Abstract
The splicing of pre-mRNAs is an essential step of gene expression in eukaryotes. Introns are removed from split genes through the activities of the spliceosome, a large ribonuclear machine that is conserved throughout the eukaryotic lineage. While unicellular eukaryotes are characterized by less complex splicing, pre-mRNA splicing of multicellular organisms is often associated with extensive alternative splicing that significantly enriches their proteome. The alternative selection of splice sites and exons permits multicellular organisms to modulate gene expression patterns in a cell type specific fashion, thus contributing to their functional diversification. Alternative splicing is a regulated process that is mainly influenced by the activities of splicing regulators, such as SR proteins or hnRNPs. These modular factors have evolved from a common ancestor through gene duplication events to a diverse group of splicing regulators that mediate exon recognition through their sequence specific binding to pre-mRNAs. Given the strong correlations between intron expansion, the complexity of pre-mRNA splicing, and the emergence of splicing regulators, it is argued that the increased presence of SR and hnRNP proteins promoted the evolution of alternative splicing through relaxation of the sequence requirements of splice junctions.
Keywords: Pre-mRNA splicing, Spliceosome, alternative splicing, splicing regulation, SR protein, hnRNP protein, evolution, intron expansion, gene duplication, multicellular eukaryote, unicellular eukaryote, exon recognition, splice site
INTRODUCTION
Pre-mRNA splicing is a fundamental process required for the expression of most metazoan genes 1. Defects in splicing lead to many human genetic diseases 2–4, and splicing mutations in a number of genes involved in growth control have been implicated in multiple types of cancer 5–10. Splicing is carried out by the spliceosome, which recognizes splicing signals and catalyzes the removal of non-coding intronic sequences to assemble protein coding sequences into mature mRNA prior to export and translation 11. Of the approximately 25,000 genes encoded by the human genome 12, more than 90% are believed to produce transcripts that are alternatively spliced 13, 14. Thus, alternative splicing of pre-mRNAs can lead to the production of multiple protein isoforms from a single pre-mRNA, significantly enriching the proteomic diversity of higher eukaryotic organisms 15–19. Because regulation of this process can determine the timing and location that a particular protein isoform is produced, changes in alternative splicing patterns modulate many cellular activities. Consequently, the process of splicing must occur with a high degree of specificity and fidelity to ensure the appropriate expression of functional mRNAs.
A critical step in pre-mRNA splicing is the recognition and correct pairing of 5′ and 3′ splice sites. While the 5′ splice site junction is defined by a single element of 9 nucleotides (nts), the 3′ splice site is defined by three sequence elements usually contained within 40 nts upstream of the 3′ intron/exon junction 20. Spliceosomal formation proceeds through four distinct complexes that can be resolved by nondenaturing gel electrophoresis 21, suggesting a sequential model of spliceosome assembly that requires the activity of more that 150 distinct protein factors and the U1, U2, U4, U5, and U6 small nuclear RNAs (snRNA) 22. In mammals, the 5′ splice site follows a degenerate consensus sequence YAG/GURAGU (where Y is a pyrimidine, R is A or G, and the / denotes the actual splice site) 20 that base pairs with U1 snRNA early during spliceosomal assembly. The three sequence elements that make up the 3′ splice site are the branch point sequence (BPS), the polypyrimidine tract (PPT), and the 3′ intron/exon junction. U2 snRNP interacts with the BPS, and the PPT functions as a binding platform for U2 snRNP auxiliary factor (U2AF) 23. The sequence complementarity of the 5′ splice site to U1 snRNA and the extent of the PPT at the 3′ splice site are used to determine the strength of splice sites. Greater complementarity to U1 snRNA and longer uninterrupted PPTs translate into higher affinity binding sites for spliceosomal components and, thus, more efficient splice site recognition 24. This concept of complementarity has been used extensively in numerous methods for deriving splicing scores 25–27. Given the complexity of higher eukaryotic genes and the relatively low level of splice site conservation 20, the precision of the splicing machinery in recognizing and pairing splice sites is impressive. Introns ranging in size from less than 100 up to 100,000 bases are removed efficiently. At the same time, a large number of alternative splicing events accompany the processing of pre-mRNAs 13.
It is known that there are many potential splice sites in the human genome that are not used and form pseudoexons 28. Interestingly, they occur more frequently than true exons by an order of magnitude 29. For pseudoexons to be ignored and for true exons to be recognized, there must be more information in a pre-mRNA molecule than the splice site strength. Indeed, biochemical and bioinformatic approaches demonstrated that exonic and intronic sequences contain additional information regarding splice site recognition 30, 31. Some of these, termed exonic splicing enhancers (ESEs), increase exon inclusion by serving as binding sites for the assembly of multi-component splicing enhancer complexes 11. Since the discovery of ESEs other classes of splicing regulatory elements (SREs) were identified. SREs recruit proteins and complexes that enhance as well as silence splicing and have been named descriptively: intronic splicing enhancers (ISEs), and exonic and intronic splicing silencers (ESSs and ISSs). These elements are important for selecting between pseudoexons and real exons, competing splice sites, and for the splicing of constitutive exons.
ESEs are usually recognized by at least one member of the essential serine/arginine (SR)-rich protein family while the best-characterized splicing silencers are recognized by heterogeneous nuclear ribonucleoprotein (hnRNPs). These RNA binding proteins appear to be ancestral components of eukaryotic life that are believed to have participated in shaping the diverse genomes that evolved within the eukaryotic lineage. While it is known that other RNA binding proteins contribute to pre-mRNA splicing regulation, the families of SR and hnRNP proteins are the most prominent mediators of splice site recognition. Here, we review their evolutionary history and to what degree their family expansion correlates with increased alternative splicing.
