Abstract
Background
Odorant binding proteins (OBPs) play important roles in insect olfaction. The brown planthopper (BPH), Nilaparvata lugens Stål (Delphacidae, Auchenorrhyncha, Hemiptera) is one of the most important rice pests. Its monophagy (only feeding on rice), wing form (long and short wing) variation, and annual long distance migration (seeking for rice plants of high nutrition) imply that the olfaction would play a central role in BPH behavior. However, the olfaction related proteins have not been characterized in this insect.
Methodology/Principal Findings
Full length cDNA of three OBPs were obtained and distinct expression profiles were revealed regarding to tissue, developmental stage, wing form and gender for the first time for the species. The results provide important clues in functional differentiation of these genes. Binding assays with 41 compounds demonstrated that NlugOBP3 had markedly higher binding ability and wider binding spectrum than the other two OBPs. Terpenes and Ketones displayed higher binding while Alkanes showed no binding to the three OBPs. Focused on NlugOBP3, RNA interference experiments showed that NlugOBP3 not only involved in nymph olfaction on rice seedlings, but also had non-olfactory functions, as it was closely related to nymph survival.
Conclusions
NlugOBP3 plays important roles in both olfaction and survival of BPH. It may serve as a potential target for developing behavioral disruptant and/or lethal agent in N. lugens.
Introduction
Chemodetection plays a key role in insect behaviors, such as locating food and mates. In insect, most chemosensillums are located on antenna, and others are on mouthparts, wings, legs and ovipositors. The odorant binding proteins (OBPs) in antennae lumen are thought to bind and transport the exogenous odorant to the odorant receptors (ORs) [1], [2], [3], [4], [5], [6], [7] located on dendrite membranes. By interaction between odorant (odorant-OBP complex) and the receptor, cascade reactions are initiated and eventually lead to the transduction of chemical signals to electric signals. After the transduction, the odorant is rapidly inactivated by odorant degrading enzymes (ODEs) to resume the sensitivity of the receptor neurons [8].
OBP is a family of small water soluble proteins consisting of around 120 to 150 amino acids. The six conserved cysteine residues were thought to be the gold standard of an OBP [9]. These six cysteines constitute three disulfide bridges, which together with some amino acids form an odorant binding pocket to bind and protect small hydrophobic ligands [10], [11]. OBP homologues have been identified in numerous species of Orders throughout the Neoptera, which represents more than 98% of all insect species [12], [13], [14]. OBPs are expressed among diverse classes of sensilla, which all have unique odor specificities.
Most OBP binding experiments reported were conducted in vitro using recombinant proteins [15], [16], [17], [18]. However, in vitro OBP binding with an odor does not necessarily means that the odor is physiologically active because a specific odorant receptor must be present in the same sensillum for the physiological response. In addition, studies in Drosophila melanogaster have demonstrated that a muted PBP (the LUSHD118A) that in configuration closely resembled to the complex of wild PBP (the LUSH)/ the sex pheromone (11-cis vaccenyl acetate) could activate the receptor [11], suggesting that not just binding but the proper OBP configuration induced by this binding is of primary importance. In addition to the binding assay in vitro, an in vivo method, the RNA interference (RNAi) was employed to explore the functions of OBPs in insects including Epiphyas postvittana [19], Spodoptera exigua [20], Culex quinquefasciatus and Anopheles gambiae [21], [22] and Drosophila melanogaster [23]. These studies showed that RNAi is a useful tool in functional studies of genes related to chemosensing.
The brown planthopper (BPH), Nilaparvata lugens Stål (Delphacidae, Auchenorrhyncha, Hemiptera), is a notorious rice pest in Asian countries, causing extensive damage by sap-sucking and virus-transmitting. BPH is a monophagous herbivore restricted to cultivated rice and its allied wild rice [24], and therefore a mechanisms to find the rice plants would be crucial in BPH. BPH nymph undergoes 5-6 nymph instars with 3-6 days for each instar, and the adult could live for 15 days or longer [25]. Like many other Delphacidae insects, BPH adults have dual wing form, short wing (brachypterous) and long wing (macropterous). Short wing BPH, unable to fly, occurs when the nature condition (mainly the nutrition quality of rice plants) is suitable for rapid population increase. In contrast, the long wing adults are produced when the population is too high to be sustained by rice plants, and thus inclines to migrate to find rice plants with higher nutritional quality. Additionally, temperature and photoperiodism also affect the wing form. The sensitive stage for wing form change is the 2nd-4th instar nymph [26]. BPH conducts annual long distance migrations from tropical areas into subtropical and temperate areas with the help of climate air circulation [27]. The biology of this insect implies a critical role of chemosensory.
Hemiptera, a subgroup of Hemipteroid Assemblage (the sister division to Neoptera Endopterygota) is classified into Sternorrhyncha, Auchenorrhyncha and Heteroptera. So far, research of OBPs in Hemiptera was restricted to few species in Sternorrhyncha such as aphids [28], [29], [30], and in Heteroptera such as Lygus lineolaris [31], [32] and Adelphocoris lineolatus [18], [33]. For insects in Auchenorrhyncha, OBPs were rarely addressed except for our identification of three putative OBP cDNA fragments in BPH [14] by using ESTs reported by Noda et al [34]. In the present study we obtained the full-length cDNA of these OBPs; revealed distinct expression profiles in terms of tissues, development stages, wing forms and genders; obtained ligands binding characteristics; and finally by using RNAi gene silencing combined with behaviour analysis, we demonstrated that NlugOBP3 plays important roles both in host seeking and in nymph survival in BPH.
Results
Full-length cDNAs of NlugOBPs
The cDNA full-lengths of three OBPs were obtained from N. lugens by using RACE strategy. These sequences bear all hallmarks of “classic OBP” subfamily, including a signal peptide, a major hydrophobic domain, six conserved Cysteine residues, and considerable identities to reported OBP homologous in amino acid sequence (Table 1 and Fig. 1). NlugOBP1 had the maximum identity of 46% with an OBP from Rhodnius prolixus (CAX63265), whereas the full-length cDNAs of NlugOBP2 and NlugOBP3 presented the highest identities with OBP19a (30%) from D. melanogaster (NP_728338) and OBP16 (36%) from Tribolium castaneum (EFA02853), respectively. However, identities among the three NlugOBPs were much lower, only 17% between NlugOBP1 and NlugOBP2, 14% between NlugOBP1 and NlugOBP3, and 24% between NlugOBP2 and NlugOBP3. NlugOBP1 encoded 173 amino acids (aa), much longer than NlugOBP2 (143aa) and NlugOBP3 (147aa).
Table 1. Sequence information of three OBP cDNAs cloned from Nilaparvata lugens.
| Gene name(Gene ID) | ORF(aa) | SP (aa) | 3′UTR (bp) | 5′UTR (bp) | Blastx result | ||
| Identity (%) | Species | Protein ID | |||||
| NlugOBP1 (FJ215305) | 173 | 23 | 591 | 48 | 46 | R. pro | CAX63265 |
| NlugOBP2 (FJ215306) | 143 | 21 | 593 | 140 | 30 | D. mel | NP_728338 |
| NlugOBP3 (FJ215307) | 147 | 22 | 512 | 68 | 36 | T. cas | EFA02853 |
ORF, open reading frame; SP, signal peptides; UTR, untranslated region; aa, amino acids. R. pro, Rhodnius prolixus; D. mel, Drosophila melanogaster; T. cas, Tribolium castaneum.
