Abstract
Given that RNA is involved in virtually all biological processes, it is perhaps not surprising that several RNA binding proteins are associated with aging and with different age related disorders. Other chapters in this volume will discuss some specific examples of diseases where RNA plays a role that are also associated with aging, such as cancer and inflammation, so here I will discuss some general aspects of how RNA changes with the aging process. I will also discuss some specific examples of RNA binding proteins that are associated with age-dependent neurological diseases as these provide an interesting framework to examine how lifetime mutations might lead to a late onset disease, although the answers to these questions are still not well understood.
Keywords: Aging, Senescence, Age-related diseases, Progeria, Neurodegeneration, Alzheimer’s disease, Parkinson’s disease, Amyotrophic Lateral Sclerosis
Age related changes in RNA
After maturation, most organisms continue to undergo a number of changes in biology that are collectively termed senescence. The proposed mechanisms involved in aging, and their underlying biology, are far outside of the scope of this article, but it is important to recognize that both genetic or programmed factors and environmental and/or stochastic events contribute to overall aging. Possibly related to this, aging consists of both generalized changes in an organ or tissue across time and a number of variable events such as age-related diseases.
As for many macromolecules in the cell, mRNA is subject to age related changes of various types. The overall gene expression profile of an aged tissue is different from that of its young, but still mature, equivalent. To name a few examples, studies in different tissues including heart 1, kidney 2, muscle 3 and brain 4, 5 have identified genes that vary along lifespan. Systematic analyses in mice suggest that there are some genes that show consistent changes with aging in all tissues although different tissues can ‘age’ at different rates 6. Between species there are also common signatures of aging, particularly in mitochondrial electron transport chain genes 6, which might reflect the role that oxidative stress plays in damaging events associated with aging.
Whether RNA expression profiles are a cause or consequence of aging is difficult to determine (see below for more discussion), but what is perhaps more interesting is that most of the changes reported in these studies above are quite modest. Re-analysis of one of these datasets suggests that there are significant numbers of changes in the relative expression of genes from being tightly correlated in younger tissues to being less highly co-expressed in aged tissues 7. Additionally, survey of gene expression in the aging heart suggests that there is an increase in variance in gene expression with aging, especially increased variability from cell to cell 1. The mechanism(s) underlying such changes are not known, but one obvious candidate is that normally repressive mechanisms change with aging, leading to an altered control of gene expression.
Possibly related to this, a few recent studies have examined expression of non-coding RNAs with aging. As discussed elsewhere in this volume, several novel types of RNA species have been found in recent years including structural RNA species and regulatory small RNAs such as small interfering RNA (siRNA) and microRNAs (miRNA). As an example of a critically important class of such molecules, recent surveys of miRNA expression with aging show an interesting pattern of increasing with age 8, 9. Upregulated miRNAs cluster in functional categories, oxidative defense, DNA repair, intermediate metabolism, cytoskeletal organization, cell cycle control, and apoptosis. As these are functional categories that one might suspect are important in aging, and overlap with the observation above that mitochondrial/oxidative metabolism mRNAs are a consistent target for age-related processes. Age-related changes in miRNA are seen in many organisms, including Caenorhabditis elegans, were one miRNA (lin4) has been shown to impact longevity 10.
Whether age-related changes in miRNA and mRNA expression profiles are related to each other is not directly known. Because miRNA generally either destabilize mRNA transcripts or block translation (although there are exceptions), we might expect mRNAs to generally be decreased for translation from those mRNA species with age. As discussed above, there is less evidence of a generalized loss of mRNA but more evidence of a relaxed control of levels. How much of this is due to miRNA is unclear, but it is a potentially tractable question. One experiment that should be performed is to examine known mRNA; miRNA target pairs with aging. One would predict that in the simplest scenario, miRNA increases should be associated with correlated changes in mRNA targets (but not other targets) that, for groups of genes, may therefore change in co-expression.
Another important age related change in RNA is the acquisition of oxidative damage. Like all macromolecules, reactive oxygen species can attack RNA leading to a number of stable modifications that be measured in tissues. In fact, because RNA may be especially susceptible to attack by oxidative stress compared to other macromolecules. For example, RNA is relatively less protected than DNA as it is not bound to histone proteins and is also single stranded compared to double stranded. Furthermore, although there probably are RNA repair mechanisms, these are apparently less extensive than DNA repair. Supporting this, there is strong evidence of oxidized RNA but less evidence of oxidized DNA in aged muscle 11.