THE EVOLUTION OF SR PROTEINS
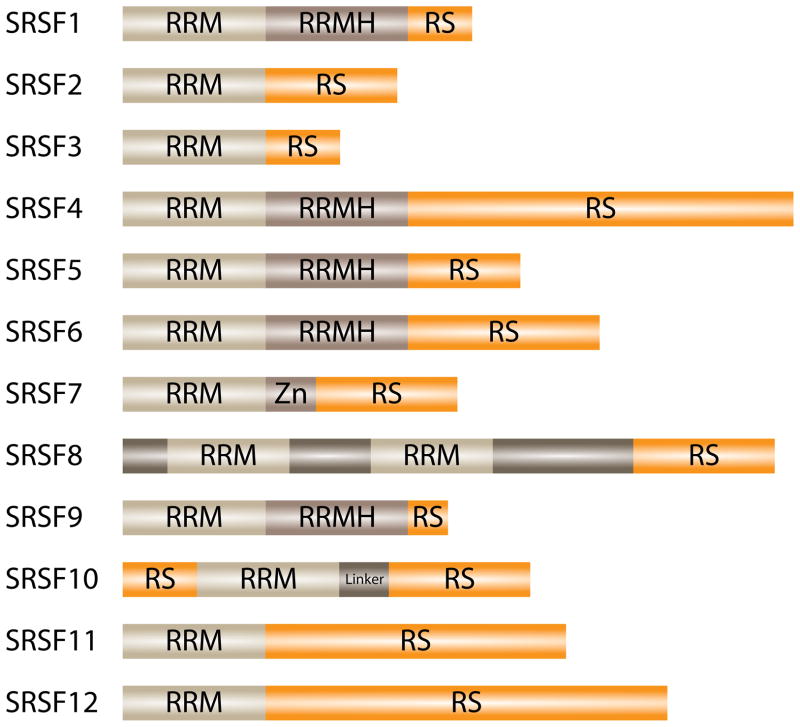
The sequence characteristics that define SR proteins are the presence of extended arginine and serine dipeptides (the arginine/serine (RS) domain) and at least one RNA binding domain of the RNA Recognition Motif (RRM)-type 32–34. The family of SR proteins has recently been redefined based on the sequence requirement for “one or two N-terminal RRMs (PF00076), followed by a downstream RS domain of at least 50 amino acids with >40% RS content, characterized by consecutive RS or SR repeats” 35. In humans, this definition results in the identification of 12 SR proteins, now designated as Serine/Arginine-rich Splicing Factor, (SRSF) 1–12 (Figure 1). Other RS domain containing factors exist that often participate in alternative or constitutive splicing 36–39. However, they either have different or no RNA binding domains, or they lack the ability to complement splicing reactions. These SR-related proteins function in multiple RNA processing pathways, expanding the SR protein family and increasing the complexity with which splicing can be regulated 40.
Figure 1.
Domain configuration of human SR proteins. SRSF1–12 are members of the canonical SR protein splicing family that is defined by N-terminal RRMs followed by a downstream RS domain 35. The RRM is responsible for RNA binding, while the RS domain mediates protein/protein interactions.
The function of SR proteins in regulating pre-mRNA splicing
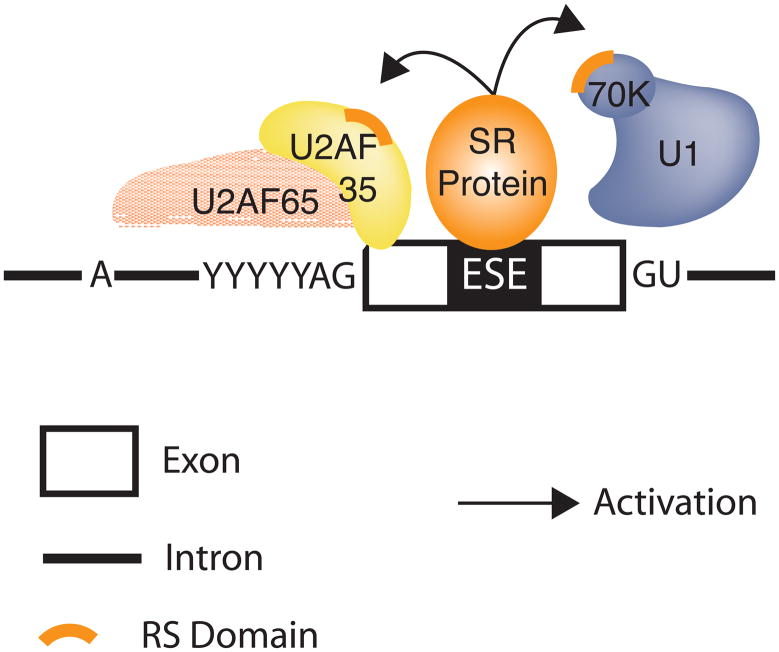
SR proteins are involved in recruiting the splicing machinery to splice sites 11, 41. It has been proposed that the RS domain of an ESE-bound SR protein interacts directly with the RS domain of other splicing factors, thereby facilitating the recruitment of spliceosomal components such as U1 snRNP to the 5′ splice site or U2AF to the 3′ splice site 41. Thus, SR proteins bound to ESEs function as general activators of exon definition 42 (Figure 2). Interestingly, SR protein binding sites are present not only within alternatively spliced exons, but also within constitutively spliced exons 43. It is therefore likely that SR proteins bind to sequences found in most exons. In summary, SR proteins bound to ESEs activate constitutive and alternative splice sites by recruiting spliceosomal components to the pre-mRNA to enhance exon recognition.
Figure 2.
Exon dependent splicing activation by SR proteins. Exon-bound SR proteins interact with components of the general splicing machinery via RS/RS domain interactions. SR protein interactions with U2AF35 (yellow) and U1 snRNP (blue) are indicated to facilitate 3′ splice site (U2AF) or 5′ splice site (U1 snRNP) recognition. The splice junctions are indicated by AG and GU.
Typically, SR protein and hnRNP binding sites are present within the vicinity of exon/intron junctions, suggesting that the interplay between activation and repression modulates the probability of exon inclusion. In vitro studies showed that the location and frequency of SR proteins or hnRNPs along the pre-mRNA alter their effectiveness. For example, as the distance between enhancer complexes and the splice site increased, the probability of exon inclusion decreased 44. Increasing the number of SR protein binding sites lessened this effect, supporting the notion that their activity depends on their context within the pre-mRNA molecule 45, 46.