Figure 1. Alignment of amino acid sequences of NlugOBPs.
Predicted signal peptides were boxed. Conserved cysteines were highlighted in red and marked with ''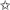 '' below the alignment, and other conserved residues were highlighted in gray.
'' below the alignment, and other conserved residues were highlighted in gray.
Expression profiles of development stage, gender, wing form and tissue
Nymphs vs adults
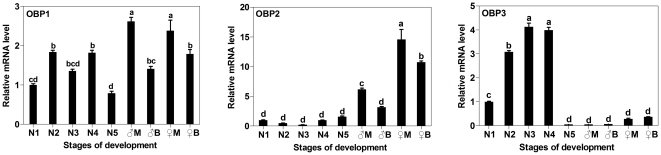
By qRT-PCR, the expression levels were determined for each OBP with respect to development stages, genders and wing forms (Table 2 and Fig. 2). Generally, NlugOBP1 in adult showed not much difference in expression level compared with those in nymph. However, NlugOBP2 expressed significantly higher in adult while in contrast NlugOBP3 expressed significantly lower than in nymph. NlugOBP3 expression displayed a sharp decrease in 4th instar compared to 5th instar, therefore, its expression was only partially nymph biased (not including 5th instar).
Table 2. Relative expression levels of NlugOBP genes in various developmental stages.
| OBP1 | OBP2 | OBP3 | |
| N1 | 679.34 | 144.11 | 840.44 |
| N2 | 1247.65 | 72.66 | 2595.87 |
| N3 | 922.88 | 31.71 | 3468.27 |
| N4 | 1238.18 | 142.12 | 3357.10 |
| N5 | 533.74 | 229.13 | 43.93 |
| ♂M | 1777.95 | 890.21 | 40.53 |
| ♂B | 959.41 | 455.40 | 59.80 |
| ♀M | 1613.52 | 2099.75 | 230.88 |
| ♀B | 1218.59 | 1551.02 | 314.52 |
N1-N5, nymph of 1st instar to nymph of 5th instar; ♂M, macropterous male; ♂B, brachypterous male; ♀M, macropterous female; ♀B, brachypterous female.
Figure 2. Relative mRNA expression level of NlugOBPs in nymphs of different developmental stages, and in adults of different wing forms and genders.
The relative expression level was expressed as mean ± SE (N = 3), with the 1st instar nymph as the calibrator. N1-N5, nymph of 1st to 5th instar; ♂M (macropterous) and ♂B (brachypterous), male adults; ♀M and ♀B, female adults.
Females vs males
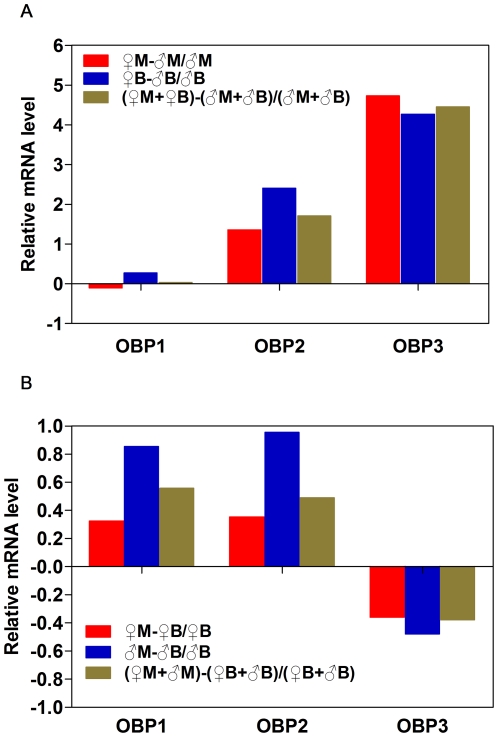
Relative expression levels between female and male adults (Fig. 2) were calculated as female / male (IF), and showed in Fig. 3A. NlugOBP2 and NlugOBP3 were obviously female biased in both short- and long-wing adults. NlugOBP1 was expressed in similar levels between females and males.
Figure 3. Relative mRNA expression level (♀/♂ or M /B) of OBP genes.
(A) For female and male adults, (B) and long and short wing form adults.
Long wing vs short wing
Similarly, long wing form / short wing form in expression levels were calculated (Fig. 3B). All three NlugOBPs showed no great differences between long and short wing adults, with the absolute IF <1.
Tissue
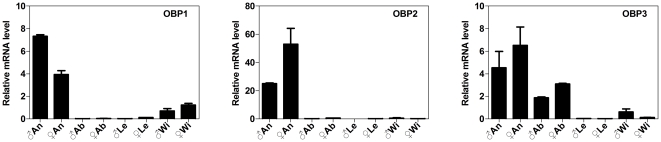
NlugOBP2 expressed high in antenna with little detected in other tissues. NlugOBP1 expressed high in antennae and also had considerable expression in wings of both sexes. NlugOBP3 expressed in high levels in antenna and in abdomen. In addition, NlugOBP1 showed obviously male biased expression, whereas NlugOBP2 was female biased (Fig. 4).
Figure 4. Relative expression levels of NlugOBPs in different tissues.
The expression levels of NlugOBPs were quantified by qRT-PCR. ♂An, male antennae; ♀An, female antennae; ♂Ab, male abdomen; ♀Ab, female abdomen; ♂Le, male legs; ♀Le, female legs; ♂Wi, male wings; ♀Wi, female wings.
In vitro expression of OBPs
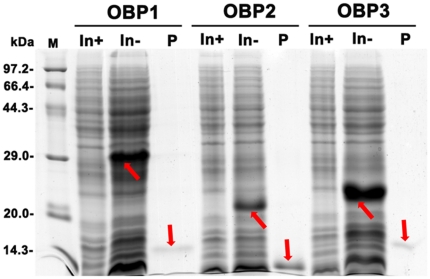
NlugOBPs were expressed in a bacterial system for ligands binding assays. High yields of recombined proteins (20 mg/L) were achieved. Unlike many other OBPs [15], [16], [30], all three NlugOBPs were not in inclusion bodies but soluble proteins. The purified OBPs were obtained by a His-tag affinity column, and the His-tag was cleavaged by enterokinase (Fig. 5).
Figure 5. Expression and purification of three NlugOBPs.
A SDS-PAGE was used to detect the OBP crude extracts from the bacterial pellets before (In-) and after (In+) induction with IPTG, and purified samples after His-tag cleavage by enterokinase (P). The target bands were marked by arrows.