Aging of experimental animals results in the accumulation of 8-OHG modified RNA that can be reversed by mitochondrial metabolites 12. If mRNA is modified in this way, it is likely to be a less than optimal template for the translational machinery. Although not directly measured in these experiments, small RNA is likely to be as susceptible to oxidative stress as messenger RNA and this will impair mRNA regulation.
As well as the above studies suggesting that RNA is a target of the aging process, aging can also affect utilization of RNA. There is substantial evidence that the translational machinery is affected in aging, probably by multiple mechanisms. Oxidative stress is a trigger for the phosphorylation of 4EBP which then binds to and sequesters eiF4E, an important component of the translation initiation complex 13. This means that under conditions of chronic oxidative stress, such as aging, translation of mRNA to protein is generally less efficient. Added to direct oxidative damage to RNA, overall translation is likely to be very different in a senescent cell than in a younger cell. In fact, there is some evidence that altered protein translation contributes to shortened lifespan in a number of species and that rapamycin can extend lifespan 14. Rapamycin inhibits the kinase mTOR (mammalian target of rapamycin) that, in combination with regulatory subunits, phosphorylates substrates involved in protein translation, autophagy and telomere length 15. Therefore, manipulating protein translation may be one way to limit senescenece.
The above data shows that the RNA component changes throughout ages in ways that are relatively subtle but cumulatively impact the structure, regulation and function of mRNA. In turn, this impacts the output of RNA, namely translation into protein.
RNA in age-related disorders
Overlaid over the patterns of ‘normal’ aging are a number of disorders with a strong age-related component that affect many individuals, particularly in humans. The number of age-related disorders is too many to cover in this review, although other chapters in this series will discuss some major diseases such as cancer and cardiovascular disease. Here, we will discuss RNA changes in late onset neurodegenerative diseases where age is the major risk factor for disease development.
One of the diseases often considered part of the spectrum of old age is Alzheimer’s disease (AD). Patients with AD have impairment in several cognitive functions, most famously in short term memory formation and retrieval, that worsen over time. Several studies have examined how mRNA expression differs between AD and similarly aged controls without neurodegeneration 16, 17, even at the level of the single cell 18. Similar studies have been reported for Parkinson’s disease (PD) 19, 20 and amyotrophic lateral sclerosis (ALS) 21–23, neurodegenerative diseases that affect neurons in the brainstem and in the spinal cord respectively, and each have generated lists of differentially expressed genes. In some cases, formal analyses of many different datsets have been performed 24. A smaller set of studies have examined miRNA expression in neurodegeneration, but there is some evidence of specific alterations in these regulatory species in neurodegeneration and under oxidative conditions in cell culture 25.
As well as differences in steady state mRNA expression levels, there is also evidence of accumulation of damaged RNA in many of these diseases. For example, in AD 26–28, PD 29 and ALS 30, oxidized RNA accumulates even at early stages of the disease. For ALS, there is some evidence that blocking oxidative stress and reversing accumulation of oxidized RNA is partially protective, although it is difficult to be certain if oxidative RNA damage alone is either necessary or sufficient for pathogenesis is age-related neurodegenerative diseases 31.
These results suggest that RNA Is one target of the disease process in age-related neurodegeneration, but interpretation is difficult for a number of reasons. First, there are shared patterns of altered RNA expression and modification in both aging and neurodegeneration but because aging is a risk factor for these diseases and thus aging and disease are not independent factors. As an example of this, in one study of the aging hippocampus, a brain area affected by AD, there was distinct upregulation of genes involved in cholesterol trafficking including ApoE 5. The cholesterol/ApoE pathway appears to have a contributory genetic effect to lifetime risk of AD 32, 33 and so whether aging affects cholesterol metabolism which affects AD risk or the aging and cholesterol metabolism effects are independent is unclear. Second, the diseased brain is very different from the normal brain in that neurons are lost and glial cells react and proliferate in response to neuronal damage and loss. There have been attempts to correct for these changes in cellularity that occur in neurodegeneration 24 but fundamentally it is difficult to tell if the cells that survive to the end of a neurodegenerative process are those that are inherently resistant to the toxic events, have adapted to the detrimental environment of the diseased brains or are those that will die eventually – or a mixture of all three.
The larger difficulty is that there is a classic difficulty of understanding if changes in RNA in either aging or in age-related diseases are a cause or a consequence of aging. Put simply, correlation does not imply causation and the long lists of RNA changes that occur do not, a priori, mean that those changes mean anything. Because changes in expression of multiple RNA molecules occur it is impractical to test whether each one makes a contribution to the aging phenotype. One might be able to test whole pathways, but this is difficult if the pathway itself is required for survival (eg mitochondrial electron transport chain identified above). Likewise, the hypothesis that RNA oxidation is critical to senescence or to age-related disease is very difficult to assess, as under circumstances where RNA is oxidized, other molecules will be oxidized as well. For example, the work showing that Vitamin E can block RNA oxidation in an ALS model does not disprove the counter hypotheses that DNA, protein, lipids or any other cellular component are responsible for ALS.