The frequency of SR proteins in eukaryotes
The intron density among eukaryotic genomes varies dramatically. For example, most human genes are interrupted by intervening sequences with an average of more than 8 introns/gene 47. By contrast, only ~4% of S. cerevisiae genes harbor introns, most of which are single intron genes. While the origin of “split genes” is still debated, it is well accepted that the complexity of alternative splicing increased with multicellular complexity 47, 48. Because SR proteins have functionally been associated with the regulation of alternative splicing, it may be anticipated that the number of SR protein family members increases with more complex alternative splicing. As illustrated in Table 1, this is exactly the case. While only two SR proteins are found in the fungus S. pombe, multiple SR proteins are expressed in plants and metazoans. Interestingly, the largest number of SR proteins is identified in the plant Arabidopsis thaliana, which is known to support extensive alternative splicing. However, classical SR proteins are missing from S. cerevisiae, which also lacks alternative splicing. Instead, three SR-like proteins have been identified in S. cerevisiae, one of which, Npl3, has been shown to modulate the efficiency of pre-mRNA splicing 49. In general, the species-specific presence of SR family proteins correlates with the presence of RS domains within other components of the general splicing machinery allowing interaction between them.
Table 1.
Comparison of SR protein abundance in eukaryotes
| Family | Human | D. melanogaster | C. elegans | A. thaliana[b] | S. pombe | S. cerevisiae |
|---|---|---|---|---|---|---|
| SRSF3/7 | SRSF7 (Q16629) SRSF3 (P84103) |
xl6 (Q9V3V0) Rbp1 (Q02427) Rbp1-like (Q9VYD8) Rsf1 (Q24491) |
rsp-6 (Q18409) | RSZ21 (O81127) RSZ22 (O81126) RSZ22a (Q9SJA6) RSZ32 (Q9FYB7) RSZ33 (Q9FYA7) |
||
| SRSF11 | SRSF11 (Q05519) | Srp54 (Q9VL71) | rsp-7 (O01159) | |||
| SRSF2/8 | SRSF2 (Q01130) SRSF8 (Q9BRL6) |
SC35 (Q9V3T8) | rsp-4 (Q09511) | SCL28 (Q1PDV2) SCL30 (Q9FYA9) SCL30a (Q9FYA8) SCL33 (Q9SEU4) SC35 (Q9FYB1) |
srp1 (Q10193) | |
| SRSF1/9 | SRSF1 (Q07955) SRSF9 (Q13242) |
SF2 (Q9V3W7) | rsp-3 (Q9GQI7) | RS31a (Q9ZPX8) SR1 (Q9SPI1) SRp34a (A2RVS6) SR34b (Q3EAC7) SRp30 (6Q9XFR5) |
||
| SRSF4/5/6 | SRSF4 (Q08170) SRSF5 (Q13243) SRSF6 (Q13247) |
B52 (P26686) | rsp-5 (Q10021) rsp-1 (Q23121) rsp-2 (Q23120) |
RSP41 (P92966) RSP31 (P92964) RSP40 (P92965) |
srp2 (P78814) | NPL3 (Q01560) |
| SRSF10/12 | SRSF10 (O75494) SRSF12 (Q8WXF0) |
[a] | [a] | [a] |
identical to SC35-family homologues
gene names were obtained either from UniProt or www.arabidopsis.org
(UniProt-identifier is given in parentheses)
Homologues were identified according to Barbosa-Morais et al 48.
To demonstrate the significance of the observed expansion within the family of SR proteins, it is useful to compare the conservation of splicing factors associated with the general splicing machinery. The five snRNPs U1, U2, U4, U5, and U6 make up the core of the spliceosome. Each snRNP is assembled with a common set of Sm proteins and additional proteins unique to each snRNP. Homologues of the vast majority of these spliceosomal core proteins are present throughout all eukaryotes (Table 2), demonstrating that the splicing machinery itself is highly conserved and that the presence of the snRNPs and their associated components is essential to support intron removal.
Table 2.
Comparison of general spliceosomal component abundance in eukaryotes
| Human | D. melanogaster | C. elegans | A. thaliana[a] | S. pombe | S. cerevisiae |
|---|---|---|---|---|---|
| U1-70K (P08621) | U1-70K (P17133) | rnp-7 (Q09584) | U1-70K (Q42404) | U1-70K (O13829) | U1-70K (Q00916) |
| U1A (P09012) | U1A (P43332) | rnp-2 (Q21322) | U1A (Q39244) | RU1A (O74968) | U1A (P32605) |
| U1C (P09234) | U1C (Q9VE17) | U1C (P90815) | U1C (Q56XE4) | U1C (Q9P794) | U1C (Q05900) |
| U2A′ (P09661) | U2A′ (Q9V4Q8) | U2A′ (Q9BLB6) | U2A′ (P43333) | U2A′ (Q9USX8) | U2A′ (Q08963) |
| U5-100KD (Q9BUQ8) | CG10333 (Q9VJ74) | ddx-23 (Q95QN2) | RH21(P93008) | PRP28 (Q9Y7T7) | PRP28 (P23394) |
| PRP4 (O43172) | CG6322 (Q9VVI0) | prp-4 (Q93339) | PRP4 (O22212) | YIDC (Q9UTC7) | PRP4 (P20053) |
| SmB/B′ (P14678) | SmB (Q05856) | SmB (P91918) | AT4G20440 (Q9SUN5) | SmB (Q10163) | SmB (P40018) |
| Sm-D2 (P62316) | Sm-D2 (Q9VI10) | Sm-D2 (Q18786) | Sm-D2 (Q8RUH0) | Sm-D2 (O14036) | Sm-D2 (Q06217) |
| SmE (P62304) | SmE (Q9VLV5) | SmE (Q9XTU6) | SmE (Q9ZV45) | SmE (Q9USZ3) | SmE (Q12330) |
| U2AF65 (P26368) | U2AF50 (Q24562) | U2AF65 (P90978) | U2AF65 (O23212) | U2AF59 (P36629) |
gene names were obtained either from UniProt or www.arabidopsis.org
(UniProt-identifier is given in parentheses)
Homologues were identified according to Barbosa-Morais et al 48.