Ligand-binding experiments
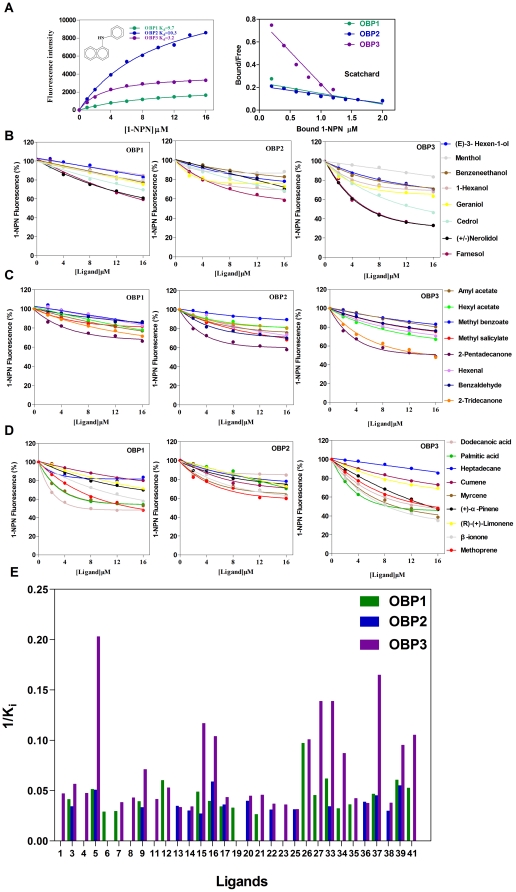
Fourty-one compounds including 17 rice plant volatiles were selected as ligands to define the binding characteristics of the three NlugOBPs. All of them bound the fluorescent probe (1-NPN) well, with dissociation constants (Kd) of 9.7±0.9, 10.3±1.3 and 3.2±0.3 µM for OBP1, OBP2 and OBP3, respectively (Fig. 6A). Typical displacement curves of 1-NPN by ligands with different function groups were showed in Fig. 6B-D. Based on the displacement curves, IC50 and Ki values for all 41 compounds were calculated and listed in Table 3 and Table 4.
Figure 6. Ligand-binding assay of three NlugOBPs.
(A) Binding curves of 1-NPN (Left) and relative Scatchard plot analyses (Right). (B-D) Competitive binding curves of three OBPs with Alcohols (B), Aldehydes, Esters and Benzoates (C), and Carboxylic acids, Alkane and Terpenes (D). (E) Comparison of binding ability (indicated by 1/Ki values) of three OBPs with 31 compounds. Numbers representing ligands were the same as in Table 3 and Table 4.
Table 3. Binding data of recombinant NlugOBPs with different plant volatiles (Alcohols, Aldehydes, Ketones, Esters and Benzoates).
| Ligand | Ligand name/ | OBP1 | OBP2 | OBP3 | |||
| Number | Structural formula | IC50 | Ki | IC50 | Ki | IC50 | Ki |
| (µM) | (µM) | (µM) | (µM) | (µM) | (µM) | ||
| Alcohols | |||||||
| 1 | 1-Hexenol /C6H14O | >50 | >50 | >50 | >50 | 29.7 | 21.2 |
| 2 | 2-Heptanol/C7H16O | >50 | >50 | >50 | >50 | >50 | >50 |
| 3 * | (+/-)Nerolidol/C15H26O | 28.4 | 24.2 | 33.9 | 29.1 | 24.8 | 17.7 |
| 4 | (E)-3-Hexen-1-ol/C6H12O | >50 | >50 | >50 | >50 | 29.6 | 21.1 |
| 5 | Farnesol/C15H26O | 22.8 | 19.5 | 23.0 | 19.8 | 6.9 | 4.9 |
| 6 | Linalool/C10H18O | 40.8 | 34.7 | >50 | >50 | >50 | >50 |
| 7 * | 2-phenylethanol /C8H10O | 39.9 | 34.0 | >50 | >50 | 36.6 | 26.1 |
| 8 | Geraniol/C10H18O | >50 | >50 | >50 | >50 | 32.6 | 23.3 |
| 9 * | Cedrol/C15H26O | 30.0 | 25.6 | 35.0 | 30.1 | 19.7 | 14.1 |
| 10 | Menthol/C10H20O | >50 | >50 | >50 | >50 | >50 | >50 |
| Aldehydes | |||||||
| 11 * | Hexenal/C6H12O | >50 | >50 | >50 | >50 | 33.8 | 24.1 |
| 12 | Dodecyl aldehyde/ C12H24O | 19.5 | 16.6 | >50 | >50 | 26.5 | 18.9 |
| 13 * | (E)-2-Hexenal/ C6H10O | >50 | >50 | 33.5 | 28.8 | 42.0 | 30.0 |
| 14 * | Benzaldehyde/ C7H6O | >50 | >50 | 38.7 | 33.3 | 41.0 | 29.3 |
| Ketones | |||||||
| 15 * | 2-Tridecanone/ C13H26O | 24.0 | 20.5 | 43.2 | 37.1 | 12.0 | 8.6 |
| 16 * | 2-Pentadecanone/ C15H30O | 29.7 | 25.3 | 19.8 | 17.0 | 13.5 | 9.6 |
| 17 * | Acetophenone/ C8H8O | 34.3 | 29.3 | 32.3 | 27.8 | 32.4 | 23.1 |
| Esters and Benzoates | |||||||
| 18 | Amyl acetate/ C7H14O2 | >50 | >50 | >50 | >50 | >50 | >50 |
| 19 | Isoamyl acetate/ C7H14O2 | 35.7 | 30.5 | >50 | >50 | >50 | >50 |
| 20 | (E)-2-Hexenyl acetate/ C8O2 | >50 | >50 | 29.3 | 25.2 | 31.3 | 22.4 |
| 21 | Hexyl acetate/ C8H16O2 | 44.4 | 37.9 | >50 | >50 | 30.8 | 22.0 |
| 22 * | (Z)-3-Hexenyl acetate/ C8O2 | >50 | >50 | 37.8 | 32.5 | 38.0 | 27.1 |
| 23 * | Ethyl benzoate/ C9H10O2 | >50 | >50 | >50 | >50 | 39.0 | 27.9 |
| 24 | Methyl benzoate/ C8H8O2 | >50 | >50 | >50 | >50 | >50 | >50 |
| 25 * | Methyl salicylate/ C8H8O3 | >50 | >50 | 37.4 | 32.2 | 44.8 | 32.0 |
*Marked ligands were identified as rice plant volatile according to literatures; “>50” for the IC50 and Ki means that IC50 or Ki cannot be accurately calculated with the ligand concentration range tested in the assay.