One way to address whether RNA changes play a causal role in aging or in age-related diseases in humans is to look for genetic conditions linked to RNA metabolism. Although it is always difficult to be certain that a rare mendelian disorder is etiologically linked to a common process such as aging, mutations are helpful in establishing sufficiency for a process in that disease. In the remainder of this article, we will discuss causal mutations for, first, accelerated aging and, second, age-related neurodegenerative diseases to try and understand whether RNA can be have a causal role in the aging process.
Accelerated aging
There are a number of rare diseases that have features reminiscent of an accelerated type of aging. These include Wiedemann-Rautenstrauch 34, Cockayne 35, Werner 36, and other rare syndromes. Several of these disorders have common deficiency in DNA repair enzymes, particularly in base excision repair, that lead to genomic instability 37. Therefore, if there are effects on RNA they are probably secondary to effects on DNA and it is very difficult to distinguish which nucleic acid is necessary or sufficient to cause progeria.
Another accelerated aging phenoytpe, Hutchinson-Gilford progeria syndrome (HGPS), can be caused by mutations in the Lamin A/C gene, LMNA 38. Because Lamins are a major component of the nuclear envelope, patients with truncating, loss of function mutations in LMNA are associated with deformation of the nuclear envelope 39. Here, again, although there are differences in the mRNA component of cells from HGPS compared to normal cells 40, 41, many of these relate to altered transcription factors or other nuclear factors rather than RNA metabolism per se. Therefore, as in several of the base excision repair enzymes, the primary effect in HGPS is probably related to DNA (specifically in the maintenance of heterochromatin) rather than on RNA itself.
Another condition linked to DNA response pathways is Seckel syndrome, which is associated in some cases with mutations in the ATR (ataxia-telangiectasia and Rad3-related protein) gene 42. In humans, the phenotype of this disorder is strongly developmental, with dwarfism, microcephaly and mental retardation 42. Interestingly, mouse models of ATR deficiency either with alleles found in human patients 43 or with a conditional knockout allele to avoid embryonic lethality seen in full knockouts 44 both show premature aging. Although ATR is a kinase associated with DNA damage repair processes, the protein is also implicated in RNA metabolism, specifically in splicing of TAF1, a subunit of RNA polymerase 45–47. Therefore, it seems reasonable to infer that ATR mutations affect RNA metabolism by a slightly indirect mechanism. However, it has not been proven that the RNA metabolism changes make a causal contribution to aging in Seckel syndrome.
The data presented above shows that while RNA changes in the aging organism, evidence that this is a primary causal change is uncertain. Furthermore, mutations that cause premature aging as part of several human syndromic conditions are generally associated with DNA stability and therefore, RNA involvement is potentially important but probably secondary to the primary event. There, are however, a number of mutations that are more directly relevant to RNA metabolism that are causal for age-related diseases, specifically the neurodegenerative conditions ALS and PD.
RNA binding proteins and age-related neurodegeneration
As discussed above, it is reasonable to infer causality of a specific gene in a disease where mutations are inherited in families. The simplest forms of this argument are for recessive mutations as in those cases we can be certain that the normal function of the protein is lost and therefore causes disease. For dominant mutations, the arguments are more complex as mutations can either enhance normal function, cause a 50% (or more) loss of normal function or generate a completely novel function for the mutant protein. Often, but certainly not always, mutations in different genes in the same pathway can cause similar phenotypes in humans and thus identify a general biological process that causes that disease.
One recent example of this logic has been the identification of mutations that cause ALS in rare families. Because familial ALS (fALS), like many neurodegenerative diseases, shows age-dependent penetrance, understanding the genes involved may provide some insight into age related processes. The two recently identified genes are TARDBP/TDP-43 (43 kDa TAR DNA-binding protein) and FUS (fused in sarcoma, also known as TLS or translocated in liposarcoma) and multiple mutations in each are found in dominant fALS reviewed in 48.