The evolutionary origin of SR proteins
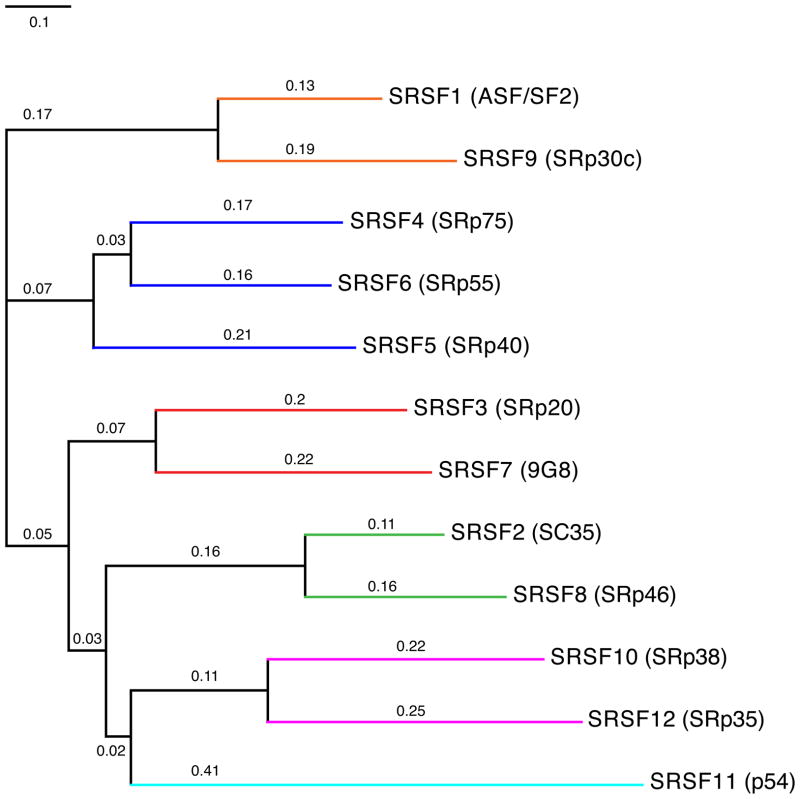
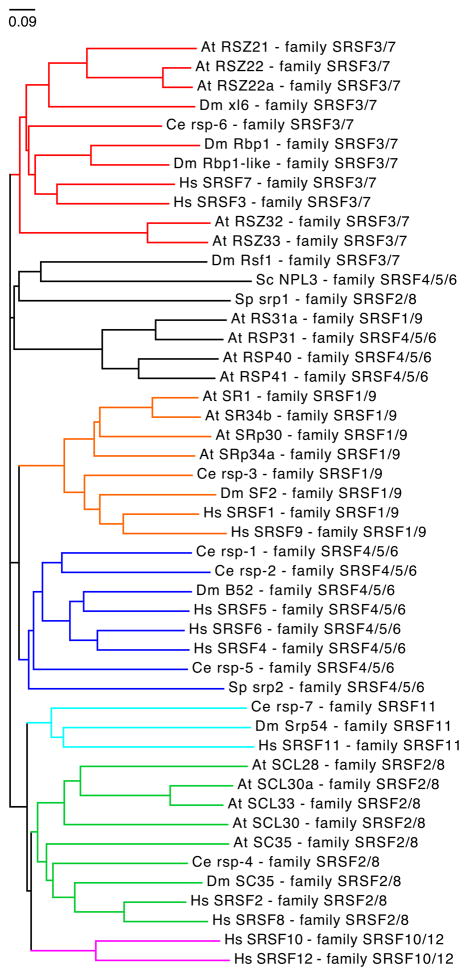
A phylogenetic tree analysis of the 12 human SR proteins indicates the presence of 6 families, one of which consists of three closely related SR protein members (Figure 3). These families are largely conserved among higher eukaryotes as illustrated by a species-specific tree comparison (Figure 4). Based on amino acid sequence alignments among several eukaryotic species it was suggested that successive gene duplications were instrumental in evolving the SR protein diversity as is observed in complex multicellular organisms today 50. These duplication events were coupled with high rates of nonsynonymous substitutions that presumably promoted positive selection, thus favoring the rapid gain of new functions for duplicated SR protein genes. These observations link SR protein evolution to the “classical” model for selective retention of gene duplicates 51, where one of the duplicate genes is expected to retain the original function, while the other accumulates a number of mutations that eventually conferred new advantageous functions (neofunctionalization) 50. An SR protein domain structure analysis further demonstrated relatively high levels of conservation. Importantly, the same study did not find sufficient evidence for domain shuffling 50. Therefore, it is unlikely that the expansion of SR protein families was accomplished by appending RS domains onto other RNA binding factors. Each SR protein appears to have changed on its own with the RRM and RS domains evolving together. Given the fact that SR proteins and SR-like proteins (Npl 3 in S. cerevisiae) are present in most single cell eukaryotes, it is very likely that SR proteins are ancestral to eukaryotes and that these ancestral SR proteins were subsequently lost independently in some lineages. Thus, it is possible that variations among the extent of alternative splicing mirrors the regulatory requirements of an organism and that these requirement differences caused a simplification of alternative splicing in some single-cell eukaryotes 52.
Figure 3.
Evolutionary relationship between human SR proteins. The phylogenetic tree is based on the alignment of all human SR proteins (Table 1). The number above each bar indicates the degree of similarity. The colored lines indicate different clusters, also referred to as SR protein families. The old names of SR proteins are given in parentheses. ClustalW was used to align protein sequences and to perform phylogenetic analysis. Phylogenetic trees were drawn by CTree using the ClustalW output.
Figure 4.
Evolutionary relationship between eukaryotic SR proteins. The phylogenetic tree is based on the alignment of Homo sapiens (Hs), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Arabidopsis thaliana (At), and Schizosaccharomyces pombe (Sp) SR protein sequences (Table 1). The sequence for Npl3, a SR-like protein from Schizosaccharomyces cerevisiae (Sc) was also included in the analysis. Homologues of the six human SR protein families are highlighted in color. Tree analysis was performed as described in Figure 3.