Table 4. Binding data of recombinant NlugOBPs with different plant volatiles (Carboxylic acids, Alkanes, Terpenes and Others).
| Ligand | Ligand name | OBP1 | OBP2 | OBP3 | ||||||||
| number | Structural formula | IC50 | Ki | IC50 | Ki | IC50 | Ki | |||||
| (µM) | (µM) | (µM) | (µM) | (µM) | (µM) | |||||||
| Carboxylic acids | ||||||||||||
| 26 | Dodecanoic acid/ C12H24O2 | 12.1 | 10.3 | >50 | >50 | 13.9 | 9.9 | |||||
| 27 * | Palmitic acid/ C16H32O2 | 25.9 | 22.1 | >50 | >50 | 10.1 | 7.2 | |||||
| Alkanes | ||||||||||||
| 28 | Eicosane/C20H42 | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| 29 | Tridecane/C13H28 | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| 30 | Heptadecane/ C17H36 | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| 31 | Octadecane/C18H38 | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| 32 | Tetradecane/ C14H30 | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| Terpenes | ||||||||||||
| 33 * | Myrcene/C10H16 | 19.0 | 16.2 | 33.9 | 29.1 | 10.1 | 7.2 | |||||
| 34 * | (+)-α-Pinene/ C10H16 | 36.4 | 31.1 | >50 | >50 | 16.1 | 11.5 | |||||
| 35 * | (R)-(+)-Limonene/ C10H16 | 32.4 | 27.6 | >50 | >50 | 33.1 | 23.6 | |||||
| 36 | β-Caryophyllene/ C15H24 | >50 | >50 | 30.1 | 25.9 | 37.4 | 26.7 | |||||
| 37 * | β- Ionone/C13H20O | 25.2 | 21.5 | 25.8 | 22.2 | 8.5 | 6.1 | |||||
| 38 | Cumene /C9H12 | >50 | >50 | 39.2 | 33.7 | 37.2 | 26.6 | |||||
| 39 | Methoprene/C19H34O3 | 19.4 | 16.6 | 21.1 | 18.1 | 14.7 | 10.5 | |||||
| Others | ||||||||||||
| 40 | Cineole/C10H18O | >50 | >50 | >50 | >50 | >50 | >50 | |||||
| 41 | 2,6-Di-tert-butylphenol/ C14H22O | 22.3 | 19.0 | >50 | >50 | 13.3 | 9.5 | |||||
*Marked ligands were identified as rice plant volatile according to literatures; “>50” for the IC50 and Ki means that IC50 or Ki cannot be accurately calculated with the ligand concentration range tested in the assay.
In general, Ketone and Terpene presented high binding affinities to the OBPs. For example, 2-Tridecanone, 2-Pentadecanone, Myrcene, (+)-α-Pinene, β-Ionone and Methoprene showed very high affinities with NlugOBP3 (Ki <12). In contrast, Alkanes showed no binding ability with OBPs, with incalculable Ki. Differences among the three OBPs were also found. Firstly, OBP1 and OBP2 could bind only 19 and 15 ligands, respectively, while OBP3 could bind 28 ligands indicating a broader binding spectrum of OBP3. Secondly, OBP3 showed higher binding abilities than OBP1 and OBP2, based on IC50 and Ki values. The averaged IC50 (25.3) and Ki (18.7) of OBP3 were obviously lower than those of OBP1 (IC50, 28.2; Ki, 24.0) and OBP2 (IC50, 27.1; Ki, 24.6). However, Linalool and Isoamyl acetate were two exceptions, exhibited some affinities with NlugOBP1, but none with OBP3 (Fig. 6, Table 3 and Table 4).
Functional investigation of NlugOBP3
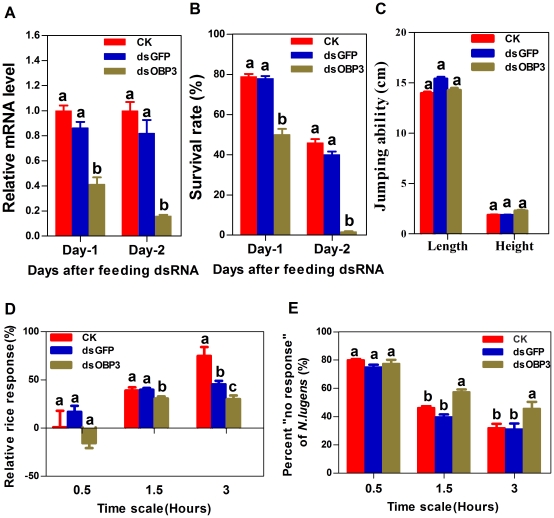
As indicated by above experiments, NlugOBP3 was the most expressed one in the nymph stage among the three OBPs, and showed a wider binding spectrum and higher binding affinities compared to the other two. Furthermore, NlugOBP3 expression was significantly nymph biased, but with a sharp decrease in 5th instar nymph, implying a possible role in BPH metamorphosis as well as in olfaction. NlugOBP3 was therefore selected for the in vivo function study by RNAi. Comparing to the insects feeding on a normal artificial diet (CK) and an exogenous dsRNA mixed diet (dsGFP), the nymph on OBP3-dsRNA diet (dsOBP3) showed a significantly reduced mRNA expression, with about 60% and 80% reduction at 1 day and 2 days after the treatment, respectively (Fig. 7A).
Figure 7. Effects of feeding NlugOBP3 dsRNA on NlugOBP3 mRNA level (A), survival rate (B), jumping ability (C), and response to rice seedlings (D-E) of BPH nymphs.
CK, nymph fed with normal diet; dsGFP, fed with diet mixed with dsRNA of green fluorescent protein (0.5 mg/ml); dsOBP3, fed with dsRNA of NlugOBP3 (0.5 mg/ml). Data topped with different letters are significant different as determined using a one-way ANOVA (Duncan’s multiple range test, P<0.05). All error bars represent the SE of the mean obtained from at least three independent replicates.
Very surprisingly, the survival rate of dsOBP3 treated nymphs was significantly decreased compared to the CK and dsGFP groups. Only 50% survival rate was observed in dsOBP3 group, while about 80% survival rates were showed in dsGFP and CK groups at 1 day after the treatment. At 2 days after the treatment, <5% survival rate was observed in dsOBP3 group (Fig. 7B). To exclude the possibility that the high mortality in dsOBP3 group was resulted from the reduced nymph vitality, a jumping assay was conducted. The results showed no difference both in jumping distance and jumping height between the dsOBP3 group and the two control groups (Fig. 7C).
Two-choice bioassays revealed that the seeking behavior of second instar nymphs for rice seedlings was significantly inhibited by OBP3 dsRNA diet (Fig. 7D). At 1.5 h and 3.0 h after placed into the olfactometer, the relative response (%) was significantly lower in dsOBP3 group than that in CK and dsGFP groups. Meanwhile, the non-response rate of dsOBP3 group was significantly higher than that of CK and dsGFP groups at 1.5 and 3.0 h.
Discussion
The amino acids sequences of OBPs in BPH
Based on the fragments obtained in our previous work [14], full-length cDNAs of the three OBPs of BPH were cloned in the present study. This is the first report of this kind, providing the basis for elucidation of mechanisms underlying chemosensing in BPH. The OBPs in BPH have conserved features of typical OBPs such as the six cyseines and some amino acid residues. The low similarity (<25%) among the three NlugOBPs also was consistent with that in other insects [30], [33], [35], which suggests that these OBPs diverged at early time or/and are still evolving.
OBP expression patterns and functional implications
Expression patterns of tissues, developmental stages, sexes and other specific phenotypes can provide functional clues of genes. All three NlugOBPs were expressed more in antennae than in other tissues (Fig. 4), suggesting an olfactory role of these OBPs. In addition, NlugOBP1 also expressed a considerable amount in wings (Fig. 4A, B), therefore it may also involve in gustatory function, as wing plays somewhat gustory role in insects.