The TDP43 protein was originally cloned by screening for protein interactors of a region of DNA generated by integration of the human immunodeficiency virus (HIV1) RNA into the host genome 49. Subsequent work suggested that TDP43 is involved in the regulation of exon-skipping of several genes including the cystic fibrosis transmembrane receptor (CFTR) 50, 51. Some of the effects on splicing are reported to be driven by RNA, not DNA, interactions 50, 52. Supporting the possibility that the protein may directly bind to RNA, there are two RNA recognition motifs (RRM) in the TDP43 protein C-terminal to a nuclear localization sequence. The structure of the more C-terminal RRM in complex with RNA has been elucidated and shows an atypical fold associated with dimerization of the domain 53. Recently published experiments have identified the RNA binding partners of TDP43 54–56.
As well as the two RRMs, a glycine-rich domain is found at the C-terminus of TDP43. Glycine-rich domains are found in a variety of protein contexts, but often with RNA binding motifs 57 and/or in proteins that interact with other ribonucleoproteins 58. TDP43 binds to heterogeneous nuclear ribonucleoproteins (hnRNP) including hnRNP A2/B1 and hnRNP A1 that have splicing inhibitory activity 59. Additional interactors include proteins involved in translation as well as splicing, and some of these interactions are RNA-dependent 60. Overall, the primary structure of TDP43 supports an important role for the protein in RNA metabolism. Furthermore, TDP43 can be recruited to cytoplasmic, RNA-rich stress granules under conditions of oxidative stress 61. Therefore, TDP43 could play important roles in translation under oxidative conditions, which as we have established are also candidates for aging.
Mutations in TDP43 were first identified in one family with dominant transmission of fALS and two apparently sporadic ALS cases 62. Various additional surveys of gene mutations throughout the world have identified a number of additional heterozygous mutations in the same gene associated with fALS and occasionally with sporadic ALS 62–66. Most of the pathogenic mutations reported to date are concentrated at the C-terminus of the protein, specifically in the glycine-rich region 48.
Importantly, there is evidence that TDP-43 may play a role in sporadic ALS and a possibly related disorder, frontotemporal dementia (FTD). FTD affects the cerebral cortex and is associated with changes in personality and cognition and might therefore be thought to be related more to AD, where neocortical areas are heavily involved, than ALS where the motor system appears to be the primary target. However, several lines of evidence from genetic, pathological and clinical studies suggest that ALS and FTD are part of the same disease spectrum 67, 68.
Further supporting this idea, TDP43 is the major protein that is found in ubiquitinated inclusions in both ALS and FTD 69. Protein inclusions are, along with cell death, one of the primary defining pathologies found in many neurodegenerative diseases and are often generated by proteins that are inappropriately modified or folded. In many cases, the primary protein deposited can also cause genetic forms of the same disease when mutated. In some cases, it may be enough that the protein is more abundant than normal to drive the disease process 70. C-terminal truncation products for TDP43 have been shown to accumulate in pathological inclusions in ALS/FTD and may be associated with a toxic gain of function 71, 72.
A second gene for ALS/FTD that is also an RNA binding protein is FUS, which was originally cloned as a potential oncogene in liposarcoma associated with chromosomal rearrangements that fuse FUS to CHOP 73, 74. Like, TDP43, FUS mutations cause ALS in a dominant manner 75, 76. The FUS protein has QGSY-rich and glycine-rich regions, followed by a series of nucleic acid binding motifs (an RRM, two RGG rich domains and a zinc finger). Pathogenic mutations again cluster at the C-terminus of FUS, somewhat away from the identified domains.
FUS interacts both with the spliceosome 77–79 and with RNA polymerases 80. Fus is proposed to influence mRNA localization in neurons 81 to influence the density of dendritic spines 82. FUS also interacts with non-coding RNAs to alter transcription from in response to DNA damage signals 83. FUS can be regulated in a number of ways, including phosphorylation by ATM, a kinase related to the Seckel syndrome kinase ATR 84 and by neuronal activity 82. Although this is a wide range of potential interactors and therefore functions, FUS links several of the themes that we have discussed as part of the aging process including DNA damage and regulation of the transcriptional response.
This leads to two critical questions about both FUS and TDP43 in relation to RNA metabolism in the context of age-related diseases. First, is RNA metabolism actually important for the effects of mutations? Although both FUS and TDP43 are clearly involved in RNA metabolism, because mutations are dominant is does not inevitably follow that the normal function of the proteins is anything to do with disease. For example, it has been proposed that C-terminal fragments of TDP43 can cause cell toxicity by mechanisms that are unrelated to normal function, at least as measured by CFTR splicing assays 72. There are caveats about this work as the toxic effects of truncations need to be confirmed in vivo and the truncations are not natural pathogenic mutations. However, the results raise an interesting question of how, and indeed whether, to assign causality to RNA in aging or age-related disorders simply because the protein happens to bind RNA. One can address this by either genetic approaches, comparing overexpressed dominant mutations with knockouts and crosses of the two, or by making hybrid molecules that contain mutations but cannot bind RNA.