The RS domains of SR proteins differ in their RS repeat density. From a functional point of view it has been established that a higher RS-repeat density correlates with increased splice site recognition potential. When comparing the composition of RS domains between eukaryotes an interesting trend was observed. SR and SR-like proteins harbor R-rich C-termini with a variable content of RX repeats. X can be S (serine), D (aspartic acid), E (glutamic acid), or G (glycine) generating diverse R-rich repeat domains 47. In metazoans these regions display a high density of RS repeats, whereas fungi are RD-rich, and Npl 3 in S. cerevisiae is RG rich. Based on comparative studies this change in RS-repeat density can be correlated with the binding potential of U2 snRNP to the branch-point sequence. The lower the intrinsic affinity of U2 snRNP to the branch point the higher the number of RS repeats in the C-terminus domain 47. These observations suggest that the emergence of more complex multicellular organisms sparked an increase in the density of RS repeats or vice versa.
Co-evolution of SR proteins and splice site recognition signals
An important question to answer is to what degree the evolution of SR proteins correlates with increased alternative splicing and/or with the evolution of other pre-mRNA splicing elements. Two not mutually exclusive models have been put forward to describe the evolution of alternative splicing 53. The first one proposes that the accumulation of mutations within splice sites rendered them suboptimal, resulting in insufficient recognition by the spliceosome and, eventually, the skipping of the exon that is flanked by one of the drifted splice sites. This would then require the evolution of splicing enhancers that would allow that site to be recognized. The second model proposes that the evolution of splicing regulatory factors (like SR-proteins or hnRNPs) with abilities to positively or negatively influence the recruitment of splicing components (like snRNPs) reduces selective pressures to maintain strong splice sites, thus permitting relaxation and weakening of splice site signals. The following observations argue in favor of the latter model. It was demonstrated that the S. pombe SR protein Srp2 could enhance the recognition of a suboptimal 3′ splice site, thus promoting intron excision 54. These results suggested an early evolutionary origin of exonic splicing enhancers. Furthermore, the hypothesis that the expression of an SR protein could activate intron removal in an organism that lacks canonical SR proteins was tested. Indeed, when introduced into S. cerevisiae, a mammalian SR protein was capable of specifically activating the recognition of a weak splice site, thus promoting intron removal 55. These results demonstrated that canonical SR proteins can promote pre-mRNA splicing activity even in an SR-free organism and suggest that SR proteins did exist before alternative splicing was prevalent. Given that SR proteins were already present before the appearance of alternative splicing and the fact that SR proteins can activate recognition of suboptimal splice sites, it is likely that during eukaryotic evolution the influence of SR proteins shifted from supporting the general splicing reaction to mediating alternative splicing decisions. The proliferation of the SR protein family is then expected to correlate with increasing complexities of alternative splicing, a proposal consistent with alternative splicing evaluations of uni- or multicellular organisms.
These proposals predict that SR protein expansion coincides with changes in the splice site consensus sequence with the expectation that an increased SR protein presence relaxes splice site sequence conservation. Recent computational tests demonstrated that for branch point sequences at the 3′ splice site, as well as for the 5′ splice site sequence, this is exactly the case 47. However, this splice site signal relaxation is not only contributed to the expansion of SR proteins. For example, changes in the PPT sequence requirement strongly correlate with accompanying structural changes in the SR-related spliceosomal factor U2AF, which binds to this sequence element. Weaker correlations were also detected between alterations within the 5′ splice site signal and the sequence of the U1 snRNA, which base pairs to it 56. Together, these analyses imply that the expansion of SR proteins and nucleotide changes in U2AF had a fundamental role in the relaxation of the splicing signals and in the evolution of regulated alternative splicing.
THE EVOLUTION OF HnRNP PROTEINS
HnRNP bound splicing silencers occur frequently and have been found to influence constitutive and alternative splicing events throughout the human genome 57. Like SR proteins hnRNPs are also modular 58. The most prevalent domain amongst the hnRNPs is the RRM that mediates specific interactions with the pre-mRNA (with the exceptions of hnRNPs E/K, which interact with RNA via the hnRNP KH (K homology) domain). Most hnRNPs also harbor RGG boxes (repeats of Arg-Gly-Gly tripeptides), and additional glycine-rich, acidic or, proline-rich domains 59. The modularity of the hnRNPs ensures structural variation that promotes functional diversity. As a result, hnRNP proteins participate in a wide range of biological functions. HnRNP proteins are not as strictly defined as SR proteins allowing flexibility in their classification. Here, we have limited our discussion to the canonical hnRNPs initially identified by Dreyfus and colleagues 60, thus omitting other hnRNP-like RNA binding factors such as CELF proteins 61, Fox proteins 62, Nova 63, or TDP-43 64.
The function of hnRNP proteins in regulating pre-mRNA splicing
Like SR proteins, hnRNP proteins direct their influence on pre-mRNA splicing through site-specific binding with the target RNA. This binding is supported by the RRM or the KH domains present in hnRNP proteins. After binding, the business end of hnRNPs (RGG boxes, glycine-rich, acidic or, proline-rich domains) then promotes protein/protein interactions that ultimately mediate splicing decisions. Unlike SR proteins, the mechanism through which hnRNPs interfere with splicing is known only for a small number of cases. These include repressing spliceosomal assembly through multimerization along exons 65, blocking the recruitment of snRNPs 66, 67, or by looping out entire exons 68 (Figure 5). Clouding our mechanistic understanding of hnRNP action is the fact that some hnRNPs can repress or activate exon recognition depending on their location relative to the regulated splice site. Clearly, additional functional work needs to be carried out to derive the common mechanisms employed by hnRNPs in modulating alternative splicing.
Figure 5.