NlugOBP3 may have functions besides olfaction as it expressed highly in the non-olfactory abdomen (Table 2 and Fig. 4). Further investigation on the temporal expression showed that OBP3 displays a nymph (before 5th instar) biased expression with a 4-fold reduction from 4th instar to 5th instar (Table 2 and Fig. 2). Since 5th instar is the final nymph stage before molting to adults, this temporal expression pattern may suggest its involvement in metamorphosis in addition to olfactory function, which is confirmed by the RNAi experiment and subsequent bioassay (see detailed discussion below).
Binding ability of NlugOBPs with odors of different structures
Locating host plant is essential for a phytophagous insect [36]. An efficient mechanism of host detection and recognition is particularly important for BPH as it is a monophagous and a long distance migrating insect. The competitive binding experiments showed that the three NlugOBPs displayed some common binding features and also some differences (Table 3, Table 4 and Fig. 6).
Four volatiles of rice plants (2-Tridecanone, 2-Pentadecanone and β-Ionone, and Acetophenone) are present in a volatile mixture that is attractive to BPH [37], [38]. In our experiments, these four volatiles also showed relatively high binding abilities to all three NlugOBPs, especially to NlugOBP3 (Ki<12, except for Acetophenone) (Fig. 6C, D, Table 3 and Table 4). Two green leaf volatiles (6-carbon compounds) Hexanal and (E)-2-Hexenal were among the most abundant volatiles of rice [39], and generate high Electroantennogram (EAG) response in BPH and some other Hemiptera insects [40], [41], [42]. As expected, NlugOBP3 could bind these two volatiles, although the Ki were not very high. However, NlugOBP2 can bind only with (E)-2-Hexenal; and NlugOBP1 binds none of the two volatiles (Fig. 6E and Table 3).
Another group of volatiles showing high affinity with the three OBPs is Terpenes. These volatiles (e.g. α-Pinene, β-Pinene, Myrcene, Limonene, Camphene and Carene) attract a number of beetle species [43]. α-Pinene and Limonene are components of rice plant volatiles [39], [44]. In our study, Myrcene and α-Pinene showed extremely high binding abilities with NlugOBP3 (Ki = 7.2 and 11.5 µmol/L, respectively) (Table 4). Limonene and β-Caryophyllene also showed considerable affinities with NlugOBP3 (Fig. 6D, E). These volatiles could be a valuable resource to identify attractants or deterrents for the control of BPH as well as other insect pests.
A very interesting finding in our binding assay is that Farnesol and Methoprene, two juvenile hormone analogues of insects [45], [46], have high binding ability with all three OBPs, especially with NlugOBP3 (Fig. 6B, D and Table 4). Combining with the expression profile (high abdomen expression, and 1st-4th instar nymph and female biased expression), we could hypothesize that OBP3 is involved in transporting juvenile hormone in BPH. The later RNAi experiments showing high mortality of OBP3 deficit nymphs partially supported this hypothesis. The possible role of OBP in juvenile hormone transport deserves further investigations.
Additionally, NlugOBP3 has obviously broader binding spectrum and higher binding ability with tested volatiles compared to NlugOBP1 and NlugOBP2. Taken together with its distinct expression profile and higher expression level, OBP3 probably is more important than the other two OBPs in odor perception and/or other functions.
Effects of RNAi of NlugOBP3
RNAi is widely used to investigate gene functions in insects as well as in other organisms, and the technology has been successfully used in BPH gene silencing using dsRNA diet or injection [47], [48]. As discussed earlier, NlugOBP3 possibly has functions in addition to olfaction. To confirm this, NlugOBP3 RNAi diet experiments were conducted. The results showed that target gene silencing rate is very high, up to about 60% at one day and about 80% at two days after treatment (Fig. 7A), providing an in vivo methodology for OBP gene silencing in BPH.
The RNAi experiments and two-choice olfactometer bioassays together revealed that NlugOBP3 plays a role in perception of rice plant volatiles, as the host-seeking behavior of nymphs was significantly inhibited after silencing NlugOBP3 expression (Fig. 7D). However, the “no response” nymphs in dsOBP3 group were also significantly increased compared to the non-dsRNA control groups. The jumping assay excluded the possibility that the reduced response rate of dsOBP3 treated nymphs is due to low viability (Fig. 7C), thus confirmed that the lower response rate is due to the reduced olfaction of the seedling volatiles.
More importantly, our RNAi experiment revealed the non-olfactory function of NlugOBP3, as silencing the gene resulted in strikingly high nymph mortality (Fig. 7B). The possibility that this mortality was induced by olfaction suppression can be excluded because the artificial liquid diets used in the RNAi experiment contain no plant volatiles. As speculated earlier, the high mortality may due to the disruption of JH binding and transporting possibly carried out by NlugOBP3.
To summarize, we obtained the full-length cDNAs coding for three OBPs in BPH for the first time and conducted experiments on expression patterns, ligand binding properties and phenotype changes after gene silencing (NlugOBP3 only). The results suggest that NlugOBP3, compared to NlugOBP1 and NlugOBP2, plays more important roles in olfaction of plant volatiles, and may possess non-olfactory functions associated with BPH survival. NlugOBP3 could be a potential target for behavioral disruptant and lethal agent for BPH.
Materials and Methods
Insect rearing and collection
BPH were collected from a rice field at Jiangsu Academy of Agricultural Science, China, and reared with rice seedlings in plastic box (40×30×40 cm) in the laboratory under 26±1°C, 16 h light : 8 h dark cycle and 70–90% relative humidity. Nymphs of different instars and virgin long and short wing adults of both sexes were collected in three replications (20 individuals per replication). Various tissues from virgin short wing adults were dissected under microscope and collected in three replications for each tissue type. Each replication contained 100 antenna or wings, or 40 legs, or 20 abdomen. Whole body and tissue samples were all frozen in liquid nitrogen and stored at -70°C until use.
RNA isolation and cDNA synthesis
Total RNA was extracted by SV 96 Total RNA Isolation System (Promega, Madison, WI, USA) following the manufacturer's instructions. RNA quality was checked with a spectrophotometer (NanoDrop™ 1000, Thermo Fisher Scientific, USA). The single-stranded cDNA templates were synthesized using 1 µg total RNAs from various samples with oligo (dT) 18 primer as the anchor primers. The M-MLV Reverse Transcriptase (M-MLV) (TaKaRa, Dalian, Liaoning, China) was used for the cDNA synthesis, with reaction conducted at 42°C for 1 h, and then stopped by heating at 70°C for 15 min.
RACE amplification and sequence alignment
To get the full-length sequence of OBPs, Rapid Amplification of cDNA End (RACE) PCR was performed using GeneRacer™ kit (Invitrogen Carlsbad, CA, USA) for 3′end and 5′ends amplification. The full-length sequences were assembled with RACE results and then confirmed by end-to-end PCR using specific primers designed at both ends. Three positive clones of each gene were sequenced to detect possible PCR mistakes. The primer sequences designed by Primer Premier 5.0 (PREMIER Biosoft International, CA, USA) were listed in Table S1. Multiple alignments of OBPs protein sequences were performed using CLUSTALX 2.0 [49] and arranged by Jalview 2.4.0 b2 [50]. The signal peptides were predicted by SignalP 3.0 http://www.cbs.dtu.dk/services/SignalP/) [51].
