR, Donovan GP, Maddon PJ, Dragic T, Olson WC. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc Natl Acad Sci U S A. 2003;100:4498–4503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cormier EG, Durso RJ, Tsamis F, Boussemart L, Manix C, Olson WC, Gardner JP, Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc Natl Acad Sci U S A. 2004;101:14067–72. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–11. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448–55. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, Butera D, Ho DD, Dustin LB, Rice CM, Balfe P. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J Virol. 2004;78:8496–505. doi: 10.1128/JVI.78.16.8496-8505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, Barth H, Baumert TF, Dubuisson J, Wakita T. Cd81 Expression Is Important For Heterogeneous Hcv Permissiveness Of Huh7 Cell Clones. J Virol. 2007 doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of Hepatitis C Virus into host cells. J Virol. 2006 doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meola A, Sbardellati A, Bruni Ercole B, Cerretani M, Pezzanera M, Ceccacci A, Vitelli A, Levy S, Nicosia A, Traboni C, McKeating J, Scarselli E. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J Virol. 2000;74:5933–8. doi: 10.1128/jvi.74.13.5933-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, Rice CM, McKeating JA. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80:11331–42. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci U S A. 2004;101:7270–4. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–50. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krieger M. Scavenger receptor class B type I is a multiligand HDL receptor that influences diverse physiologic systems. J Clin Invest. 2001;108:793–7. doi: 10.1172/JCI14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada E, Montoya M, Schuettler CG, Hickling TP, Tarr AW, Vitelli A, Dubuisson J, Patel AH, Ball JK, Borrow P. Analysis of the binding of hepatitis C virus genotype 1a and 1b E2 glycoproteins to peripheral blood mononuclear cell subsets. J Gen Virol. 2005;86:2507–12. doi: 10.1099/vir.0.81169-0. [DOI] [PubMed] [Google Scholar]
- 57.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–3. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 58.Vishnyakova TG, Kurlander R, Bocharov AV, Baranova IN, Chen Z, Abu-Asab MS, Tsokos M, Malide D, Basso F, Remaley A, Csako G, Eggerman TL, Patterson AP. CLA-1 and its splicing variant CLA-2 mediate bacterial adhesion and cytosolic bacterial invasion in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:16888–93. doi: 10.1073/pnas.0602126103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset FL. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology. 2005;41:265–74. doi: 10.1002/hep.20542. [DOI] [PubMed] [Google Scholar]
- 60.Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM, Cosset FL. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79:6023–34. doi: 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217–29. doi: 10.1128/JVI.79.13.8217-8229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793–9. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 63.von Hahn T, Lindenbach BD, Boullier A, Quehenberger O, Paulson M, Rice CM, McKeating JA. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology. 2006;43:932–42. doi: 10.1002/hep.21139. [DOI] [PubMed] [Google Scholar]
- 64.Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–9. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 65.Beglova N, Blacklow SC. The LDL receptor: how acid pulls the trigger. Trends Biochem Sci. 2005;30:309–17. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Thomssen R, Bonk S, Propfe C, Heermann KH, Koechel HG, Uy A. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol. 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 67.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919–28. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wunschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055–62. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz FX, Allison SL. Flavivirus structure and membrane fusion. Adv Virus Res. 2003;59:63–97. doi: 10.1016/s0065-3527(03)59003-0. [DOI] [PubMed] [Google Scholar]
- 70.Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237–42. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- 71.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. Embo J. 2004;23:728–38. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80:6964–72. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol. 2006;80:11571–8. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Codran A, Royer C, Jaeck D, Bastien-Valle M, Baumert TF, Kieny MP, Pereira CA, Martin JP. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J Gen Virol. 2006;87:2583–93. doi: 10.1099/vir.0.81710-0. [DOI] [PubMed] [Google Scholar]
- 75.Yagnik AT, Lahm A, Meola A, Roccasecca RM, Ercole BB, Nicosia A, Tramontano A. A model for the hepatitis C virus envelope glycoprotein E2. Proteins. 2000;40:355–66. doi: 10.1002/1097-0134(20000815)40:3<355::aid-prot20>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 76.Heinz FX, Stiasny K, Allison SL. The entry machinery of flaviviruses. Arch Virol Suppl. 2004:133–7. doi: 10.1007/978-3-7091-0572-6_11. [DOI] [PubMed] [Google Scholar]
- 77.Voisset C, Dubuisson J. Functional hepatitis C virus envelope glycoproteins. Biol Cell. 2004;96:413–20. doi: 10.1016/j.biolcel.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 78.Lavillette D, Bartosch B, Nourrisson D, Verney G, Cosset FL, Penin F, Pecheur EI. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J Biol Chem. 2006;281:3909–17. doi: 10.1074/jbc.M509747200. [DOI] [PubMed] [Google Scholar]
- 79.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–41. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walker MA. Hepatitis C virus: an overview of current approaches and progress. Drug Discov Today. 1999;4:518–529. doi: 10.1016/s1359-6446(99)01414-2. [DOI] [PubMed] [Google Scholar]
