Abstract
Background
Smyd1b is a member of the Smyd family that plays a key role in sarcomere assembly during myofibrillogenesis. Smyd1b encodes two alternatively spliced isoforms, smyd1b_tv1 and smyd1b_tv2, that are expressed in skeletal and cardiac muscles and play a vital role in myofibrillogenesis in skeletal muscles of zebrafish embryos.
Methodology/Principal Findings
To better understand Smyd1b function in myofibrillogenesis, we analyzed the subcellular localization of Smyd1b_tv1 and Smyd1b_tv2 in transgenic zebrafish expressing a myc-tagged Smyd1b_tv1 or Smyd1b_tv2. The results showed a dynamic change of their subcellular localization during muscle cell differentiation. Smyd1b_tv1 and Smyd1b_tv2 were primarily localized in the cytosol of myoblasts and myotubes at early stage zebrafish embryos. However, in mature myofibers, Smyd1b_tv1, and to a small degree of Smyd1b_tv2, exhibited a sarcomeric localization. Double staining with sarcomeric markers revealed that Smyd1b_tv1was localized on the M-lines. The sarcomeric localization was confirmed in zebrafish embryos expressing the Smyd1b_tv1-GFP or Smyd1b_tv2-GFP fusion proteins. Compared with Smyd1b_tv1, Smyd1b_tv2, however, showed a weak sarcomeric localization. Smyd1b_tv1 differs from Smyd1b_tv2 by a 13 amino acid insertion encoded by exon 5, suggesting that some residues within the 13 aa insertion may be critical for the strong sarcomeric localization of Smyd1b_tv1. Sequence comparison with Smyd1b_tv1 orthologs from other vertebrates revealed several highly conserved residues (Phe223, His224 and Gln226) and two potential phosphorylation sites (Thr221 and Ser225) within the 13 aa insertion. To determine whether these residues are involved in the increased sarcomeric localization of Smyd1b_tv1, we mutated these residues into alanine. Substitution of Phe223 or Ser225 with alanine significantly reduced the sarcomeric localization of Smyd1b_tv1. In contrast, other substitutions had no effect. Moreover, replacing Ser225 with threonine (S225T) retained the strong sarcomeric localization of Smyd1b_tv1.
Conclusion/Significance
Together, these data indicate that Phe223 and Ser225 are required for the M-line localization of Smyd1b_tv1.
Introduction
Smyd1, also known as Bop, is a member of the Smyd family that plays a key role in muscle cell differentiation [1]–[4]. Smyd1 encodes two alternatively spliced isoforms, smyd1_tv1 and smyd1_tv2, that are expressed in skeletal and cardiac muscles [3], [4]. Smyd1_tv1 differs from Smyd1_tv2 by containing a 13 amino acid insertion encoded by the smyd1_tv1-specific exon 5 [3], [4]. Targeted deletion of smyd1 in mice resulted in defective ventricle formation and early embryonic lethality at E10.5, suggesting a vital role of Smyd1 in cardiomyogenesis [3]. Knockdown of smyd1b expression in zebrafish resulted in paralyzed zebrafish larvae with defective myofibril assembly in skeletal myofibers [4].
The molecular mechanism by which Smyd1 controls the myofibrillogenesis is not clear. Biochemical studies indicate that Smyd1b could methylate histone H3 proteins in vitro [4]. Consistent with its potential function in transcriptional regulation, Smyd1 is initially localized in the nucleus of C2C12 myoblasts [5]. In vitro studies have revealed that smyd1 represses gene transcription in a histone deacetylase (HDAC) dependent fashion [3]. However, it has been reported that Smyd1 undergoes a nucleus to cytoplasm translocation during myoblast differentiation into myotubes [5], suggesting that Smyd1 may have additional function in the cytoplasm.
A better characterization of Smyd1b localization is critical for the mechanistic understanding of Smyd1b function in regulating muscle cell differentiation. In this study, we analyzed the subcellular localization Smyd1b_tv1 and Smyd1b_tv2 during muscle development in zebrafish embryos as well as in adult skeletal muscles. The data showed that Smyd1b_tv1 and Smyd1b_tv2 were primarily localized in the cytosol of myoblasts and myotubes of zebrafish embryos at the early stage. However, in mature myofibers of late stage embryos, a sarcomeric localization was evident for Smyd1b_tv1 and Smyd1b_tv2 although Smyd1b_tv2 appeared to be weaker. Double immunostaining with M- or Z-line markers revealed that Smyd1b_tv1 was localized on the M-line of sarcomeres. The strong M-line localization requires Phe223 and Ser225 located within the Smyd1b_tv1-specific 13 aa insertion. Mutation of Phe223 or Ser225 to alanine significantly reduced the sarcomeric localization of Smyd1b_tv1. In contrast, replacing Ser225 with threonine had no effect on the Smyd1b_tv1 sarcomeric localization
Results
Characterization of Smyd1b_tv1 and Smyd1b_tv2 subcellular localization during muscle development in zebrafish embryos
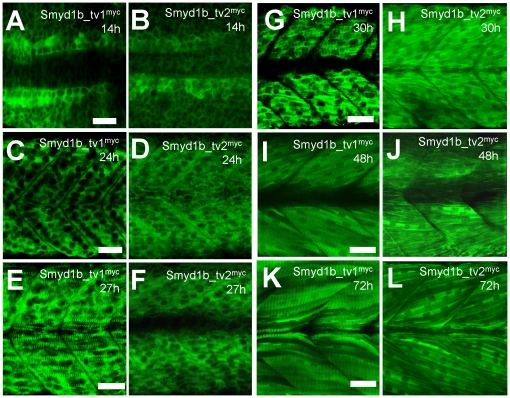
Previous studies have shown that Smyd1 undergoes a nucleus to cytoplasm translocation during C2C12 myoblast differentiation in vitro [5]. It is not clear whether the two isoforms, Smyd1b_tv1 and Smyd1b_tv2, from alternative splicing have similar or distinct subcellular localization in muscle cells during muscle development. To better understand Smyd1b function in myofibril assembly, we analyzed the subcellular localization of Smyd1b_tv1 and Smyd1b_tv2 during muscle development using transgenic zebrafish models that expressed a myc-tagged Smyd1b_tv1 or Smyd1b_tv2 under the control of its own promoter (Fig. 1). The results showed a dynamic subcellular localization of Smyd1b_tv1myc and Smyd1b_tv2myc during muscle development. In early stage embryos at 14 and 24 hpf, Smyd1b_tv1 and Smyd1b_tv2 were primarily localized in the cytosol of myoblast and myotubes with little or no nuclear localization (Fig. 2A–D). However, as embryos develop into late stages, a clear sarcomeric localization was detected for Smyd1b_tv1 in differentiated myofibers at 27 hpf (Fig. 2E). The sarcomeric localization appeared earlier for Smyd1b_tv1 than Smyd1b_tv2 (Fig. 2F). The sarcomeric localization of Smyd1b_tv1myc was maintained throughout early development (Fig. 2G, I, K). In contrast, a weak sarcomeric localization of Smyd1b_tv2 could be detected in zebrafish embryos at 72 hpf (Fig. 2L).