The second question is how aging and mutation interact to cause an age-related phenotype. Because ALS and FTD are not developmental phenotypes and the effects of mutations are not recognized until the fifth decade of life or later, it is reasonable to infer that aging plays an important role. However, the exact relationship is hard to understand. As RNA components change during the aging process as discussed above, the RNA species that FUS or TDP43 can interact with will also change. Thus, the function of the RNA binding proteins would be the same at different ages but dysfunction might be revealed as aging changes the cellular landscape. It is also possible that there is a more stochastic component of aging that makes TDP43 or FUS more important and oxidative and/or DNA damage accumulate. Overlaid over this complex set of interactions is a second parameter, which is the tissue specific nature of the phenotypes of TDP43/FUS mutations, expressed predominantly in CNS tissues. There are no simple answers to these questions, which are raised to demonstrate just how difficult it is to provide an integrative answer to understanding age-related disorders.
ALS is generally an adult-onset disease where symptoms manifest in the 5th and 6th decades of life. There are, however, juvenile onset motor neuron diseases that also involve RNA metabolism proteins. One well characterized example is spinal muscular atrophy (SMA), a relatively common disorder where degeneration of lower motor neurons leads to paralysis and death in affected infants. A known for SMA was cloned by Lefebvre et al and termed SMN1 85. SMN1 has a homologue, SMN2, that is nearby on Chromosome 5 that is normally expressed at low levels, and it has been suggested that increasing SMN2 protein levels might rescue the effects of SMN1 deficiency 86. Interestingly in the consideration of RNA metabolism and neurodegeneration, SMN1 acts together with partner proteins to regulate splicing of other genes 87. The mechanisms by which loss of SMN1 lead to motor neuron disease, and the distinctness from adult-onset ALS, are unclear. Another example of an RNA metabolism is found in fetal motor neuron disese 88. GLE1, mutated in fetal MND, encodes a protein component of the export complex for RNA from the nucleus to the cytoplasm, which occurs after splicing and editing before translation. Taken together, the observations that alteration in a splicing protein (SMN1) and a nuclear export protein (GLE1) suggest that normal RNA metabolism is important in at least some forms of very early onset (fetal or juvenile) neurodegenerative disease.
The examples of early onset motor neuron diseases show how a neurodegenerative disease can result from loss of function of the protein product of genes. There are some examples where the RNA itself can be toxic to neurons in an age-related fashion. One form of myotonic dystrophy is caused by an expansion in a (CTG) repeat sequence in the 3-prime untranslated region of the DMPK gene 89. Current models suggest that the expanded RNA binds to splice factors, sequestering them from their normal function of metabolism of other RNA species, thus leading to a toxic gain of function effect 90. A similar mechanism may occur in fragile X tremor ataxia syndrome, a neurological syndrome seen in persons carrying expanded repeats in the untranslated region of the frataxin gene 91.
One final example of an age-related neurodegenerative disease that may be relevant to RNA is DJ-1. Mutations in DJ-1 are associated with autosomal recessive parkinsonism 92 and therefore the normal function of the DJ-1 protein is critical for understanding this disease. Several lines of evidence show that DJ-1 responds to oxidative stress by the formation of a specific cysteine modification 93–98 and this modification is critical for normal function in a number of systems 94, 99, 100. DJ-1 was originally cloned as a regulator of RNA-protein interactors 101 and we have suggested that it may bind specific subsets of RNA species rich in GG/CC motifs including components of the mitochondrial electron transport chain 102, although this work has not yet been replicated. DJ-1 may therefore link RNA metabolism to oxidative stress, which is again a potentially important contributor to the aging process. However, whether RNA binding is really critical to DJ-1 function remains to be addressed.
These examples show that RNA binding proteins can be critical in at least some contexts that are relevant to human aging, namely age-related neurodegenerative diseases. Exactly how such mutations interact with aging to produce tissue and cell-specific changes remains to be clarified.
Conclusion
As in many aspects of general biology, RNA is critically involved in the aging process and in age-related diseases. Changes in the transcribed RNA profile of cells and tissues are seen with aging and, like other macromolecules, RNA Is subject to cumulative oxidative stress. General themes include alterations in oxidative stress handling and, specifically, the mitochondrial electron transport chain. There is also evidence of altered utilization of RNA via protein translation mediated at least in part by the mTOR pathway and making a causal contribution to age-related phenotypes. Mutations associated with accelerated aging in humans are associated more strongly with DNA repair pathways and thus affect RNA probably through indirect mechanisms, although ATR, which causes Seckel syndrome in humans and accelerated aging in mice, may provide a link between DNA damage and RNA splicing. Finally, age-related neurodegenerative conditions provide a more direct link between RNA and aging, as several mutations are directly in RNA binding proteins.