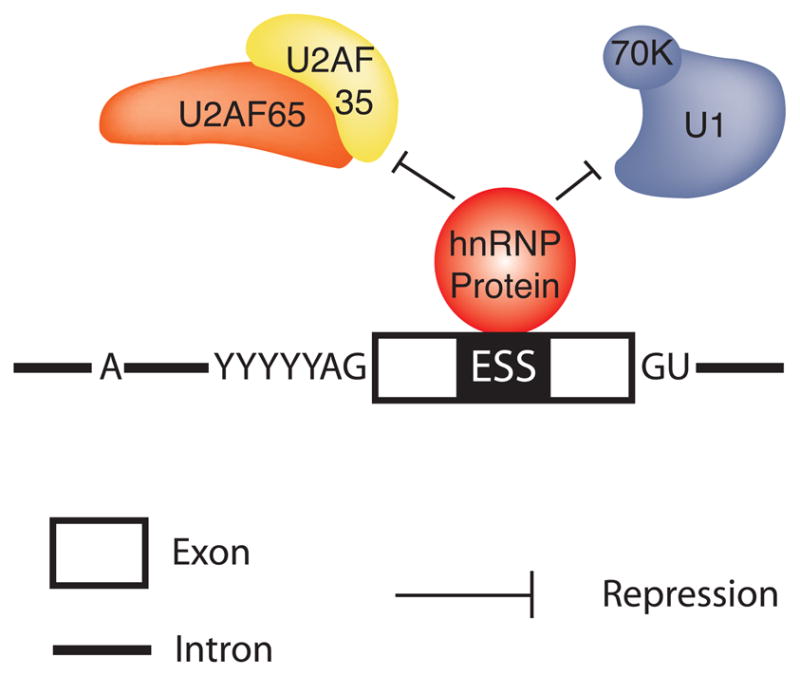
Exon dependent splicing repression by hnRNP proteins. Exon-bound hnRNP proteins interfere with the association of the general splicing machinery with the pre-mRNA. AG and GU indicate the splice junctions.
Over the last years it has become increasingly clear that exon selection is influenced by a number of activating and inhibitory elements. Given the divergent sequences and architectures of eukaryotic genes, every exon is expected to have a specific set of identity elements that permit its recognition by the spliceosome. Each exon is flanked by a unique pair of splice site signals and contains a unique group of SR and hnRNP protein binding sites. The sum of contributions from these SR and hnRNP protein binding sites ultimately defines the overall recognition potential of an exon, or the overall binding affinity for the spliceosome 24.
The frequency and evolution of hnRNP proteins in eukaryotes
The difference in hnRNP abundance between unicellular and multicellular organisms is more striking than that observed for SR proteins as the number of families and family members is higher in multicellular organisms (Table 3). These observations suggest that hnRNP genes were either subjected to more duplications events, or that different selective pressures existed for duplicated hnRNP genes. Ultimately this results in a higher degree of functional hnRNP diversification. As was observed for SR proteins, only one clear hnRNP homologue is detected in S. pombe (Musashi). This hnRNP is conserved across most eukaryotes (Table 3); however, it is apparently missing in Arabidopsis. Another interesting difference between the nature of hnRNP and SR protein expansion is the fact that Arabidopsis hosts the most plentiful SR protein ensemble while only a limited number of hnRNPs are detected (compare Tables 1 and 3). It is possible that these differences may reflect properties of how alternative pre-mRNA splicing is achieved in each species. Clear homologues of the hnRNP family are missing in S. cerevisiae. Instead, S. cerevisiae expresses Hrp1, an hnRNP-like protein involved in 3′ end processing that correlates with the expression of the SR-like Npl3 69. Thus, a distantly related gene exists in S. cerevisiae. However, it is unclear whether it originated from the ancestral gene that gave birth to the hnRNP protein family.
Table 3.
Comparison of hnRNP protein abundance in eukaryotes
| Family | Human | D. melanogaster | C. elegans | A. thaliana | S. pombe | S. cerevisiae |
|---|---|---|---|---|---|---|
| hnRNP-A | HNRNPA0 (Q13151) HNRNPA1 (P09651) HNRNPA2 (P22626) HNRNPA3 (P51991) |
Hrb87F (P48810) Rb97D (Q02926) Hrb98DE (P07909) |
HNRNPAa (Q8WSM6) hrp-1 (Q22037) |
rnp1 (Q8W034) HNRNPAa (Q8W555) HNRNPAb (O22791) |
||
| hnRNP-C | RALY (Q9UKM9) RALYL (Q8N1C2) HNRNPC (P07910) HNRNPCL1 (O60812) |
|||||
| hnRNP-D | HNRNPAB (Q99729) HNRNPD (Q14103) HNRPDL (O14979) |
sqd (Q08473) | sqd-1 (Q8MXR6) | |||
| hnRNP-E | PCBP1 (Q15365) PCBP2 (Q15366) PCBP3 (P57721) PCBP4 (P57723) |