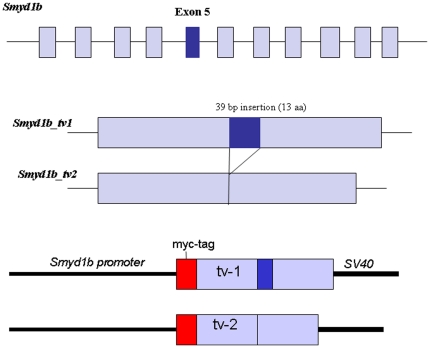
Figure 1. Generation of Smyd1b_tv1 and Smyd1b_tv2 by alternative splicing and construction of the Smyd1b_tv1myc and Smyd1b_tv2myc transgenes.
Smyd1b_tv1 and Smyd1b_tv2 transcripts are generated by alternative splicing. Their cDNA sequences are identical, with the exception of the 39 bp insertion encoded by exon 5. It translates into a 13 amino acids insertion in Smyd1b_tv1. Smyd1b_tv1myc and Smyd1b_tv2myc transgenes are constructed by fusing with an in frame myc-tag at the N-terminus. Expression of the transgenes are directed by its own promoter. A SV40 PolyA signal was included at the 3′ end.
Figure 2. Smyd1b_tv1myc and Smyd1b_tv2myc show dynamic localizations during muscle cell differentiation in zebrafish embryos.
A–D. Whole-mount immunostaining with anti-myc antibody shows the cytosolic localization of Smyd1b_tv1myc (A, C) or Smyd1b_tv2myc (B, D) in myoblasts of transgenic zebrafish embryos at 14 and 24 hours-post-fertilization (hpf), respectively. A and B, dorsal views. C and D, side views. E, G, I, K. Immunostaining with anti-myc antibody shows the sarcomeric localization of Smyd1b_tv1myc in myofibers of transgenic fish embryos at 27, 30, 48, and 72 hpf, respectively. F, H, J, L. Immunostaining with anti-myc antibody shows the cytosolic (F, H, J) and sarcomeric (L) localization of Smyd1b_tv2myc in myofibers of transgenic fish embryos at 27, 30, 48, and 72 hpf, respectively. Scale bars: 30 µm.
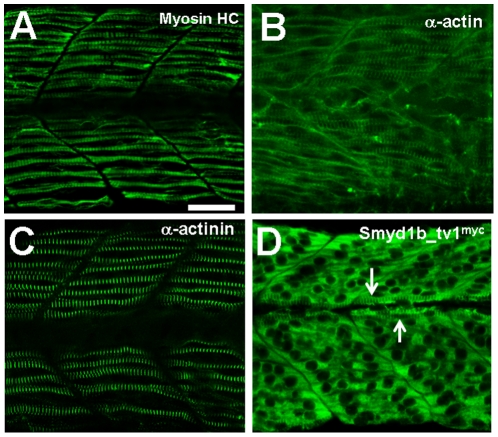
The timing of Smyd1b_tv1myc sarcomeric localization was compared with other sarcomeric proteins in trunk muscles of the same stage zebrafish embryos. The results showed that the sarcomeric localization of myosin heavy chain, α-actin and α-actinin occurred before the Smyd1b_tv1 sarcomeric localization (Fig. 3). Thick and thin filaments as well as Z-lines were clearly organized in myofibers of zebrafish embryos at 24 hpf (Fig. 3A–C). In contrast, there was little sarcomeric localization of Smyd1b-tv1myc in trunk muscles of the same stage embryos, except muscle pioneer cells in the myoseptum region representing the first group of muscle cells to differentiate in zebrafish embryos (Fig. 3D). Collectively, these data indicate that the sarcomeric localization of Smyd1b_tv1 occurred after that of myosin, α-actin and α-actinin, suggesting that although Smyd1b is required for myofibril assembly, the sarcomeric localization of Smyd1b_tv1 was not required in this initial process.
Figure 3. The sarcomeric localization of Smyd1b_tv1 occurs after the sarcomere formation in myofibers of zebrafish embryos.
A–C. Immunostaining using sarcomeric specific antibodies against MyHC (A), α-actin (B), and α-actinin (C) in the trunk muscles of zebrafish embryos at 24 hpf. D. Immunostaining using anti-myc antibody shows the primary cytoplasmic localization of Smyd1b_tv1 in the trunk muscles of smyd1b_tv1myc transgenic fish embryos at 24 hpf. Muscle pioneer cells with the sarcomeric localization are indicated by arrows. Scale bar: 30 µm.
Characterization of Smyd1b_tv1 and Smyd1b_tv2 subcellular localization using GFP fusion proteins
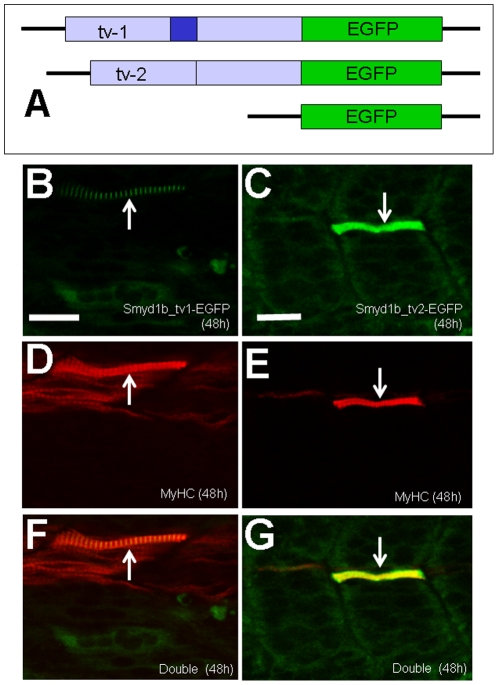
To better follow the dynamic localization of Smyd1b_tv1 and _tv2 during embryonic muscle development, we generated two DNA constructs, pTol2-smyd1b_tv1-EGFP and pTol2-smyd1b_tv2-EGFP, that express the GFP tagged Smyd1b_tv1 and Smyd1b_tv2 fusion proteins, respectively (Fig. 4A). Rescue assay by co-injecting Smyd1b morpholino (MO) with pTol2-smyd1b_tv1-EGFP or pTol2-smyd1_tv2-EGFP construct revealed that Smyd1b_tv1-EGFP and Smyd1b_tv2-EGFP fusion proteins are biologically active. Both Smyd1b_tv1-EGFP and Smyd1_tv2-EGFP could rescue the myofibril defects from Smyd1b knockdown (Fig. 4B–G). Expression of Smyd1b_tv1-EGFP or Smyd1_tv2-EGFP resulted in a mosaic pattern of normal myofibers in 90% (n = 60) of smyd1b knockdown embryos (Fig. 4F, G). In contrast, embryos co-injected with the Smyd1b MO and EGFP vector control showed no normal myofibers (data not shown). Similar to myc-tagged proteins, a sarcomeric localization was detected for Smyd1b_tv1-EGFP but not for Smyd1_tv2-EGFP (Fig. 4B, C). Collectively, these data indicate that Smyd1b_tv1-EGFP and Smyd1b_tv2-EGFP fusion proteins are biologically active and exhibit similar localization with the respective myc-tagged proteins.
Figure 4. Rescue of myofibril organization defect in smyd1b knockdown embryos by expression of Smyd1b_tv1-EGFP or Smyd1b_tv2-EGFP fusion protein.