The key future direction for understanding the role of RNA in the aging process is to disentangle primary causes from secondary and reactive events. When is RNA really driving the aging process and when is a downstream mediator of other initiating events such as DNA damage? Given that biological systems are dynamic and usually have layers of redundancy to support homeostasis, these are difficult questions to answer. For neurodegenerative diseases, as an example of age-related events thought of as distinct from ‘normal’ or healthy aging, it is important to determine whether RNA binding is actually the critical function of mutant proteins, as well as to determine why these are expressed in a tissue-specific manner.
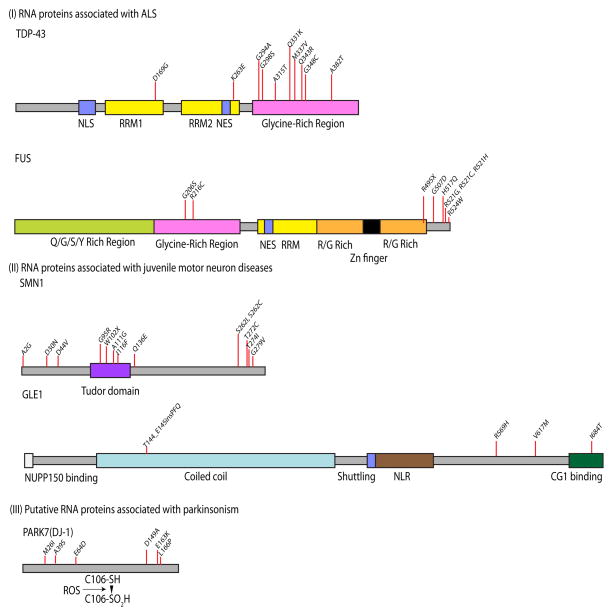
Figure 1.
RNA metabolism genes associated with age-related neurodegenerative conditions. Inherited neurodegenerative diseases can be associated with mutations in genes encoding several RNA binding proteins, including TDP43 and FUS (associated with adult onset amyotrophic lateral sclerosis), SMN1 and GLE1 (associated with juvenile onset motor neuron diseases and DJ-1 (also known as PARK7, associated with recessive parkinsonism). Domains and individual regions of the proteins are indicated below the ideograms for each protein, which are presented at the same scale. Selected examples of allelic variants found in each disease are shown above each protein; in some cases there are also known duplication or insertion alleles. NLS, nuclear localization sequence; RRM, RNA recognition motif; NES, nuclear export sequence; NLR, nuclear localization region.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Cross-References
Other age-related diseases in this collection include:
RNA-binding proteins & disease
RNA and cancer – splicing, translation etc, musashi cardiovascular disease
References
- 1.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 2.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh S, Tseng GC, Sibille E. Reciprocal phylogenetic conservation of molecular aging in mouse and human brain. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, Landfield PW. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates DJ, Liang R, Li N, Wang E. The impact of noncoding RNA on the biochemical and molecular mechanisms of aging. Biochim Biophys Acta. 2009;1790:970–979. doi: 10.1016/j.bbagen.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Lanceta J, Prough RA, Liang R, Wang E. MicroRNA group disorganization in aging. Exp Gerontol. 2009 doi: 10.1016/j.exger.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Ibanez-Ventoso C, Driscoll M. MicroRNAs in C. elegans Aging: Molecular Insurance for Robustness? Curr Genomics. 2009;10:144–153. doi: 10.2174/138920209788185243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer T, Marzetti E, Xu J, Seo AY, Gulec S, Knutson MD, Leeuwenburgh C, Dupont-Versteegden EE. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp Gerontol. 2008;43:563–570. doi: 10.1016/j.exger.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM, Cotman CW, Ames BN. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc Natl Acad Sci U S A. 2002;99:2356–2361. doi: 10.1073/pnas.261709299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhoads RE. eIF4E: new family members, new binding partners, new roles. J Biol Chem. 2009;284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy BK, Kaeberlein M. Hot topics in aging research: protein translation, 2009. Aging Cell. 2009;8:617–623. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagida M. Cellular quiescence: are controlling genes conserved? Trends Cell Biol. 2009;19:705–715. doi: 10.1016/j.tcb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Bronner IF, Bochdanovits Z, Rizzu P, Kamphorst W, Ravid R, van Swieten JC, Heutink P. Comprehensive mRNA expression profiling distinguishes tauopathies and identifies shared molecular pathways. PLoS One. 2009;4:e6826. doi: 10.1371/journal.pone.0006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan MG, Chua WT, Esiri MM, Smith AD, Vinters HV, Lai MK. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2009 doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- 18.Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller RM, Federoff HJ. Altered gene expression profiles reveal similarities and differences between Parkinson disease and model systems. Neuroscientist. 2005;11:539–549. doi: 10.1177/1073858405278330. [DOI] [PubMed] [Google Scholar]
