Abstract
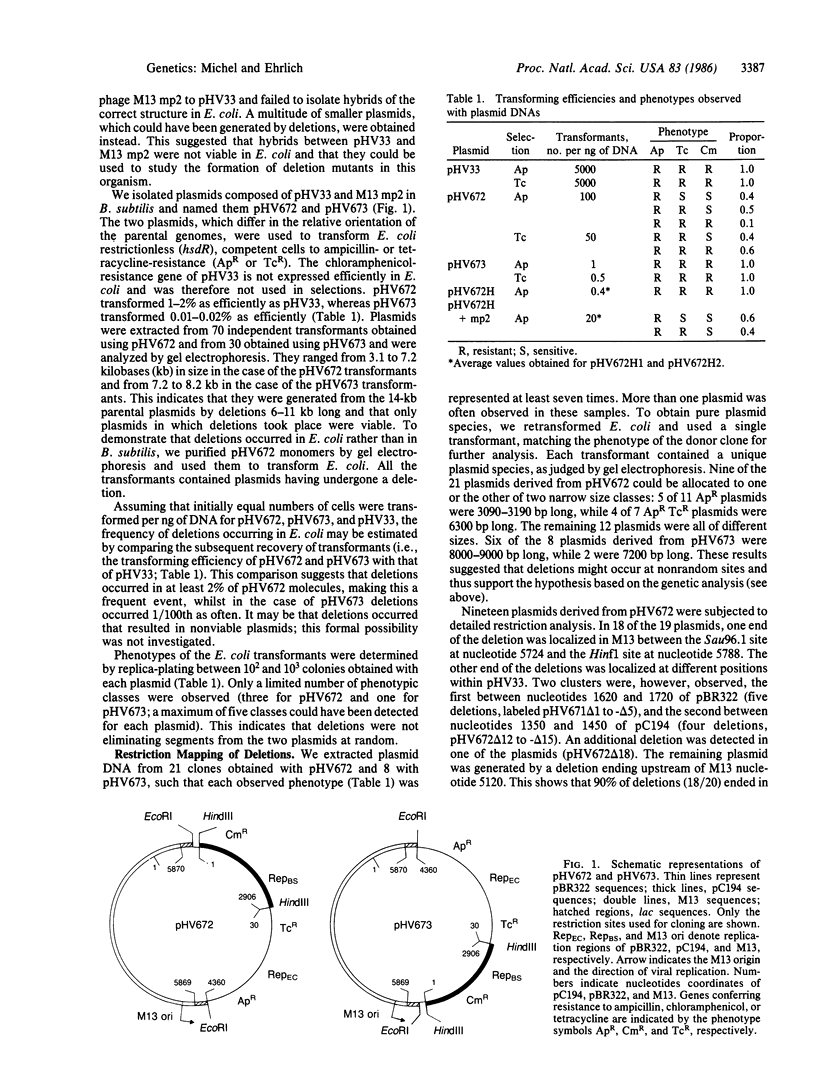
Hybrids composed of phage M13 and plasmid pHV33 were used to study the formation of deletions in Escherichia coli. Eighty to ninety percent of the deletion endpoints were at the position of the nick introduced into the M13 replication origin by the phage gene II protein. This suggests the existence of a novel mechanism of illegitimate recombination.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Backman K., Boyer H. W. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene. 1983 Dec;26(2-3):197–203. doi: 10.1016/0378-1119(83)90190-7. [DOI] [PubMed] [Google Scholar]
- Been M. D., Champoux J. J. Breakage of single-stranded DNA by eukaryotic type 1 topoisomerase occurs only at regions with the potential for base-pairing. J Mol Biol. 1984 Dec 15;180(3):515–531. doi: 10.1016/0022-2836(84)90025-1. [DOI] [PubMed] [Google Scholar]
- Been M. D., Champoux J. J. DNA breakage and closure by rat liver type 1 topoisomerase: separation of the half-reactions by using a single-stranded DNA substrate. Proc Natl Acad Sci U S A. 1981 May;78(5):2883–2887. doi: 10.1073/pnas.78.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D. One and two codon insertion mutants of bacteriophage f1. Mol Gen Genet. 1981;181(3):288–291. doi: 10.1007/BF00425599. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Vovis G. F., Zinder N. D. Insertion mutant of bacteriophage f1 sensitive to EcoRI. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2699–2702. doi: 10.1073/pnas.76.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. Instability of palindromic DNA in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):409–416. doi: 10.1101/sqb.1981.045.01.055. [DOI] [PubMed] [Google Scholar]
- Colman A., Byers M. J., Primrose S. B., Lyons A. Rapid purification of plasmid DNAs by hydroxyapatite chromatography. Eur J Biochem. 1978 Nov 2;91(1):303–310. doi: 10.1111/j.1432-1033.1978.tb20966.x. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Transitory recombination between plasmid pHV33 and phage M13. EMBO J. 1983;2(12):2117–2122. doi: 10.1002/j.1460-2075.1983.tb01711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Jakes K. S., Zinder N. D. Signals for the initiation and termination of synthesis of the viral strand of bacteriophage f1. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):717–722. doi: 10.1101/sqb.1983.047.01.082. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. The functional origin of bacteriophage f1 DNA replication. Its signals and domains. J Mol Biol. 1984 Feb 5;172(4):507–521. doi: 10.1016/s0022-2836(84)80020-0. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Zinder N. D. Reduction of the minimal sequence for initiation of DNA synthesis by qualitative or quantitative changes of an initiator protein. Nature. 1984 Sep 20;311(5983):279–280. doi: 10.1038/311279a0. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn B., Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978 Mar 23;272(5651):375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Halligan B. D., Davis J. L., Edwards K. A., Liu L. F. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J Biol Chem. 1982 Apr 10;257(7):3995–4000. [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Ravetch J. V., Zinder N. D. DNA replication of bacteriophage f1 in vivo. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):389–399. doi: 10.1101/sqb.1979.043.01.045. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Moriya K., Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Shiozaki M. Nonhomologous recombination mediated by Escherichia coli DNA gyrase: possible involvement of DNA replication. Cold Spring Harb Symp Quant Biol. 1984;49:401–409. doi: 10.1101/sqb.1984.049.01.046. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J Bacteriol. 1974 Nov;120(2):805–814. doi: 10.1128/jb.120.2.805-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. F., Geider K. Enzymatic synthesis of bacteriophage fd viral DNA. Nature. 1982 Apr 29;296(5860):828–832. doi: 10.1038/296828a0. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Recombination efficiency is a quadratic function of the length of homology during plasmid transformation of Bacillus subtilis protoplasts and Escherichia coli competent cells. EMBO J. 1984 Dec 1;3(12):2879–2884. doi: 10.1002/j.1460-2075.1984.tb02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Ehrlich S. D. In vitro genetic labeling of Bacillus subtilis cryptic plasmid pHV400. Plasmid. 1979 Jan;2(1):48–58. doi: 10.1016/0147-619x(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Nilsson S. V., Magnusson G. Sealing of gaps in duplex DNA by T4 DNA ligase. Nucleic Acids Res. 1982 Mar 11;10(5):1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. Role of the F factor oriV1 region in recA-independent illegitimate recombination. Stable replicon fusions of the F derivative pOX38 and pBR322-related plasmids. J Mol Biol. 1984 May 25;175(3):263–284. doi: 10.1016/0022-2836(84)90348-6. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. Site-specific recombination in the oriV1 region of the F factor. Cold Spring Harb Symp Quant Biol. 1984;49:421–434. doi: 10.1101/sqb.1984.049.01.048. [DOI] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Schaller H. The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):401–408. doi: 10.1101/sqb.1979.043.01.046. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Ehrlich S. D. Use of the T4 polynucleotide ligase in the joining of flush-ended DNA segments generated by restriction endonucleases. Eur J Biochem. 1978 May 16;86(2):531–537. doi: 10.1111/j.1432-1033.1978.tb12336.x. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Horiuchi K. Multiregulatory element of filamentous bacteriophages. Microbiol Rev. 1985 Jun;49(2):101–106. doi: 10.1128/mr.49.2.101-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]