A. DNA constructs encoding Smyd1b_tv1-EGFP, or Smyd1b_tv2-EGFP fusion proteins or EGFP control were generated and injected into zebrafish embryos. B and C. Myofibers expressing Smyd1_tv1-EGFP (B) or Smyd1_tv2-EGFP (C) was directly observed by GFP. D and E. Myosin thick filaments organization was determined by F59 antibody staining in Smyd1_tv1-EGFP (D) or Smyd1_tv2-EGFP (E) co-injected embryos. F and G. Double staining shows the colocalization of normal fibers with Smyd1_tv1-EGFP (F) or Smyd1_tv2-EGFP (G) expression. Scale bars: 20 µm.
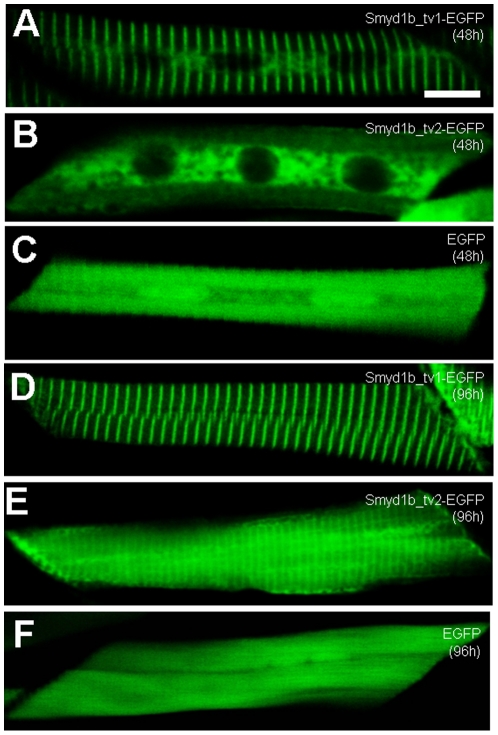
The expression and subcellular localization of Smyd1b_tv1-EGFP and Smyd1_tv2-EGFP were further characterized in zebrafish embryos at 48 and 96 hpf. Consistent with the data from the myc-tagged proteins, Smyd1b_tv1-EGFP was mainly localized on the sarcomeres of skeletal muscles at 48 and 96 hpf (Fig. 5A, D). In contrast, Smyd1b_tv2-EGFP (Fig. 5B), like the GFP control (Fig. 5C), showed little or no sarcomeric localization in skeletal myofibers of zebrafish embryos at 48 hpf. However, a weak sarcomeric localization was observed with Smyd1b_tv2-EGFP in 96 hpf embryos (Fig. 5E). Together, these studies suggest a dynamic subcellular localization of Smyd1b_tv1 and tv_2 during muscle development.
Figure 5. Characterization of the sarcomeric localization using Smyd1b_tv1-EGFP and Smyd1b_tv2-EGFP fusion proteins.
DNA constructs encoding Smyd1b_tv1-EGFP, or Smyd1b_tv2-EGFP fusion proteins or EGFP control injected into zebrafish embryos. Their expression and localization was determined in myofibers of the injected zebrafish embryos at 48 (A–C) and 96 (D–F) hpf. A and D, Smyd1b_tv1-EGFP; B and E, Smyd1b_tv2-EGFP; C and F, EGFP control. Scale bar: 8 µm.
Smyd1b_tv1 is localized on the M-line of sarcomeres
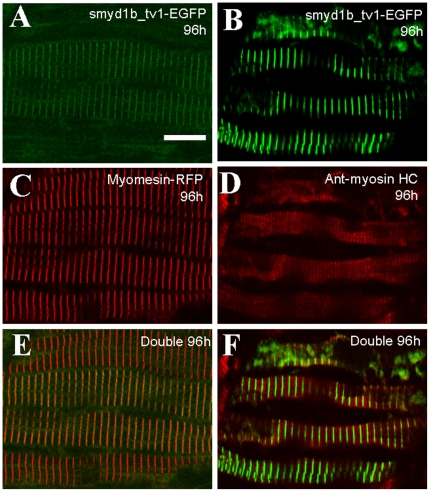
To better define the sarcomeric localization of Smyd1b_tv1 and _tv2, we localized Smyd1b_tv1-EGFP in a zebrafish embryos expressing a myomesin-RFP fusion protein at the M-line. The pTol2-smyd1b_tv1-EGFP construct was microinjected into the myomesin-RFP zebrafish embryos at 1–2 cells stages. The sarcomeric localization of Smyd1b_tv1-EGFP (green) and myomesin-RFP (red) was determined by confocal microscopy at 96 hpf. A clear co-localization of Smyd1b_tv1-EGFP and myomesin-RFP was observed in myofibers expressing the Smyd1b_tv1-EGFP fusion protein in zebrafish embryos (Fig. 6A, C, E). Moreover, co-staining with anti-MyHC antibody revealed that Smyd1b_tv1-GFP was localized in the middle of the A-band (Fig. 6B, D, F), consistent with the M-line localization of Smyd1b_tv1.
Figure 6. Smyd1b_tv1-EGFP is localized on the M-line of zebrafish skeletal muscles.
Smyd1b_tv1-EGFP construct was injected into Myomesin-RFP (A, C, E) or wild type (B, D, F) zebrafish embryos at 1–2 cell stages. Smyd1b_tv1-EGFP localization was determined together with M-line marker (Myomesin-RFP) and A-band marker (Myosin heavy chain) at 96 hpf. A, B and C. Co-localization of Smyd1b_tv1-EGFP and Myomesin-RFP was observed in myofibers of the injected embryos. D, E and F. Immunostaining with anti-MyHC antibody (F59) shows the localization of Smyd1b_tv1-EGFP in the middle of the A-bands in myofibers of zebrafish embryos. Scale bar: 12 µm.
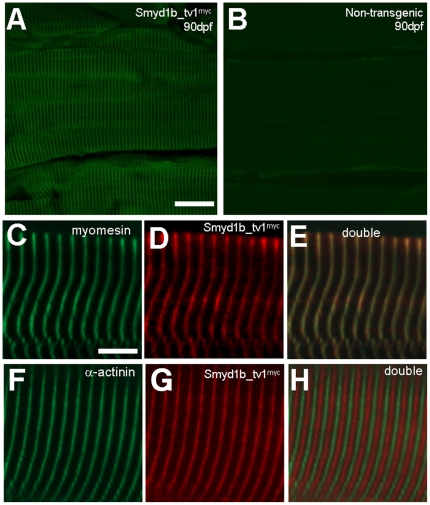
To determine whether the sarcomeric localization of smyd1b_tv1myc is maintained in adult skeletal muscles, we carried out an anti-myc antibody staining on adult skeletal muscles of transgenic zebrafish expressing the myc-tagged Smyd1b_tv1myc. A clear sarcomeric localization was detected for Smyd1b_tv1myc (Fig. 7A). Double staining with the M-line (myomesin) and Z-line (α-actinin) specific antibodies further confirmed that Smyd1b_tv1myc was co-localized with myomesin on the M-line (Fig. 7E), but not with α-actinin on the Z-line in adult skeletal muscles (Fig. 7H). The M-line localization is consistent with the results from the embryonic muscles. Together, these data indicate that Smyd1b_tv1 is localized on the M-lines of skeletal muscles, and may be involved in M-line organization.
Figure 7. Smyd1b_tv1myc is localized on the M-line of adult zebrafish skeletal muscles.