- 81.Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A. 1994;91:7792–6. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter HJ, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A. 1996;93:15394–9. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zibert A, Dudziak P, Schreier E, Roggendorf M. Characterization of antibody response to hepatitis C virus protein E2 and significance of hypervariable region 1-specific antibodies in viral neutralization. Arch Virol. 1997;142:523–34. doi: 10.1007/s007050050098. [DOI] [PubMed] [Google Scholar]
- 84.Christie JM, Healey CJ, Watson J, Wong VS, Duddridge M, Snowden N, Rosenberg WM, Fleming KA, Chapel H, Chapman RW. Clinical outcome of hypogammaglobulinaemic patients following outbreak of acute hepatitis C: 2 year follow up. Clin Exp Immunol. 1997;110:4–8. doi: 10.1046/j.1365-2249.1997.5081412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chapel HM, Christie JM, Peach V, Chapman RW. Five-year follow-up of patients with primary antibody deficiencies following an outbreak of acute hepatitis C. Clin Immunol. 2001;99:320–4. doi: 10.1006/clim.2001.5036. [DOI] [PubMed] [Google Scholar]
- 86.Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, Feinstone SM, Alter H, Rice CM, McKeating JA. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A. 2004;101:10149–54. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, Emerson SU, Cosset FL, Purcell RH, Bukh J. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A. 2005;102:4560–5. doi: 10.1073/pnas.0501275102. Epub 2005 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hadlock KG, Gish R, Rowe J, Rajyaguru SS, Newsom M, Warford A, Foung SK. Cross-reactivity and clinical impact of the antibody response to hepatitis C virus second envelope glycoprotein (E2) J Med Virol. 2001;65:23–9. [PubMed] [Google Scholar]
- 89.Netski DM, Mosbruger T, Depla E, Maertens G, Ray SC, Hamilton RG, Roundtree S, Thomas DL, McKeating J, Cox A. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis. 2005;41:667–75. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- 90.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, Patel AH, Meisel H, Baumert J, Viazov S, Rispeter K, Blum HE, Roggendorf M, Baumert TF. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104:6025–30. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J, Bartenschlager R, Lavillette D, Cosset FL. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285–95. doi: 10.1074/jbc.M602706200. [DOI] [PubMed] [Google Scholar]
- 92.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 93.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 94.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79:11095–104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schofield DJ, Bartosch B, Shimizu YK, Allander T, Alter HJ, Emerson SU, Cosset FL, Purcell RH. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology. 2005;42:1055–62. doi: 10.1002/hep.20906. [DOI] [PubMed] [Google Scholar]
- 96.Keck ZY, Op De Beeck A, Hadlock KG, Xia J, Li TK, Dubuisson J, Foung SK. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol. 2004;78:9224–32. doi: 10.1128/JVI.78.17.9224-9232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJ, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 2006;43:592–601. doi: 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- 98.Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O’Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH, 2nd, Buckheit RW, Jr, Nara PL, Pannell LK, Sowder RC, 2nd, Henderson LE. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–30. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O’Keefe BR, Smee DF, Turpin JA, Saucedo CJ, Gustafson KR, Mori T, Blakeslee D, Buckheit R, Boyd MR. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob Agents Chemother. 2003;47:2518–25. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shenoy SR, O’Keefe BR, Bolmstedt AJ, Cartner LK, Boyd MR. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J Pharmacol Exp Ther. 2001;297:704–10. [PubMed] [Google Scholar]
- 101.Helle F, Wychowski C, Vu-Dac N, Gustafson KR, Voisset C, Dubuisson J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J Biol Chem. 2006;281:25177–83. doi: 10.1074/jbc.M602431200. [DOI] [PubMed] [Google Scholar]
- 102.Balzarini J. Carbohydrate-binding agents: a potential future cornerstone for the chemotherapy of enveloped viruses? Antivir Chem Chemother. 2007;18:1–11. doi: 10.1177/095632020701800101. [DOI] [PubMed] [Google Scholar]
- 103.Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. Viral and Cellular Determinants of Hepatitis C Virus Envelope-Heparan Sulfate Interaction. J Virol. 2006 doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee E, Pavy M, Young N, Freeman C, Lobigs M. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antiviral Res. 2006;69:31–8. doi: 10.1016/j.antiviral.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 105.Vaillant A, Juteau JM, Lu H, Liu S, Lackman-Smith C, Ptak R, Jiang S. Phosphorothioate oligonucleotides inhibit human immunodeficiency virus type 1 fusion by blocking gp41 core formation. Antimicrob Agents Chemother. 2006;50:1393–401. doi: 10.1128/AAC.50.4.1393-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.VanCompernolle SE, Wiznycia AV, Rush JR, Dhanasekaran M, Baures PW, Todd SC. Small molecule inhibition of hepatitis C virus E2 binding to CD81. Virology. 2003;314:371–80. doi: 10.1016/s0042-6822(03)00406-9. [DOI] [PubMed] [Google Scholar]
- 107.Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, Remaley AT, Csako G, Thomas F, Eggerman TL, Patterson AP. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J Biol Chem. 2005;280:8031–40. doi: 10.1074/jbc.M405009200. [DOI] [PubMed] [Google Scholar]
- 108.Cai L, de Beer MC, de Beer FC, van der Westhuyzen DR. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J Biol Chem. 2005;280:2954–61. doi: 10.1074/jbc.M411555200. [DOI] [PubMed] [Google Scholar]
- 109.Lavie M, Voisset C, Vu-Dac N, Zurawski V, Duverlie G, Wychowski C, Dubuisson J. Serum amyloid A has antiviral activity against hepatitis C virus by inhibiting virus entry in a cell culture system. Hepatology. 2006;44:1626–34. doi: 10.1002/hep.21406. [DOI] [PubMed] [Google Scholar]
- 110.Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7:139–47. [PubMed] [Google Scholar]