mub (P91632) | pes-4 (Q95Y67) | |||
| hnRNP-F/H | GRSF1 (Q12849) HNRNPF (P52597) HNRNPH1 (P31943) HNRNPH2 (P55795) HNRNPH3 (P31942) |
glo (Q9VGH5) | hrpf-1 (Q9BIB7) hrpf-2 (Q8MXR2) |
HNRNPFa (Q9FKY1) HNRNPFb (Q9LT32) |
||
| hnRNP-G | HNRPG (P38159) HNRNPGT (O75526) |
|||||
| hnRNP-I | PTBP1 (P26599) PTBP2 (Q969N9) ROD1 (O95758) |
heph (Q8WR53) | ptb-1 (Q18999) | PTBP1 (Q9MAC5) PTBP2 (Q9FGL9) PTBP3 (Q6ICX4) |
||
| hnRNP-K | HNRNPK (P61978) | HNRNPK (Q9V948) | HNRNPK (P91277) | |||
| hnRNP-L | HNRNPL (P14866) HNRPLL (Q8WVV9) |
HNRNPL (Q7JMZ7) | HNRNPL (Q95QR5) | |||
| hnRNP-M | MYEF2 (C9JI55) HNRNPM (P52272) |
rump (Q9VHC7) | HNRNPM (Q9XVS2) | |||
| hnRNP-R | HNRPQ (O60506) HNRNPR (O43390) |
HNRPQ (Q95TW4) | hrp-2 (Q9NLD1) | HNRPQa (Q9ASP6) HNRPQb (Q8RWQ1) F3C22_6o (Q9LXJ8) |
||
| hnRNP-U | HNRNPUL1 (Q9BUJ2) HNRNPU (Q00839) HNRNPUL2 (Q1KMD3) |
HNRNPU (A1ZBB4) | HNRNPU (Q9U2H8) | |||
| Musashi | MSI1 (O43347) MSI2 (Q96DH6) |
msi (Q8MS04) Rbp6 (Q8MQZ1) |
msi-1 (Q21911) | YHKF (O94432) | HRP1 (Q99383) |
gene names were obtained either from UniProt or www.arabidopsis.org
(UniProt-identifier is given in parentheses)
Homologues were identified according to Barbosa-Morais et al 48
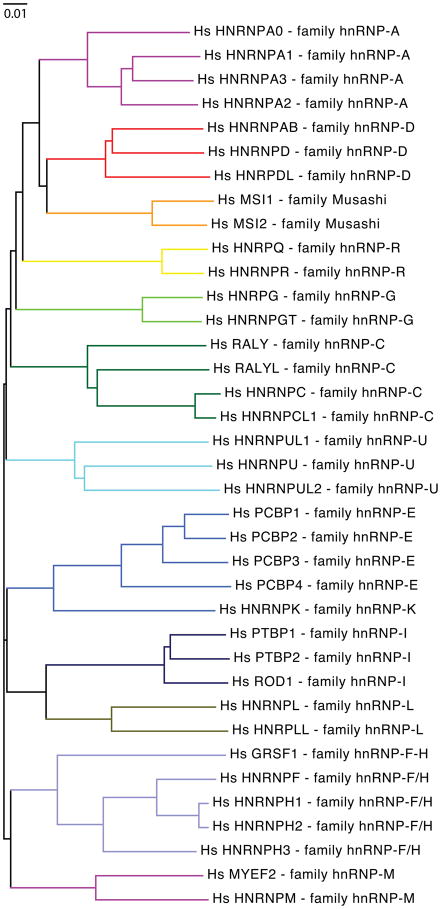
The phylogenetic tree analysis of the human hnRNP proteins indicates the presence of 13 families consisting of multiple closely related hnRNP members (Figure 6). The members of each family are most closely related among higher eukaryotes as illustrated by a species-specific tree comparison (Figure 7). HnRNP family diversity appears to have derived from successive gene duplication events. For example, it was shown that the intron/exon architecture of the hnRNP A2 gene is near identical to that of hnRNP A1, in agreement with the “common origin by gene duplication” model 70. Similar arguments can be made for the other hnRNP families. If Hpr1 in S. cerevisiae is considered a relative of the hnRNP family, it can be argued that hnRNPs are ancestral to eukaryotes just like SR proteins are. As pre-mRNA splicing diversified in multicellular organisms to include more and more complex alternative splicing patterns, a striking expansion of hnRNPs was promoted. By contrast, simplification of pre-mRNA splicing may have resulted in the loss of hnRNP diversity.
Figure 6.
Evolutionary relationship between human hnRNP proteins. The phylogenetic tree is based on the alignment of all human hnRNP proteins (Table 3). The colored lines indicate different hnRNP families. Tree analysis was performed as described in Figure 3.
Figure 7.
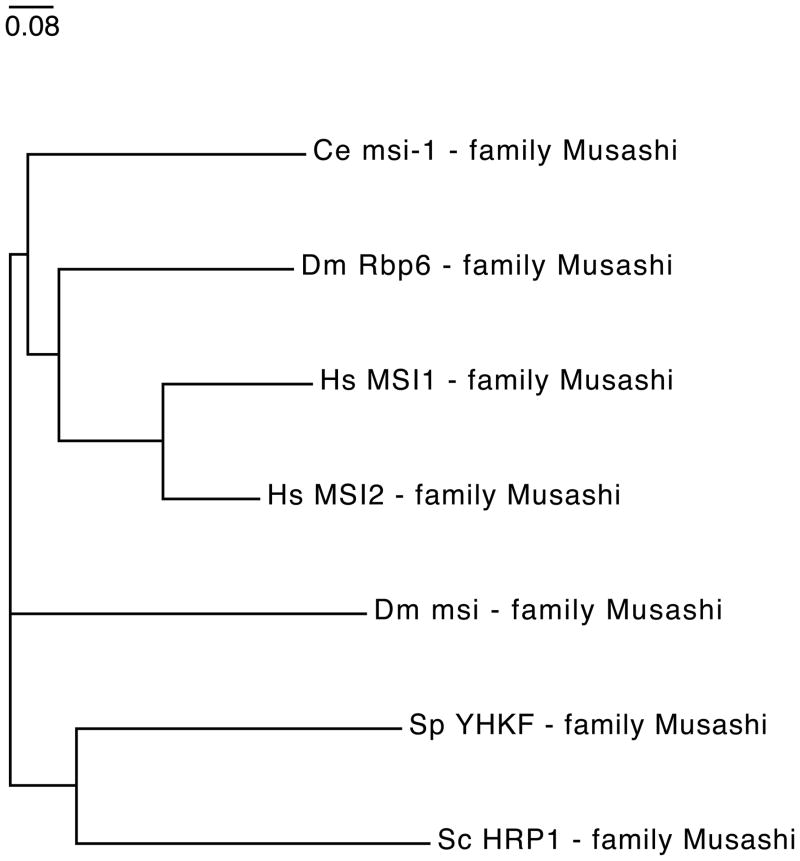
Evolutionary relationship between eukaryotic hnRNP proteins. The phylogenetic tree is based on the alignment of Homo sapiens (Hs), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), and Schizosaccharomyces pombe (Sp) Musashi protein sequences (Table 3). The sequence for Hrp1, an hnRNP-like protein from Schizosaccharomyces cerevisiae (Sc) was also included in the analysis. Tree analysis was performed as described in Figure 3.