Quantitative RT-PCR
Before quantitative RT-PCR (qRT-PCR), normal RT-PCR were done with each primer pair to ensure that the correct products were being amplified and no primer dimer was present using rTaq DNA polymerase (TaKaRa, Dalian, Liaoning, China) and 1.5% (w/v) electrophoresis agarose gel. The qRT-PCR reactions for each sample were performed on an ABI 7300 (Applied Biosystems, Foster City, CA, USA) using 20 ng of cDNA template and 10 µM gene-specific primers designed by Beacon Designer 7.6 (PREMIER Biosoft International, CA, USA), which were listed in Table S1. The reactions were 10 s at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 31 s. The mRNA levels were measured by qRT-PCR using the SYBR Premix Ex Taq™ (TaKaRa, Dalian, Liaoning, China). This was followed by the measurement of fluorescence during a 55 to 95°C melting curve in order to detect a single gene-specific peak and to check the absence of primer dimer peaks. Under these conditions, a single and discrete peak was detected for all primers tested. Negative controls were non-template reactions (replacing cDNA with H2O). Ten-fold dilution series were used to construct a relative standard curve to determine the PCR efficiencies and for further quantification analysis. In all experiments, all primers gave amplification efficiencies of 90-100%. Each reaction was run in triplicate (technical replicate). mRNA level was quantified in relation to the expression of β-actin (EU179846 [52] ) that was used as the control gene. The primer pair for each gene was designed to amplify a 70-200 bp product, which was verified by nucleotide sequencing. Means and standard errors were obtained from the average of three independent biological replicates. The relative copy numbers of OBP genes were calculated according to the 2−ΔΔCt method [53]. The mRNA level in different samples was analyzed using the ABI 7300 analysis software SDS 1.4.
E. coli expression and purification of the recombinant protein
The three NlugOBP sequences encoding mature proteins were amplified by primers including BamH I and Xho I restriction enzyme sites (Table S1). The purified PCR products were ligated into pEASY T3 cloning vector (TransGen, Beijing, China) for 15 min, and then the products were transformed to E.coli Trans1-T1 competent cells. Three positive clones were sequenced by Genscript Biology Company (Nanjing, Jiangsu, China). The plasmids contained appropriate OBP sequences and the empty pET30a plasmid were digested with BamH I and Xho I FastDigest® restriction enzymes (Fermentas, Thermo Fisher Scientific, USA) for 10 min at 37°C. The purified double-enzymes digested products of OBPs were ligated into digested empty pET30a with T4 ligase (Fermentas, Thermo Fisher Scientific, USA) for 1 h at 22°C. After ligation, the products were transformed into BL-21 DE3 pLys E. coli cells. The positive clones were validated by PCR and sequencing. The expression of recombinant proteins were induced by addition of IPTG to a final concentration of 2 mM when the LB medium culture had reached a value of OD600 = 0.5. Cells were grown overnight with 200 rpm and 37°C, harvested by centrifugation (8000 g×20 min 4°C) and sonicated. All the protein was dissoluble. The proteins were purified by XK-16 Column with Ni Sepharose High performance (GE Healthcare Life Sciences). The His-tag was cleavaged by enterokinase (Genscript Biology Company, Nanjing, China). The cleavaged proteins were purified again by column mentioned above with naturing agent. The purified proteins without His-tag were desalinated by dialysis against NaCl. The resulted proteins were kept at -70°C after freeze-dry.
Competitive fluorescence binding assay
To measure the affinity of OBPs to the fluorescent probe N-phenyl-1-naphthyl-amine (1-NPN), a 2 M solution of the protein in 50 mM Tris–HCl, pH 7.4, was titrated with aliquots of 1 mM ligand in methanol to achieve various concentrations. The affinity to other ligands was measured in competitive binding assays, using both the protein and the fluorescent reporter 1-NPN at 2 µM concentration. The excitation wavelength was 337 nm and the emission spectrum was recorded between 370 and 460 nm by a fluorescence spectrometer (Hitachi F-7000 Fluorescence Spectrophotometer, Japan) with a 1 cm light path quartz cuvette and 10 nm slits for both excitation and emission. Dissociation constants for 1-NPN were calculated from Scatchard plots of the binding data, and the relative to other ligands were calculated from the corresponding IC50 values, using the equation: Ki = [IC50]/1+ [1-NPN]/K1-NPN, where [1-NPN] is being the free concentration of 1-NPN and K1-NPN being the dissociation constant of the complex protein/1-NPN.
Double strand RNA synthesis
The full cDNA sequence NlugOBP3 (FJ215307) and partial sequence of a green fluorescent protein (GFP ACY56286) were sub-cloned into pEASY-T3 vector, and the dilution plasmid was used as template for amplification of the target sequences. The sequence of NlugOBP3 was amplified by RT-PCR using specific primers conjugated with 23 bases of the T7 RNA polymerase promoter (Table S1).The PCR products of 487 bp for NlugOBP3 and 460 bp for GFP were purified using Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and used as templates for dsRNA synthesis using the T7 RibomaxTM Express RNAi System (Promega, Madison, WI, USA). The synthesized dsRNA was isopropanol precipitated, resuspended in Nuclease-free water, and quantified by a spectrophotometer (NanoDrop™ 1000, Thermo Fisher Scientific, USA) at 260 nm. The purity and integrity were determined by agarose gel electrophoresis. It was kept at -70°C until use.
Double strand RNA diet and gene expression analysis
dsRNA rearing procedure reported by Fu et al [25] and Chen et al [48] was used with small modifications. Briefly, nymphs of second instar that were pre-reared on artificial diets for one day were used. Glass cylinders, 12 cm in length and 2.8 cm in internal diameter, were used as feeding chambers. Twenty nymphs were carefully transferred into each chamber. The dsRNA was added to the artificial diet at 0.5 µg/µl [48]. This mixed diet (40 µl) was held between two layers of stretched Parafilm M (Pechiney Plastic Packaging Company, Chicago, IL, USA) that was placed at both ends of the chamber. One more piece of Parafilm M was covered to avoid entering of hydrosphere which may stick the nymphs to the cylinder wall. The chamber bodies were covered with black and wet cotton cloth exposing only the two ends (the diet) to the light. Fresh diet was provided daily and dead nymphs were cleaned.
Three treatments including non-dsRNA diet (CK), GFP dsRNA diet (dsGFP) and OBP3 dsRNA diet (dsOBP3) were set up and replicated six times (6 chambers). The experiment last for two days. On each day, one nymph from each chamber (totally 6 nymphs) was collected. Two nymphs were designed as one replicate (3 replicates for each treatment at each sampling time) for RNA isolation. The qRT-PCR method and reaction conditions were same as described above. Primers used in qRT-PCR were listed in Table S1.
The mortality effect of dsRNA diet on nymphs was evaluated in a separate experiment, where the method was the same as the dsRNA feeding experiment, but replicated 8 times (8 chambers). Mortality was recorded on both days.