- 20.Miller RM, Federoff HJ. Microarrays in Parkinson’s disease: a systematic approach. NeuroRx. 2006;3:319–326. doi: 10.1016/j.nurx.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 22.Offen D, Barhum Y, Melamed E, Embacher N, Schindler C, Ransmayr G. Spinal cord mRNA profile in patients with ALS: comparison with transgenic mice expressing the human SOD-1 mutant. J Mol Neurosci. 2009;38:85–93. doi: 10.1007/s12031-007-9004-z. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka F, Niwa J, Ishigaki S, Katsuno M, Waza M, Yamamoto M, Doyu M, Sobue G. Gene expression profiling toward understanding of ALS pathogenesis. Ann N Y Acad Sci. 2006;1086:1–10. doi: 10.1196/annals.1377.011. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland GT, Matigian NA, Chalk AM, Anderson MJ, Silburn PA, Mackay-Sim A, Wells CA, Mellick GD. A cross-study transcriptional analysis of Parkinson’s disease. PLoS One. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda K, Smith MA, Zhu X, Baus D, Merrick WC, Tartakoff AM, Hattier T, Harris PL, Siedlak SL, Fujioka H, et al. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J Biol Chem. 2005;280:20978–20986. doi: 10.1074/jbc.M500526200. [DOI] [PubMed] [Google Scholar]
- 27.Ding Q, Markesbery WR, Cecarini V, Keller JN. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 28.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CL. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong Q, Shan X, Chang Y, Tashiro H, Lin CL. RNA oxidation: a contributing factor or an epiphenomenon in the process of neurodegeneration. Free Radic Res. 2008;42:773–777. doi: 10.1080/10715760802311187. [DOI] [PubMed] [Google Scholar]
- 32.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 34.Arboleda G, Ramirez N, Arboleda H. The neonatal progeroid syndrome (Wiedemann-Rautenstrauch): a model for the study of human aging? Exp Gerontol. 2007;42:939–943. doi: 10.1016/j.exger.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Weidenheim KM, Dickson DW, Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev. 2009;130:619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Muftuoglu M, Oshima J, von Kobbe C, Cheng WH, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 38.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Csoka AB, English SB, Simkevich CP, Ginzinger DG, Butte AJ, Schatten GP, Rothman FG, Sedivy JM. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Robinson JF, O’Neil CH, Edwards JY, Williams CM, Huff MW, Pickering JG, Hegele RA. Ankyrin G overexpression in Hutchinson-Gilford progeria syndrome fibroblasts identified through biological filtering of expression profiles. J Hum Genet. 2006;51:934–942. doi: 10.1007/s10038-006-0042-0. [DOI] [PubMed] [Google Scholar]
- 42.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 43.Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandris P, Giannouli CC, Panayotou G, Kletsas D. Compromise in mRNA processing machinery in senescent human fibroblasts: implications for a novel potential role of Phospho-ATR (ser428) Biogerontology. 2010;11:421–436. doi: 10.1007/s10522-010-9261-z. [DOI] [PubMed] [Google Scholar]
- 46.Katzenberger RJ, Marengo MS, Wassarman DA. ATM and ATR pathways signal alternative splicing of Drosophila TAF1 pre-mRNA in response to DNA damage. Mol Cell Biol. 2006;26:9256–9267. doi: 10.1128/MCB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzenberger RJ, Marengo MS, Wassarman DA. Control of alternative splicing by signal-dependent degradation of splicing-regulatory proteins. J Biol Chem. 2009;284:10737–10746. doi: 10.1074/jbc.M809506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 51.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 52.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao S, Sanelli T, Dib S, Sheps D, Findlater J, Bilbao J, Keith J, Zinman L, Rogaeva E, Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011;47:167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Mousavi A, Hotta Y. Glycine-rich proteins: a class of novel proteins. Appl Biochem Biotechnol. 2005;120:169–174. doi: 10.1385/abab:120:3:169. [DOI] [PubMed] [Google Scholar]
- 58.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 59.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 60.Freibaum BD, Chitta RK, High AA, Taylor JP. Global Analysis of TDP-43 Interacting Proteins Reveals Strong Association with RNA Splicing and Translation Machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 62.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 65.Kuhnlein P, Sperfeld AD, Vanmassenhove B, Van Deerlin V, Lee VM, Trojanowski JQ, Kretzschmar HA, Ludolph AC, Neumann M. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Arch Neurol. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackenzie IR. The neuropathology of FTD associated With ALS. Alzheimer Dis Assoc Disord. 2007;21:S44–49. doi: 10.1097/WAD.0b013e31815c3486. [DOI] [PubMed] [Google Scholar]