Why are there significantly more hnRNP members in human compared to the number of human SR protein members? As mentioned above, the human genome is littered with reasonably good 5′ and 3′ splice sites that are not used. Computer predictions even suggest that pseudoexons outnumber real exon by almost an order of magnitude 29. Clearly, the generation of such pseudoexons is in direct relationship with the relaxation of the splice site consensus sequence. The more variations within intron/exon junctions were permissible, the greater the probability that alternate sequences existed nearby that were similar enough to provide sufficient binding potential for spliceosomal components. When this relaxation in splice site sequences is coupled with intron gain and intron expansion 47, a readily apparent trend can be observed between unicellular or multicellular organisms. It becomes clear that pseudo splice sites can easily outnumber real splice sites, potentially causing a multitude of detrimental pre-mRNA processing events. As hnRNP proteins generally repress splice site recognition, it is possible that their presence curbs the execution of such possible mis-splicing decisions. The differentiation between a real and a pseudo splice site may then be accomplished through the opposing actions of SR and hnRNP proteins that bind to the pre-mRNA in close vicinity of a splice site. Detailed biochemical experiments recently demonstrated that splicing repressors (presumably mediated by hnRNPs) are more powerful in negating splice site recognition when directly competing with SR protein splicing enhancer complexes that attempt to promote exon inclusion 46. These results suggest that repressing potential splice sites may be the default pathway for organisms with more complex exon/intron architectures. Promoting a repressed splice site to an actively used splice site then requires the additional activity of multiple splicing enhancers.
Did hnRNPs and SR proteins originate from a common ancestor?
When debating the origin of SR and hnRNP proteins, arguments center on the possible make-up of the common ancestor of eukaryotes. The consensus here is that the common ancestor was unicellular, but it is unclear whether it contained multiple introns. Based on the observations that SR-like and hnRNP-like factors are present in S. cerevisiae and that SR and hnRNP homologues exist in S. pombe, it can be argued that both protein classes are ancestral. The commonalities between these factors consist of the presence of the RRM and their modular domain structure. The RS domain analysis mentioned above further indicates that an ancestral arginine-rich C-terminus eventually evolved into the canonical RS domain with variable RS repeat densities 47. While the protein/protein interaction domains of hnRNPs are not as well defined as SR proteins, they do contain in many cases RGG boxes. It is therefore possible that hnRNP and SR proteins may have shared a common ancestor. Environmental pressures then promoted divergent evolution of functionally related, but antagonistic splicing regulatory factors.
Conclusions
SR and hnRNP proteins mediate pre-mRNA splicing decisions in higher eukaryotes. While their origin is likely to date back to a common ancestor of eukaryotes, their diversification and expansion parallels that of increasing pre-mRNA splicing complexity. With increasing numbers of introns and intron length per gene a significant expansion in the number of SR and hnRNP protein families is observed. Importantly, the correlation between increased alternative splicing by relaxation of splice site signals and the expansion of SR and hnRNP proteins strongly argues for the model that SR and hnRNP proteins reduced evolutionary pressures to maintain highly conserved splice sites. As a consequence, alternative splicing became more prominent, enriching proteomic diversification. Thus, the interplay between the presence of these classical splicing regulators and the spliceosomal requirements for splice site recognition is a major driving force in the ongoing evolution of alternative pre-mRNA splicing.
Acknowledgments
The authors wish to thank William Mueller for comments on the manuscript and acknowledge support from the NIH (RO1 GM62287 and R21 CA149548 to K.J.H.) and from a fellowship within the Postdoc Programme of the German Academic Exchange Service, DAAD (A.B.).
References
- 1.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 3.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 4.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q, Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Lo HS, Yang H, Gere S, Hu Y, Buetow KH, Lee MP. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 7.Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2007 doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Xing Y. Genomic analysis of RNA alternative splicing in cancers. Front Biosci. 2007;12:4034–4041. doi: 10.2741/2369. [DOI] [PubMed] [Google Scholar]
- 10.Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 11.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 12.INTERNATIONAL HUMAN GENOME SEQUENCING CONSORTIUM. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 13.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc Natl Acad Sci U S A. 2009;106:1766–1771. doi: 10.1073/pnas.0813128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 16.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 17.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 19.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 20.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RFCTR, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor, New York: CSHL Press; 1999. pp. 525–560. [Google Scholar]
- 21.Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 22.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 23.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Gen Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 24.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 25.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 28.Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Mol Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XH, Heller KA, Hefter I, Leslie CS, Chasin LA. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 2003;13:2637–2650. doi: 10.1101/gr.1679003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 32.Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 33.Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 34.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- 37.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 38.Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 40.Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. A genome-wide survey of RS domain proteins. RNA. 2001;7:1693–1701. [PMC free article] [PubMed] [Google Scholar]
- 41.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam BJ, Hertel KJ. A general role for splicing enhancers in exon definition. RNA. 2002;8:1233–1241. doi: 10.1017/s1355838202028030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaal TD, Maniatis T. Multiple Distinct Splicing Enhancers in the Protein-Coding Sequences of a Constitutively Spliced Pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre- mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhang XH, Arias MA, Ke S, Chasin LA. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plass M, Agirre E, Reyes D, Camara F, Eyras E. Co-evolution of the branch site and SR proteins in eukaryotes. Trends Genet. 2008;24:590–594. doi: 10.1016/j.tig.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res. 2006;16:66–77. doi: 10.1101/gr.3936206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escobar AJ, Arenas AF, Gomez-Marin JE. Molecular evolution of serine/arginine splicing factors family (SR) by positive selection. In Silico Biol. 2006;6:347–350. [PubMed] [Google Scholar]
- 51.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 54.Webb CJ, Romfo CM, van Heeckeren WJ, Wise JA. Exonic splicing enhancers in fission yeast: functional conservation demonstrates an early evolutionary origin. Genes Dev. 2005;19:242–254. doi: 10.1101/gad.1265905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han SP, Kassahn KS, Skarshewski A, Ragan MA, Rothnagel JA, Smith R. Functional implications of the emergence of alternative splicing in hnRNP A/B transcripts. RNA. 16:1760–1768. doi: 10.1261/rna.2142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 60.Choi YD, Dreyfuss G. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci U S A. 1984;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 64.Buratti E, Baralle FE. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- 65.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 66.Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. Embo J. 2001;20:5748–5758. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- 68.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biamonti G, Ruggiu M, Saccone S, Della Valle G, Riva S. Two homologous genes, originated by duplication, encode the human hnRNP proteins A2 and A1. Nucleic Acids Res. 1994;22:1996–2002. doi: 10.1093/nar/22.11.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]