A and B. Immunostaining using anti-myc antibody shows the sarcomeric localization of Smyd1b_tv1myc on longitudinal sections of skeletal muscles from adult transgenic zebrafish expressing a myc-tagged Smyd1b_tv1 (A) or non-transgenic control (B). C–E. Double immunostaining with anti-myomesin and anti-myc antibodies shows the colocalization of Smy1b_tv1myc with myomesin on the M-lines. F–H. Double immunostaining with anti-α-actinin and anti-myc antibodies shows the non-overlapping localization of Smy1b_tv1myc with α- actinin. Scale bars: A = 22 µm. C = 6 µm.
The enhanced sarcomeric localization of Smyd1b_tv1 requires Phe223 and Ser225 within the Smyd1b_tv1-specific 13 aa insertion
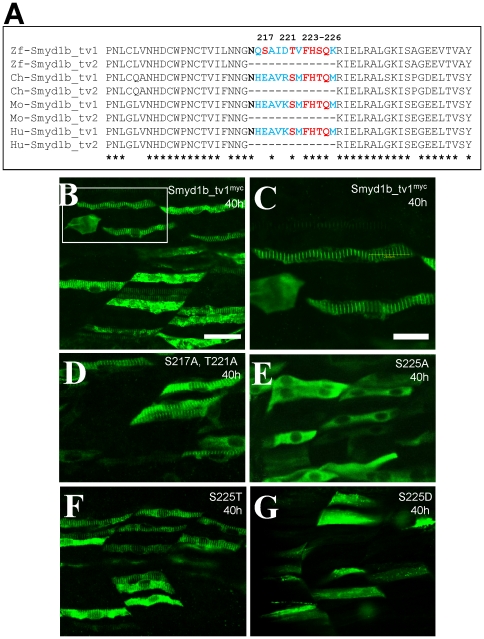
Compared with Smyd1b_tv2, the stronger sarcomeric localization of Smyd1b_tv1 suggested that the 13 amino acid insertion in the Smyd1b_tv1 might contribute to its increased sarcomeric localization. To identify the key amino acid residue(s) required for the enhanced sarcomeric localization, we compared the protein sequence within the 13 aa insertion among Smyd1b_tv1 orthologs from several vertebrate species. Several potential phosphorylation sites at Ser217, Thr221 and Ser225, were identified (Fig. 8A). To test directly whether these three Ser/Thr residues are required for the sarcomeric localization of Smyd1b_tv1, substitutions were made at these three positions by replacing them with alanine. The mutant proteins were expressed in zebrafish embryos by DNA microinjection. The subcellular localization of mutant proteins was carefully examined in the injected zebrafish embryos by antibody staining. Compared with the control (Fig. 8B, C), substitution of Ser217 and Thr221 with alanine had no effect on the sarcomeric localization of Smyd1b_tv1myc (Fig. 8D). However, substitution of Ser225 with alanine (S225A) abolished the sarcomeric localization of Smyd1b_tv1myc (Fig. 8E). Together, these results indicate that Ser225 is required for the enhanced sarcomeric localization of Smyd1b_tv1.
Figure 8. The Serine 225 is required for the enhanced sarcomeric localization of Smyd1b_tv1.
A. Sequence comparison shows that the alternative splicing of smyd1b in various vertebrates and the conserved serine and threonine residues within the 13 aa insertion. B and C. Immunostaining using anti-myc antibody shows the sarcomeric localization of Smyd1b_tv1myc in myofibers of zebrafish embryos at 38 hpf. C represents the highlighted box area in A. D–G. Immunostaining using anti-myc antibody shows the sarcomeric localization of Smyd1b_tv1myc mutant proteins that carry substitutions at S217A and T221A (D), S225A (E), S225T (F), S225D (G). Scale bars: B = 40 µm; C = 20 µm.
Sequence comparison revealed that Ser225 was replaced by threonine in Smyd1b_tv1 from chick, mouse and human (Fig. 8A). Serine and threonine are similar type of amino acids that are potential sites for post-translational modification by phosphorylation or glycosylation. To determine whether substitution of S225 with threonine (S225T) had an effect on its sarcomeric localization, we generated the S225T mutant and analyzed the sarcomeric localization of S225T in zebrafish embryos. The results showed that Ser225T substitution had no effect on the sarcomeric localization of Smyd1b_tv1 (Fig. 8F). To determine whether potential phosphorylation of Ser225 could be involved in sarcomeric localization of Smyd1b_tv1, we substituted Ser225 with aspartic acid (S225D). It has been reported that substitution of serine residues with aspartic acid mimics serine phosphorylation [6], [7]. Our results showed that S225D substitution abolished the Smyd1b_tv1 sarcomeric localization (Fig. 8G), indicating that post-translational modification by phosphorylation may not be involved in the increased sarcomeric localization of Smyd1b_tv1.
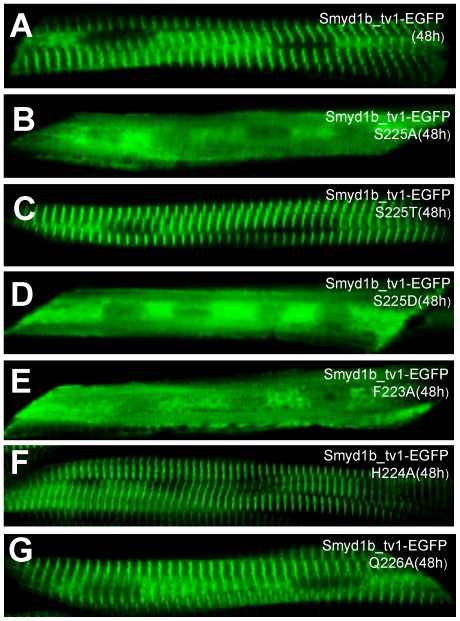
To confirm the data from the myc-tagged Smyd1b mutant proteins, we generated DNA constructs expressing Smy1b_tv1-EGFP fusion proteins with the S225A, S225T or S225D substitutions, and determined the effects on their sarcomeric localization in zebrafish embryos. The data showed that S225A substitution significantly reduced the sarcomeric localization of Smyd1b_tv1-EGFP (S225A) fusion protein (Fig. 9B). In contrast, S225T mutation had little or no effect on the sarcomeric localization of Smyd1b_tv1-EGFP (S225T) (Fig. 9C). Moreover, S225D mutation dramatically reduced the sarcomeric localization of Smyd1b_tv1-EGFP (S225D) fusion protein (Fig. 9D). These data reinforce the idea that the S225 residue is critical for the enhanced sarcomeric localization of Smyd1b_tv1. However, because the phosphomimetic Smyd1b variant (S225D) also leads to abolished M-line translocation, phosphorylation of these amino acids cannot be the cause for the M-line translocation. The S225A substitution might lead to a different protein shape masking or changing the translocation signal required for M-line localization. To evaluate the possibility that the highly conserved amino acids surrounding the Ser225 might be involved in M-line localization, we carried out additional mutations at F223, H224 and Q226. The data showed that mutating the conserved Phe223 (F223) dramatically diminished the M-line localization (Fig. 9E). In contrast, mutating two other conserved residues (H224, Q226) had no effect (Fig. 9F, G). Collectively, the data indicate that Phe223 and Ser225 are required for the sarcomeric localization. S225A or F223A substitution might lead to a different protein shape masking or changing the translocation signal or protein motif required for M-line localization.