- 68.Talbot K, Ansorge O. Recent advances in the genetics of amyotrophic lateral sclerosis and frontotemporal dementia: common pathways in neurodegenerative disease. Hum Mol Genet. 2006;15(Spec No 2):R182–187. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- 69.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 70.Singleton A, Myers A, Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum Mol Genet. 2004;13(Spec No 1):R123–126. doi: 10.1093/hmg/ddh093. [DOI] [PubMed] [Google Scholar]
- 71.Arai T, Hasegawa M, Nonoka T, Kametani F, Yamashita M, Hosokawa M, Niizato K, Tsuchiya K, Kobayashi Z, Ikeda K, et al. Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology. 2010;30:170–181. doi: 10.1111/j.1440-1789.2009.01089.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes-Casey M, Ash P, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 74.Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 75.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 76.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 79.Meissner M, Lopato S, Gotzmann J, Sauermann G, Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp Cell Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 80.Tan AY, Manley JL. TLS inhibits RNA polymerase III transcription. Mol Cell Biol. 2010;30:186–196. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 82.Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 83.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem J. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- 85.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 86.Lorson CL, Rindt H, Shababi M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum Mol Genet. 2010;19:R111–118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harley HG, Brook JD, Rundle SA, Crow S, Reardon W, Buckler AJ, Harper PS, Housman DE, Shaw DJ. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- 90.Mulders SA, van Engelen BG, Wieringa B, Wansink DG. Molecular therapy in myotonic dystrophy: focus on RNA gain-of-function. Hum Mol Genet. 2010;19:R90–97. doi: 10.1093/hmg/ddq161. [DOI] [PubMed] [Google Scholar]
- 91.Garcia-Arocena D, Hagerman PJ. Advances in understanding the molecular basis of FXTAS. Hum Mol Genet. 2010;19:R83–89. doi: 10.1093/hmg/ddq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 93.Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ito G, Ariga H, Nakagawa Y, Iwatsubo T. Roles of distinct cysteine residues in S-nitrosylation and dimerization of DJ-1. Biochem Biophys Res Commun. 2006;339:667–672. doi: 10.1016/j.bbrc.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 96.Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 97.Witt AC, Lakshminarasimhan M, Remington BC, Hasim S, Pozharski E, Wilson MA. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47:7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc Natl Acad Sci U S A. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aleyasin H, Rousseaux MW, Phillips M, Kim RH, Bland RJ, Callaghan S, Slack RS, During MJ, Mak TW, Park DS. The Parkinson’s disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc Natl Acad Sci U S A. 2007;104:18748–18753. doi: 10.1073/pnas.0709379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hod Y, Pentyala SN, Whyard TC, El-Maghrabi MR. Identification and characterization of a novel protein that regulates RNA-protein interaction. J Cell Biochem. 1999;72:435–444. [PubMed] [Google Scholar]
- 102.van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, Martindale J, Xie C, Ahmad R, Thomas KJ, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- Most of the human conditions discussed here can be found in Online Mendelian Inheritance in Man (OMIM)
- Wiedemann-Rautenstrauch syndrome. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=264090.
- Cockayne syndrome. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=216400 and http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=133540.
- Werner syndrome. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=277700.
- Hutchinson-Gilford progeria syndrome. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=176670.
- Seckel Syndrome. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=210600.
- ALS6, associated with FUS mutations. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=608030.
- ALS10, associated with mutations in TDP43. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=612069.
- Spinal Muscular Atrophy, associated with the SMN1 gene. http://www.ncbi.nlm.nih.gov/omim/253300.
- GLE1 mutations associated with a number of fetal motor neuron diseases. http://www.ncbi.nlm.nih.gov/omim/603371.
- Myotonic dystrophy. http://www.ncbi.nlm.nih.gov/omim/160900.
- Fragile X Tremor/Ataxia Syndrome. http://www.ncbi.nlm.nih.gov/omim/300623.
- PARK7, autosomal recessive parkinsonism caused by mutations in DJ-1. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=606324.