Nymph vitality bioassay
To test the possible influence of dsRNA feeding on BPH vitality, a jumping ability assay was conducted at one day after the feeding. One BPH nymph (2nd instar) was placed in a glass cylinder (same as above) with both ends sealed by Parafilm. Holding vertically with one hand, the bottom end was gently knocked by a finger to induce active jumping. The heights of the jumps were recorded. The jumping distance (length) was measured by stimulating a nymph in an open dish with a gently touch at the abdomen using an insect pin. Each nymph was stimulated 2-4 times in jumping height or length measurement and the averages were used. Thirty nymphs were tested for each treatment group.
Two-choice behavioral bioassay
The behavioral responses of BPH to rice volatiles was tested by a two-choice bioassay using an H-shaped olfactometer similar to that used by Khan and Saxena [54] and Lou and Chen [55] (Fig. S1). The olfactometer mainly comprises of two glass tubes (arms) (10 cm diameter×30 cm long) with gauze at its top end connected at their center point by another smaller glass tube (8 cm diameter×20 cm long) with nylon mesh at two ends and a small hole (1 cm diameter) at its half way point for releasing BPH.
About 20 g fresh rice seedlings with no insect damage were caged in one arm of the olfactometer. Twenty nymphs (2nd instars) were introduced, and then the hole was sealed with absorbent cotton. BPH in “B” and “C” area were counted at 0.5, 1.5 and 3.0 h.
Three treatments of BPH nymphs (one day after dsOBP3 feeding, one day after dsGFP feeding, and non-dsRNA feeding CK) were tested in four replications. After 2 tests (replications), the olfactometers were washed with 75% alcohol and the rice seedlings were placed in another arm to complete the other two replicates. At any given time, nymphs in two treatments were tested using two olfactometers. Totally 12 bioassays were performed in two days in a darkroom at (26±1)° and 70–90% relative humidity.
The relative response percentages (RRP) were calculated as[(number of nymph responded to rice seedlings (NRC) – numbers of nymph responded to control air (NRA)/(NRC+NRA)]×100%.
Data analysis
All data (mean ± SE) were compared with one-way nested analysis of variance (ANOVA Duncan's multiple range test) using SPSS Statistics 18.0 (SPSS Inc., Chicago, IL, USA).
Supporting Information
Schematic diagram of H-shaped olfactometer used for behavioral assay. A, Release hole; B, The area defined as response to rice seedlings; C, The area defined as response to air; D, Pot with rice plants; E, Pot with nothing.
(TIF)
Primers used in RACE, qRT-PCR, Vector construction and dsRNA synthesis.
(DOC)
Acknowledgments
We thank Prof. Long Zhang and Xiang Yu (China Agricultural University, Beijing, China) and Prof. Paolo Pelosi (University of Pisa, Italy) for help in competitive fluorescence binding assay; Doctoral student Yue-Liang Zhang, Master students Jian-Hua Gu, Lin-Lin Zhang, Zhan-Feng Ye and Zhen Liao (Nanjing Agricultural University, China) for help in collecting brown planthopper and tissues; Jing Chen (Sun Yat-Sen University, China) for suggestion in dsRNA feeding experiment. We also thank Xiao-Mang Ma for help in using AutoCAD software.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was in part supported by the National 973 Project (2010CB126200), National Natural Science Foundation (31071978) of China, and the Priority Academic Program Development of Jiangsu Higher Education Institutions, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pelosi P. Odorant-binding proteins: structural aspects. Annals of the New York Academy of Sciences. 1998;855:281–293. doi: 10.1111/j.1749-6632.1998.tb10584.x. [DOI] [PubMed] [Google Scholar]
- 2.Blomquist G, Vogt RG. In Insect Pheromone Biochemistry and Molecular Biology. In Insect Pheromone Biochemistry and Molecular Biology. (eds. GJ Blomquist and RG Vogt), pp 3-18. Elsevier Academic Press, London; 2003. Biosynthesis and detection of pheromones and plant volatiles: introduction and overview. [Google Scholar]
- 3.Vogt RG. Endocrinology. (LI Gilbert, K Iatro, S Gill eds). pp 753-804. Elsevier, London. ISBN; 2005. Molecular Basis of Pheromone Detection in Insects. In Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology.Volume 3.044451516X [Google Scholar]
- 4.Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annual Review of Entomology. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- 6.Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biology. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forstner M, Breer H, Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int J Biol Sci. 2009;5:745–757. doi: 10.7150/ijbs.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt RG, Riddiford LM, Prestwich GD. Kinetic properties of a sex pheromone-degrading enzyme: the sensillar esterase of Antheraea polyphemus. Proc Natl Acad Sci U S A. 1985;82:8827–8831. doi: 10.1073/pnas.82.24.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Letters. 1999;464:85–90. doi: 10.1016/s0014-5793(99)01683-x. [DOI] [PubMed] [Google Scholar]
- 10.Wojtasek H, Leal WS. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. Journal of Biological Chemistry. 1999;274:30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]
- 11.Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogt RG, Rogers ME, Franco MD, Sun M. A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera). Journal of Experimental Biology. 2002;205:719–744. doi: 10.1242/jeb.205.6.719. [DOI] [PubMed] [Google Scholar]
- 13.Bohbot J, Vogt RG. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem Mol Biol. 2005;35:961–979. doi: 10.1016/j.ibmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Xu YL, He P, Zhang L, Fang SQ, Dong SL, et al. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics. 2009;10:632. doi: 10.1186/1471-2164-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao HL, He XL, Schymura D, Ban LP, Field L, et al. Cellular and Molecular Life Sciences; 2010. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He XL, Tzotzos G, Woodcock C, Pickett JA, Hooper T, et al. Journal of Chemical Ecology; 2010. Binding of the General Odorant Binding Protein of Bombyx mori BmorGOBP2 to the Moth Sex Pheromone Components. [DOI] [PubMed] [Google Scholar]
- 17.Dani FR, Iovinella I, Felicioli A, Niccolini A, Calvello MA, et al. Mapping the expression of soluble olfactory proteins in the honeybee. Journal of Proteome Research. 2010;9:1822–1833. doi: 10.1021/pr900969k. [DOI] [PubMed] [Google Scholar]
- 18.Gu SH, Wang WX, Wang GR, Zhang XY, Guo YY, et al. Archives of Insect Biochemistry and Physiology; 2011. Functional characterization and immunolocalization of odorant binding protein 1 in the lucerne plant bug, Adelphocoris lineolatus (GOEZE). [DOI] [PubMed] [Google Scholar]
- 19.Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, et al. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Molecular Biology. 2006;15:383–391. doi: 10.1111/j.1365-2583.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou YZ, Xiu WM, Dong SL. Functional study of pheromone binding protein by using RNAi in male beet armyworm, Spodoptera exigua Hübner. Journal of Nanjing Agricultural University. 2009;32:58–62. [Google Scholar]