Figure 9. Effect of S225A, S225T, S225D, F223A, H224A and Q226A substitution on the sarcomeric localization of Smyd1_tv1-EGFP in zebrafish embryos.
DNA construct expressing Smyd1_tv1-EGFP or its derived mutants of S225A, S225T, S225D, F223A, H224A and Q226A was injected into zebrafish embryos. Their localization was analyzed in myofibers of the injected embryos at 48 hpf. A, Smyd1_tv1-EGFP; B, Smyd1_tv1-EGFP(S225A); C, Smyd1_tv1-EGFP(S225T); D, Smyd1_tv1-EGFP(S225D), E, Smyd1_tv1-EGFP(F223A), F, Smyd1_tv1-EGFP(H224A), and Smyd1_tv1-EGFP(Q226A).
Knockdown of endogenous Smyd1b advances the timing of Smyd1b_tv1myc sarcomeric localization in zebrafish embryos
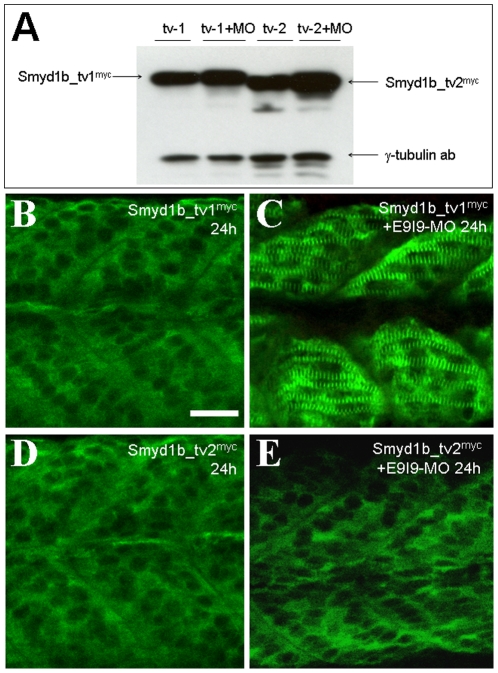
To determine whether knockdown of endogenous Smyd1b expression could affect the expression and sarcomeric localization of Smyd1b_tv1myc and Smyd1b_tv2myc from the transgene, the splicing MO (E9I9-MO) was injected into smyd1b_tv1 transgenic zebrafish embryos expressing the Smyd1b_tv1myc or Smyd1b_tv2myc fusion proteins. Western blot analysis revealed that the E9I9-MO could not knock down the expression of the myc-tagged smyd1b_tv1or smyd1b_tv2 from the transgene because they were constructed using the cDNA coding sequence and the expression does not require RNA splicing. Compared with the uninjected respective transgenic control, E9I9-MO injected embryos showed similar or slightly increased levels of Smyd1b_tv1myc and Smyd1b_tv2myc protein expression (Fig. 10A).
Figure 10. Knockdown of endogenous Smyd1b advances the timing of sarcomeric localization of Smyd1b_tv1myc in zebrafish embryos.
A. Smyd1b E9I9-MO was injected into Smyd1b_tv1myc or Smyd1b_tv1myc transgenic zebrafish embryos at 1–2 cell stages. Western blot analysis shows the expression of myc-tagged Smyd1b_tv1myc and Smyd1b_tv1myc in un-injected control or E9I9-MO injected transgenic zebrafish embryos at 24 hpf. γ-Tubulin was used as loading control. B and C. Immunostaining using anti-myc antibody shows the cytoplasmic (B) or sarcomeric localization (C) of smyd1b_tv1myc in control (B) or E9I9-MO injected (C) transgenic zebrafish embryos at 24 hpf. D and E. Immunostaining using anti-myc antibody shows the cytoplasmic localization of smyd1b_tv2myc in control or E9I9-MO injected transgenic zebrafish embryos at 24 hpf. Scale bar: 30 µm.
The subcellular localization of Smyd1b_tv1myc or Smyd1b_tv2myc was analyzed in the smyd1b knockdown embryos. The results showed that knockdown of endogenous smyd1b expression advanced the timing of Smyd1b_tv1 sarcomeric localization (Fig. 10C). Compared with the uninjected smyd1b_tv1myc embryos that showed little or no sarcomeric localization at 24 hpf (Fig. 10B), the E9I9-MO injected embryos showed a clear sarcomeric localization of Smyd1b_tv1myc at 24 hpf (Fig. 10C). In contrast, knockdown of endogenous smyd1b had no effect on cytoplasmic localization of Smyd1b_tv2 (Fig. 10E). Similar to control embryos (Fig. 10D), Smyd1b_tv2 remained in cytoplasm of smyd1b knockdown embryos at 24 hpf (Fig. 10D, E). Together, these data indicate that knockdown of the endogenous Smyd1b expression could enhance the sarcomeric localization of Smyd1b_tv1myc in zebrafish embryos.
The Smyd1b-tv1 sarcomeric localization was disrupted in hsp90α1 mutant embryos
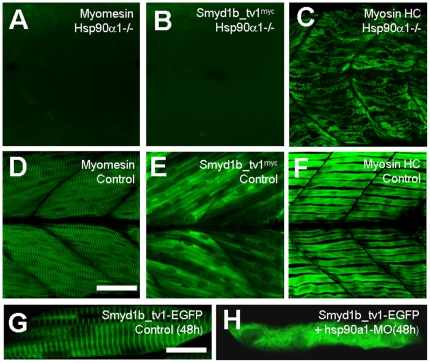
It has been reported that hsp90α1 knockdown or mutation severely disrupts the myofibril organization in skeletal muscles of zebrafish embryos [8]–[10]. To determine the effect of hsp90α1 mutation on Smyd1b_tv1 sarcomeric localization, we generated Smyd1b_tv1myc transgenic fish in the hsp90α1 mutant background and characterized Smyd1b_tv1myc localization in hsp90α1 homozygous mutant Smyd1b_tv1myc transgenic zebrafish embryos. The result showed that hsp90α1 mutation which disrupts M-line organization (Fig. 11A) completely abolished the sarcomeric localization of Smyd1b_tv1myc (Fig. 11B) despite some disorganized thick filaments remained (Fig. 11C).
Figure 11. The effect of hsp90α1 mutation or knockdown on Smyd1b_tv1 sarcomeric localization.
A and D. Immunostaining using anti-myomesin antibody shows the organization of myomesin in hsp90α1 mutant (A), or control (D) smyd1b_tv1myc transgenic embryos at 72 hpf. B and E. Immunostaining using anti-myc antibody shows the localization of Smyd1b_tv1myc in hsp90α1 mutant (B), or control (E) smyd1b_tv1myc transgenic embryos at 72 hpf. C and F. F59 staining shows the organization of slow muscle myosin in hsp90α1 mutant (C), or control (F) smyd1b_tv1myc transgenic embryos at 72 hpf. G and H. DNA construct Smyd1_tv1-EGFP was injected alone or together with hsp90α1 ATG-MO into zebrafish embryos. Smyd1_tv1-EGFP localization was determined in myofibers of the control (G), or hsp90α1 knockdown (H) at 48 hpf. Scale bars: D = 40 µm; G = 15 µm.
To confirm the requirement of Hsp90α1 on Smyd1b_tv1 localization, we analyzed Smyd1b_tv1-EGFP localization in hsp90α1 knockdown zebrafish embryos. Compared with the uninjected control (Fig. 11G), knockdown of hsp90α1 completely blocked the sarcomeric localization of Smyd1b_tv1-EGFP in myofibers of zebrafish embryos (Fig. 11H). Collectively, these data indicate that the M-line structure is essential for the Smyd1b sarcomeric localization, reinforcing the idea that Smyd1b is localized on the M-line of sarcomeres.
Discussion
In this study, we analyzed the subcellular localization of Smyd1b_tv1 and Smyd1b_tv2 during muscle development in zebrafish embryos. We found that Smyd1b_tv1 and Smyd1b_tv2 were primarily localized in the cytosol of myoblasts and myotubes in early stage embryos. However, they showed a sarcomeric localization in the differentiated myofibers of late stage zebrafish embryos and adult skeletal muscles. Compared with Smyd1b_tv2, Smyd1b_tv1 showed a stronger sarcomeric localization, and the sarcomeric localization is restricted to the M-line. The stronger sarcomeric localization of Smyd1b_tv1 requires Phe223 and Ser225 located within the Smyd1b_tv1 specific 13 aa insertion.
Cytosolic localization of Smyd1b_tv1 and Smyd1b_tv2 in myoblasts
It has been reported that Smyd1 is localized in the nucleus of C2C12 myoblasts and undergoes a nucleus-to-cytoplasm translocation during myoblast differentiation in vitro [5]. In this study, we found that Smyd1b_tv1 and Smyd1b_tv2 were primarily localized in the cytosol of myoblasts and early myotubes of zebrafish embryos. Very little or no nuclear localization could be detected in myoblasts and myotubes of zebrafish embryos in vivo. The discrepancy between these two studies is not clear. It could be due to the use of two different model systems in these two studies, the in vitro C2C12 cell culture system versus the in vivo developing zebrafish embryos. C2C12 myoblast cells in culture may not fully resemble the process of muscle cell differentiation in vivo. It takes 1–2 days for C2C12 cells to differentiate in vitro whereas only a few hours are required for myoblast cell differentiation into myofibers in vivo in zebrafish embryos. It is possible that the nuclear localization is low and transient in developing muscles of zebrafish embryos that are below the sensitivity of detection by immunostaining. Consistent with this possibility, we found a weak nuclear localization when zebrafish Smyd1b_tv1 was expressed in C2C12 cells by DNA transfection (Data not shown). Moreover, the nuclear localization of zebrafish Smyd1b_tv1 was enhanced in C2C12 cells when incubated with a nuclear export blocker Leptomycin B (unpublished data). Collectively, these studies argue that Smyd1b_tv1 and Smyd1b_tv2 may exhibit a dynamic subcellular localization during muscle cell differentiation.
Sarcomeric localization of Smyd1b_tv1 and Smyd1b_tv2 in myofibers
We showed that Smyd1b_tv1, and to some extent Smyd1b_tv2, were localized on the sarcomere of differentiated myofibers. Their sarcomeric localization appeared in a progressive fashion from anterior to posterior myotome during muscle development, correlating with the progression of myotome formation and muscle cell differentiation in zebrafish embryos. Double staining with sarcomere specific markers demonstrated that Smyd1b_tv1 was localized at the M-line. Disruption of M-line organization by Hsp90α1 knockdown or mutation completely abolished the sarcomeric localization of Smyd1b. Our findings are consistent with recent report by Just and colleagues showing the M-line localization of Smyd1b_tv1 in skeletal muscle fibers of zebrafish embryos [11]. Because the M-line is essential for stable contractions of the sarcomeres, the M-line localization of Smyd1b could be of biologically significant.
Interestingly, we showed that the M-line localization of myc-tagged Smyd1b_tv1 appeared earlier when the endogenous Smyd1b is knockdown. One possible explanation is that myc-tagged Smyd1b_tv1 proteins compete with the endogenous Smyd1b_tv1 proteins for the M-line localization. When endogenous Smyd1b_tv1 was knocked down, more myc-tagged Smyd1_tv1 proteins could be localized to the M-lines and thus become visible at an earlier stage of development. We do not expect to see a functional consequence of an advanced sarcomeric localization of the myc-tagged Smyd1b_tv1, because the myc-tagegd Smyd1b_tv1 could functionally replace the endogenous Smyd1b_tv1 [4].
Given that subcellular distribution of both Smyd1b splice forms is indeed different, what are the common functions of the isoforms since both isoforms are capable to rescue the loss of Smyd1b? It has been reported that Smyd1b_tv1 binds to myosin [11]. The myosin binding domain has been mapped to the C-terminal region between residues 278–390. This region is identical between Smyd1b_tv1 and _tv2, suggesting that both Smyd1b_tv1 and _tv2 are capable of binding to myosin, consistent with previous findings that both isoforms are capable to rescue the loss of Smyd1b [4].
Previous studies have indicated that Smyd1b is a histone methyltransferase which could methylate H3K9 in vitro [4]. Recent studies, however, have demonstrated that members of the Smyd family are able to methylate other non-histone proteins [12]. Protein methylation is a reversible post-translational modification, which controls biological activities and stability of proteins [13]–[15]. We speculate that Smyd1b could methylate structural or regulatory proteins at the M-line involved in sarcomere assembly and stability. Several muscle proteins including myosin, α-actin and creatine kinase are known to be methylated in skeletal muscles [16]–[18]. However, it remains to be determined whether any of them are the Smyd1 target proteins.
It should be noted that although sarcomeric localization of Smyd1b occurred after the α-actinin, α-actin, and MyHC, knockdown of Smyd1b completely abolished the sarcomere formation, thick and thin filament organization as well as Z-line and M-line in skeletal muscles of zebrafish embryos ([4], and unpublished data). These data suggest that Smyd1b is required for sarcomere assembly before their localization on the sarcomeres. However, it does not rule out the possibility that SmyD1b has additional function at the M-lines. It has been reported that myosin chaperones Unc45b and Hsp90α could be localized on sarcomeres of myofibers. Moreover, they shuttle between the A band and the Z line in response to stress or damage to the myofiber [19]. It has been suggested that the sarcomeric localization of myosin chaperones and their translocation within different parts of the muscle cells could be involved in the response of muscle cells to mount efficient physiological responses to muscle stress, load requirements, and/or stretch. It remains to be determined whether the sarcomeric localization of Smyd1b_tv1 is involved in sarcomere remodeling and response to muscle stress.
Regulation of sarcomeric localization by Ser225 and Phe223
Although both Smyd1b_tv1 and Smyd1b_tv2 showed the sarcomeric localization, they differ significantly in timing and intensity of sarcomeric localization. Smyd1b_tv1 appeared earlier than Smyd1b_tv2 on the sarcomeres, and the sarcomeric localization is stronger than Smyd1b_tv2. Data from this study demonstrated that the enhanced sarcomeric localization of Smyd1b_tv1 requires Phe223 and Serine 225. Substitution of Phe223 or Ser225 with alanine significantly reduced its sarcomeric localization to a similar level as Smyd1b_tv2. Serine phosphorylation is a common post-translational modification involved in the regulation of protein subcellular localization. Phosphorylation of NFAT has been shown to play a vital role in its shuttling between the nucleus and sarcomeres [20]–[22]. Post-translation modification on Ser225 of Smyd1b_tv1 could be a potential mechanism regulating its sarcomeric localization. Consistent with the potential regulation by phosphorylation, substitution of Ser225 with threonine had no effect on its sarcomeric localization. However, substitution of Ser225 with aspartic acid, which mimics phosphorylation, significantly reduced Smyd1b_tv1 sarcomeric localization, arguing against the idea that phosphorylation of Ser225 is involved in regulation of Smyd1b_tv1 sarcomeric localization. The mechanism by which Ser225 is involved in Smyd1b_tv1 sarcomeric localization remains to be determined. In addition to being a potential phosphorylation site, Serine residues could also serve as target sites for O-glycosylation. However, Smyd1b appears to be an intracellular protein that makes it an unlikely target for O-glycosylation, suggesting that post-translational modification by other mechanisms may be involved in the increased sarcomeric localization of Smyd1b_tv1. Alternatively, the Phe223 and Ser225 might be part of translocation signal or protein motif required for the M-line localization. S225A or F223A substitution might result in a different protein shape masking or changing the translocation signal and lead to disruption of the M-line localization.