- 21.Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Journal of Chemical Ecology; 2010. Knockdown of a Mosquito Odorant-binding Protein Involved in the Sensitive Detection of Oviposition Attractants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, et al. The Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) Mediates Indole Recognition in the Antennae of Female Mosquitoes. PLoS One. 2010;5:e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swarup S, Williams TI, Anholt RR. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain and Behavior. 2011;10:648–657. doi: 10.1111/j.1601-183X.2011.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sōgawa K. The Rice Brown Planthopper: Feeding Physiology and Host Plant Interactions. Annual Review of Entomology. 1982;27:49–73. [Google Scholar]
- 25.Fu Q, Zhang Z, Hu C, Lai F, Sun Z. A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae). Applied Entomology and Zoology. 2001;36:111–116. [Google Scholar]
- 26.Zhang ZQ. A study on the development of wing dimorphism in the rice brown planthopper, Nilaparvata lugens Stål. Acta Entomol Sin. 1983;26:260–267. [Google Scholar]
- 27.Zhai BP. What have we seen by entomological radar? Entomological Knowledge. 2005;42:217–226. [Google Scholar]
- 28.Zhou JJ, Vieira FG, He XL, Smadja C, Liu R, et al. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid (Acyrthosiphon pisum). Insect Molecular Biology. 2010;19:113–122. doi: 10.1111/j.1365-2583.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- 29.Vandermoten S, Francis F, Haubruge E, Leal WS. Conserved odorant-binding proteins from aphids and eavesdropping predators. PLoS One. 2011;6:e23608. doi: 10.1371/journal.pone.0023608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao HL, Tuccori E, He XL, Gazzano A, Field L, et al. Discrimination of alarm pheromone (E)-beta-farnesene by aphid odorant-binding proteins. Insect Biochemistry and Molecular Biology. 2009;39:414–419. doi: 10.1016/j.ibmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Vogt RG, Callahan F, Rogers M, Dickens J. Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera). Chemical Senses. 1999;24:481. doi: 10.1093/chemse/24.5.481. [DOI] [PubMed] [Google Scholar]
- 32.Dickens JC, Callahan FE, Wergin WP, Murphy CA, Vogt RG. Intergeneric distribution and immunolocalization of a putative odorant-binding protein in true bugs (Hemiptera, Heteroptera). Journal of Experimental Biology. 1998;201:33–41. doi: 10.1242/jeb.201.1.33. [DOI] [PubMed] [Google Scholar]
- 33.Gu SH, Wang SP, Zhang XY, Wu KM, Guo YY, et al. Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze). Insect Biochemistry and Molecular Biology. 2011;41:254–263. doi: 10.1016/j.ibmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Noda H, Kawai S, Koizumi Y, Matsui K, Zhang Q, et al. Annotated ESTs from various tissues of the brown planthopper Nilaparvata lugens: a genomic resource for studying agricultural pests. BMC Genomics. 2008;9:117. doi: 10.1186/1471-2164-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Zhang YJ, Su HH, Gao XW, Guo YY. Identification and expression pattern of putative odorant-binding proteins and chemosensory proteins in antennae of the Microplitis mediator (Hymenoptera: Braconidae). Chemical Senses. 2009;34:503–512. doi: 10.1093/chemse/bjp027. [DOI] [PubMed] [Google Scholar]
- 36.Visser JH. Host Odor Perception in Phytophagous Insects. Annual Review of Entomology. 1986;31:121–144. [Google Scholar]
- 37.Obata T, Koh HS, Kim M, Fukami H. Planthopper atractants in rice plant. Applied Entomology and Zoology. 1981;25:47–51. [Google Scholar]
- 38.Obata T, Koh HS, Kim M, Fukami H. Constituents of planthopper atractant in rice plant. Applied Entomology and Zoology. 1983;18:161–169. [Google Scholar]
- 39.Hernandez HP, Hsieh TCY, Smith CM, Fischer NH. Foliage volatiles of two rice cultivars. Phytochemistry. 1989;28:2959–2962. [Google Scholar]
- 40.Visser JH, Fu shun Y. Electroantennogram responses of the grain aphids Sitobion avenae (F.) and Metopolophium dirhodum (Walk.) (Hom., Aphididae) to plant odour components. Journal of Applied Entomology. 1995;119:539–542. [Google Scholar]
- 41.Visser JH, Piron PGM, Hardie J. The aphids' peripheral perception of plant volatiles. Entomologia Experimentalis et Applicata. 1996;80:35–38. [Google Scholar]
- 42.Youn YN. Electroantennogram responses of Nilaparvata lugens (Homoptera: Delphacidae) to plant volatile compounds. Journal of Economic Entomology. 2002;95:269–277. doi: 10.1603/0022-0493-95.2.269. [DOI] [PubMed] [Google Scholar]
- 43.Chénier JVR, Philogène BJR. Field responses of certain forest Coleoptera to conifer monoterpenes and ethanol. Journal of Chemical Ecology. 1989;15:1729–1745. doi: 10.1007/BF01012261. [DOI] [PubMed] [Google Scholar]
- 44.Yan F, Wang X, Lv J, Pang BP, Lou YG. Comparison of the volatiles from rice plants infested by rice striped stem borer, Chilo suppressalis and rice leaf folder, Cnapholocrocis medinalis. CHINESE BULLETIN OF ENTOMOLOGY. 2010;47:96–101. [Google Scholar]
- 45.Yamamoto RT, Jacobson M. Juvenile hormone activity of isomers of farnesol. Nature. 1962;196:908–909. [Google Scholar]
- 46.Van Sambeek JW, Bridges JR. Influence of the juvenile hormone analogue, methoprene, on development of the southern pine beetle, Dendroctonus frontalis Zimm. (Col. Scolytidae). Sonderdruck. 1980;89:479–488. [Google Scholar]
- 47.Liu SH, Ding ZP, Zhang CW, Yang BJ, Liu ZW. Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochemistry and Molecular Biology. 2010;40:666–671. doi: 10.1016/j.ibmb.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Zhang D, Yao Q, Zhang J, Dong X, et al. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Molecular Biology. 2010;19:777–786. doi: 10.1111/j.1365-2583.2010.01038.x. [DOI] [PubMed] [Google Scholar]
- 49.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 52.Liu SH, Yang BJ, Gu JH, Yao XM, Zhang YX, et al. Molecular cloning and characterization of a juvenile hormone esterase gene from brown planthopper, Nilaparvata lugens. Journal of Insect Physiology. 2008;54:1495–1502. doi: 10.1016/j.jinsphys.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan ZR, Saxena RC. Effect of stream distillate extracts of resistant and susceptible rice cultivars on behaviour of Sogatella furcifera (Homoptera: Delphacidae). Journal of Economic Entomology. 1986;79:928–935. [Google Scholar]
- 55.Lou YG, Cheng JA. Role of rice volatiles in the foraging behaviour of the predator Cyrtorhinus lividipennis for the rice brown planthopper Nilaparvata lugens. BioControl. 2003;48:73–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic diagram of H-shaped olfactometer used for behavioral assay. A, Release hole; B, The area defined as response to rice seedlings; C, The area defined as response to air; D, Pot with rice plants; E, Pot with nothing.
(TIF)
Primers used in RACE, qRT-PCR, Vector construction and dsRNA synthesis.
(DOC)