Materials and Methods
Zebrafish lines and maintenance
Mature zebrafish were raised at the zebrafish facility of the Aquaculture Research Center, Institute of Marine and Environmental Technology. The fish were maintained at 28°C with a photoperiod of 14 h light and 10 h dark, in 8-gallon aquaria supplied with freshwater and aeration. The Tg(smyd1-smyd1bmyc_tv1)mb6 and Tg(smyd1-smyd1bmyc_tv2)mb7 transgenic zebrafish lines were generated as described [4]. The minigenes were constructed using cDNA encoding the myc-tagged Smyd1b_tv1 or myc-tagged SmyD1b_tv2 cloned after the 5.3 kb muscle-specific zebrafish smyd1b promoter and its 5′-flanking sequence [23]. The slotu44c mutant zebrafish line was obtained from Tubingen Zebrafish Stock Center. The slotu44c mutant carries a nonsense mutation in the hsp90α1 gene resulting in truncated molecules missing the C-terminal domain, which is important for both homo- and heterodimerization [10]. The smyd1-smyd1bmyc_tv1 transgenic fish was crossed with slotu44c heterozygous mutant to generate smyd1-smyd1bmyc_tv1/slotu44c transgenic line. The myomesin-RFP zebrafish line (GBT0067) was obtained from Steve Ekker's laboratory at Mayo Clinic. It carries a RFP insertion in myomesin 3 [24]. Additional information can be found at http://www.zfishbook.org/index.php?topic=GBT0067#.
Synthesis of morpholino-modified antisense oligos
The Smyd1b splicing blocker (E9I9-MO, 5′-CGTCACCTCTAGGTCTTTAGTGATG-3′) was based on the sequence of splicing site at the exon-9 and intron-9 junction of zebrafish smyd1b gene [4]. The hsp90α1 translation blocker (ATG-MO, 5′- CGACTTCTCAGGCATCTTGCTGTGT- 3′) was targeted to sequence near the ATG start codon of zebrafish hsp90α1 mRNA transcript [9].
Morpholino and DNA microinjection in zebrafish embryos
Morpholino antisense oligos were dissolved in 1×Danieau buffer [25] to a final concentration of 0.5 mM. DNA plasmid was dissolved in water at 50 ng/µl. 1–2 nl of MO or DNA was injected into zebrafish embryos at 1 or 2 cell stages. For morpholino and DNA co-injection, morpholino (1 mM) and DNA (100 ng/µl) were mixed at 1∶1 ratio and 1–2 nl of the mixture was injected into zebrafish embryos at 1 or 2 cell stages.
Immunostaining of cryostat sections and whole mount zebrafish embryos
For immunostaining with cross sections, skeletal muscles were dissected from trunk muscles of transgenic zebrafish (90 dpf) expressing Smyd1b_tv1myc and Smyd1b_tv2myc. The muscle tissues were fixed in 4% paraformaldehyde for 1 h at room temperature. The fixed samples were washed with 1×PBS-Tween (PBS, 0.1% Tween) 2×10 min, and then soaked in 30% sucrose for 2 h. The samples were transferred into an embedding chamber filled with OCT cryostat embedding medium (Tissue Tek). The embedding chambers were frozen on dry ice, and frozen blocks were cut on a cryostat at −20°C to produce 15 µm sagittal sections. Sections were transferred to subbed slides and allowed to dry completely at 37°C for 1 h. Sections were rehydrated in PBS-Tween, and non-specific staining was blocked using 10% goat serum in PBS-Tween for 10 min. Sections were then incubated overnight at 4°C in primary antibodies diluted in PBS-Tween. They were then washed with PBS-Tween for 5×5 min and incubated with fluorescence-labeled secondary antibodies, appropriate for the primary antibody isotype, for 1 h at room temperature. Sections were coverslipped in 50% Vector shield (Invitrogen) and observed under fluorescence microscopy (Axioplan-2, Zeiss).
Immunostaining with whole mount zebrafish embryos was carried out as previously described [4]. Tg(smyd1b_tv1)mb6 or Tg(smyd1b_tv2)mb7 transgenic zebrafish embryos were fixed at 14, 24, 27, 30, 48 and 72 hpf. The subcellular localization of Smyd1b_tv1 or Smyd1b_tv2 in myoblasts, myotubes, and myofibers was determined by anti-myc antibody staining. The following antibodies were used for both cryostat sections and whole mount zebrafish embryos: anti-myc monoclonal antibody (9E10, DSHB), anti-myc polyclonal antibody (Cell Signaling Technology, #2272), anti-α-actinin (clone EA-53, #A7811, Sigma), anti-MHC for slow muscles (F59, DSHB), anti-myomesin (mMaC myomesin B4, DSHB). Secondary antibodies were FITC or TRITC-conjugates anti-mouse or anti- rabbit antibodies (Sigma).
Mutagenesis
To generate Smyd1b_tv1 constructs with S217A, S225A, S225T, S225D, T221A, F223A, H224A and Q226A mutations, we carried out the mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). The smyd1-smyd1bmyc_tv1 plasmid was used as DNA template. The following PCR primers were used.
S217A+T221A-f: 5′-AATCAGGCGGCCATCGATGCTGTfGTTT-3′
S217A+T221A-r: 5′-AAACACAGCATCGATGGCCGCCTGATT-3′
S225A-f: 5′-GTGTTTCACGCTCAGAAGAGG-3′
S225A-r: 5′-CCTCTTCTGAGCGTGAAACAC-3′
A225T-f: 5′-GATACTGTGTTTCACACTCAGAAGAGGATTG-3′
A225T-r: 5′-CAATCCTCTTCTGAGTGTGAAACACAGTATC-3′
S225D-f: 5′-ATACTGTGTTTCACGATCAGAAGAGGATTGA-3′
S225D-r: 5′-TCAATCCTCTTCTGATCGTGAAACACAGTAT-3′
F223A-f: 5′-CATCGATACTGTGGCTCACTCTCAGAAG-3′
F223A-r: 5′-CTTCTGAGAGTGAGCCACAGTATCGATG-3′
H224A-f: 5′-CGATACTGTGTTTGCCTCTCAGAAGAGG-3′
H224A-r: 5′-CCTCTTCTGAGAGGCAAACACAGTATCG-3′
Q226A-f: 5′-TGTGTTTCACTCTGCGAAGAGGATTGAG-3′
Q226A-r: 5′-CTCAATCCTCTTCGCAGAGTGAAACACA-3′
Construction of Tol2-smyd1_tv1-EGFP, Tol2-smyd1_tv2-EGFP and their derived mutant constructs
pTol2-smyd1b_tv1-EGFP and pTol2-smyd1b_tv2-EGFP constructs were generated by cloning the smyd1b_tv1 and smyd1b_tv2 coding sequence in frame upstream of the EGFP coding sequence in the Tol2 vector. Briefly, the smyd1b_tv1 and smyd1b_tv2 coding sequence without the stop codon were generated by PCR using pfu DNA polymerase (Stratagene). DNA plasmid smyd1-smyd1bmyc_tv1 and smyd1-smyd1bmyc_tv2 were used as template amplified using the smyd1b-F (5′-CGGGATCCATGGAGTTTGTGGAAGTTTTTGA-3′) and smyd1b-R (5′-CGGGATCCTTCCTGCGGAACAGGTTCTTGAT-3′) primers. A BamHI site was introduced at the 5′ and 3′ ends of the smyd1b_tv1 or smyd1b_tv2 coding sequence via the PCR primers. The PCR products were digested with BamHI and then cloned into the BamHI site of the T2A200R150G vector [26]. The DNA sequence at the smyd1b and EGFP junction was confirmed by sequencing.
pTol2-smyd1b_tv1-EGFP(S225A), pTol2-smyd1b_tv1-EGFP(S225T), pTol2-smyd1b_tv1-EGFP(S225D), pTol2-smyd1b_tv1-EGFP(F223A), pTol2-smyd1b_tv1-EGFP(H224A), and pTol2-smyd1b_tv1-EGFP(Q226A) mutant constructs were generated by PCR using the QuikChange site-directed mutagenesis kit (Stratagene) as described previously. DNA plasmid pTol2-smyd1b_tv1-EGFP was used as template amplified using the S225A-f/r, S225T-f/r, S225D-f/r, F223A-f/r, H224A-f/r and Q226A-f/r primer sets described previously.
Analysis of protein expression by Western blot
Western blot analysis was performed with control and smyd1b knockdown zebrafish embryos as described [27]. Chorions were removed from control, or MO-injected embryos (50 embryos for each group) at 24 hpf. Yolk sacs were removed by gently pipeting embryos through a glass pipet in 1 ml of PBS buffer. The embryos were collected by centrifugation at 5000 rpm for 20 seconds. The pellet of embryos was dissolved in 150 µl of 2× SDS loading buffer (3 µl for each embryo), and homogenized with a 23 gauge needle. 2 µl of PMSF was added to reduce the bubbles. The sample was boiled for 3 min at 100C. 20 µl of protein sample was analyzed on a 7.5% SDS PAGE. The proteins were transferred onto an Immobilon-P membrane (Millipore) and immunostaining was carried out using anti-myc (9E10; DSHB), and anti-γ-tubulin (T6557; Sigma) antibodies.
Acknowledgments
We thank Suzanne Hughes for editing and constructive discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by research grant No IS-8713-08 from the Israel Binational Agricultural Research and Development Fund, the United States (BARD), and an intercenter collaboration grant (Du-Fang) from University of Maryland Biotechnology Institute. (http://www.bard-isus.com/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hwang I, Gottlieb PD. Bop: a new T-cell-restricted gene located upstream of and opposite to mouse CD8b. Immunogenetics. 1995;42:353–361. doi: 10.1007/BF00179396. [DOI] [PubMed] [Google Scholar]
- 2.Hwang I, Gottlieb PD. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J Immunol. 1997;158:1165–1174. [PubMed] [Google Scholar]
- 3.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 4.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci U S A. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sims RJ, Weihe EK, Zhu L, O'Malley S, Harriss JV, et al. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- 6.Leger J, Kempf M, Lee G, Brandt R. Conversion of serine to aspartate imitates phosphorylation-induced changes in the structure and function of microtubule-associated protein tau. J Biol Chem. 1997;272:8441–8446. doi: 10.1074/jbc.272.13.8441. [DOI] [PubMed] [Google Scholar]
- 7.Saad JS, Kim A, Ghanam RH, Dalton AK, Vogt VM, et al. Mutations that mimic phosphorylation of the HIV-1 matrix protein do not perturb the myristyl switch. Protein Sci. 2007;16:1793–1797. doi: 10.1110/ps.072987607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, et al. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol. 2007;308:133–143. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Du SJ, Li H, Bian Y, Zhong Y. Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc Natl Acad Sci U S A. 2008;105:554–559. doi: 10.1073/pnas.0707330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkins TA, Haramis AP, Etard C, Prodromou C, Vaughan CK, et al. The ATPase-dependent chaperoning activity of Hsp90α regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development. 2008;135:1147–1156. doi: 10.1242/dev.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Just S, Meder B, Berger IM, Etard C, Trano N, et al. The myosin-interacting protein SMYD1 is essential for sarcomere organization. J Cell Sci. 2011;124(Pt 18):3127–3136. doi: 10.1242/jcs.084772. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 13.Paik WK, Kim S. Protein methylation. Science. 1971;174:114–119. doi: 10.1126/science.174.4005.114. [DOI] [PubMed] [Google Scholar]
- 14.Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Yang XD, Lamb A, Chen LF. Methylation, a new epigenetic mark for protein stability. Epigenetics. 2009;4:429–433. doi: 10.4161/epi.4.7.9787. [DOI] [PubMed] [Google Scholar]
- 16.Hardy MF, Harris CI, Perry SV, Stone D. Occurrence and formation of the N epsilon-methyl-lysines in myosin and the myofibrillar proteins. Biochem J. 1970;120:653–660. doi: 10.1042/bj1200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huszar G, Elzinga M. Epsilon-N-methyl lysine in myosin. Nature. 1969;223:834–835. doi: 10.1038/223834a0. [DOI] [PubMed] [Google Scholar]
- 18.Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5:4653–4664. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 19.Etard C, Roostalu U, Strahle U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J Cell Biol. 2008;180:1163–75. doi: 10.1083/jcb.200709128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Cozar FJ, Okamura H, Aramburu JF, Shaw KT, Pelletier L, et al. Two-site interaction of nuclear factor of activated T cells with activated calcineurin. J Biol Chem. 1998;273:23877–23883. doi: 10.1074/jbc.273.37.23877. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Cseresnyes Z, Randall WR, Schneider MF. Activity-dependent nuclear translocation and intranuclear distribution of NFATc in adult skeletal muscle fibers. J Cell Biol. 2001;155:27–39. doi: 10.1083/jcb.200103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du SJ, Rotllant J, Tan X. Muscle-specific expression of the smyd1 gene is controlled by its 5.3-kb promoter and 5′-flanking sequence in zebrafish embryos. Dev Dyn. 2006;235:3306–3315. doi: 10.1002/dvdy.20984. [DOI] [PubMed] [Google Scholar]
- 24.Clark KJ, Balciunas D, Pogoda HM, Ding Y, Westcot SE, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 26.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codina M, Li J, Gutiérrez J, Gao JK, Du SJ. Loss of myosin or Hsp90α1 function results in similar defects on thick and thin filaments but different effects on M-line organization in skeletal muscles of zebrafish embryos. PLoS ONE. 2009;5(1):e8416. doi: 10.1371/journal.pone.0008416. [DOI] [PMC free article] [PubMed] [Google Scholar]