Abstract
The intrahepatic biliary ducts transport bile produced by the hepatocytes out of the liver. Defects in biliary cell differentiation and biliary duct remodeling cause a variety of congenital diseases including Alagille Syndrome and polycystic liver disease. While the molecular pathways regulating biliary cell differentiation have received increasing attention (Lemaigre, 2010), less is known about the cellular behavior underlying biliary duct remodeling. Here, we have identified a novel gene, claudin 15-like b (cldn15lb), which exhibits a unique and dynamic expression pattern in the hepatocytes and biliary epithelial cells in zebrafish. Claudins are tight junction proteins that have been implicated in maintaining epithelial polarity, regulating paracellular transport, and providing barrier function. In zebrafish cldn15lb mutant livers, tight junctions are observed between hepatocytes, but these cells show polarization defects as well as canalicular malformations. Furthermore, cldn15lb mutants show abnormalities in biliary duct morphogenesis whereby biliary epithelial cells remain clustered together and form a disorganized network. Our data suggest that Cldn15lb plays an important role in the remodeling process during biliary duct morphogenesis. Thus, cldn15lb mutants provide a novel in vivo model to study the role of tight junction proteins in the remodeling of the biliary network and hereditary cholestasis.
Keywords: Claudin, liver development, zebrafish, biliary duct remodeling, biliary cells, biliary duct morphogenesis, tight junctions, cholestasis
Introduction
The liver is a vital organ with many essential functions, one of which is bile production for lipid metabolism. Hepatocytes are the primary cells in the liver and they produce bile that is secreted into the intestinal tract where it emulsifies fats. The intrahepatic biliary system consists of a network of tubes that permeates the liver. These tubes carry bile away from the hepatocytes and collectively drain into the intestinal tract. The vasculature is the other tubular network within the liver. These two networks must be organized into a functional three-dimensional structure to ensure proper liver function (Si-Tayeb et al., 2010). Disrupted liver architecture is often found in diseased states such as hepatitis and hepatocellular carcinomas (Lee and Luk, 2010). Rows of hepatocytes form a polarized epithelium where their basal surfaces are lined by the sinusoidal endothelial cells and their apical surfaces are lined by the biliary epithelial cells (BECs) (Sakaguchi et al., 2008). On the apical surface of the hepatocytes are the canaliculi which are the terminal ductules of the biliary network through which bile acids are secreted (Gatmaitan and Arias, 1995). Tight junctions in hepatocytes circumscribe these canaliculi and serve two functions – they convey barrier properties to the hepatic epithelium to create a blood-biliary barrier, and they serve as a physical scaffold to separate the apical and basolateral domains to maintain hepatic polarity (Montesano et al., 1975; Tsukita et al., 2001). Tight junctions are also present between neighboring BECs (Vroman and LaRusso, 1996) to connect the cells together and form the biliary network. Furthermore, tight junctions are present between hepatocytes and BECs along the canals of Hering. These canals serve as conduits from the canaliculi to the main biliary ducts (Saxena and Theise, 2004).
Claudins are one of the core components of tight junctions. They are tetraspanning membrane proteins that regulate the paracellular passage of small molecules and ions. There are 24 Claudin genes in the mammalian genome, 56 in the pufferfish Takifugu, at least 29 in the zebrafish, and they exhibit unique expression patterns and play important roles in development and physiology (Loh et al., 2004; Van Itallie and Anderson, 2006). Previously, we have found Claudin-15 to be necessary for the formation of a single lumen in the zebrafish gut by regulating the electrochemical and osmotic gradients across the gut epithelium (Bagnat et al., 2007). Claudins are also expressed in the liver. In vitro studies using the rat hepatoma/human fibroblast hybrid cell line WIF-B9 showed that Claudin-2 is required for the formation of canaliculi and maintenance of hepatic polarity (Son et al., 2009). In addition, expression profiles of Claudins in intrahepatic BECs have been generated, and changes in these profiles have been proposed to be useful markers of biliary tract cancer diagnosis and prognosis (Nemeth et al., 2009). Mutations in genes encoding Claudin-1 and tight junction protein 2 (TJP2) (also known as ZO-2) were identified in two forms of intrahepatic cholestatic disease, and leaky tight junctions were postulated to underlie disease manifestation (Carlton et al., 2004). In patients with Claudin-1 mutations, it has been proposed that loss of bile flow from leaky junctions is associated with sclerosing cholangitis (Baala et al., 2002; Hadj-Rabia et al., 2004), a condition where bile ducts are inflamed and scarred. Patients carrying TJP2 mutations exhibit elevated serum bile acid concentration, possibly due to an impaired blood-biliary barrier (Carlton et al., 2003). These data indicate that tight junction proteins, including Claudins, play important roles in the onset and progression of intrahepatic cholestatic diseases.
In addition to their role in controlling paracellular permeability and forming a blood-biliary barrier, Claudin-containing tight junctions also mediate the establishment and maintenance of apicobasal polarity (Shin et al., 2006). Polarization of hepatocytes occurs during liver morphogenesis and is regulated by the transcription factor HNF4α (Parviz et al., 2003). In Hnf4α mutant mice, the hepatocytes do not form normal cell-cell contacts, and expression of various junction proteins, including tight junction proteins, is downregulated (Parviz et al., 2003; Battle et al., 2006). The resulting liver contains small lesions and exhibits a lack of cohesive architecture (Parviz et al., 2003). Detailed analysis of hepatocyte polarization in zebrafish also revealed that it coincides with the development of the vascular and biliary networks, suggesting that polarization of hepatocytes is linked to the formation of the two tubular networks (Sakaguchi et al., 2008). Furthermore, disruption of planar cell polarity proteins in zebrafish via antisense technology caused defects in intrahepatic biliary development (Cui et al., 2011). Clearly, polarization of hepatocytes is a key step in liver morphogenesis, including the establishment of the biliary ducts.
Biliary duct morphogenesis in both mammals and teleosts has been shown to be regulated by Notch and TGFβ signaling (Lorent et al., 2004; Antoniou et al., 2009; Zong et al., 2009; Lorent et al., 2010). These signaling pathways regulate the expression of transcription factor genes such as those of the onecut family and hnf1b (also known as tcf2 or vhnf1) to direct biliary differentiation (Clotman et al., 2002; Coffinier et al., 2002; Matthews et al., 2004; Clotman et al., 2005; Matthews et al., 2008). However, after BECs have been specified, little is known about the cellular behaviors that give rise to the three-dimensional architecture of the biliary network. Live imaging of developing livers cultured from Tg(Tp1bglob:eGFP)um14 zebrafish (Parsons et al., 2009), where differentiated BECs are labeled by a Notch-responsive fluorescent transgenic reporter, revealed that the founder population of BECs undergoes extensive and dynamic remodeling to generate a functional ductal system (Lorent et al., 2010). These cells continuously extend and retract filopodia to sense and connect with their neighbors, and cell bodies of BECs are constantly rearranging to remodel the network (Lorent et al., 2010). Molecules that mediate this dynamic remodeling process are unknown.
In this study, we identified a novel gene encoding a member of the Claudin family of tight junction proteins, claudin 15-like b (cldn15lb), which exhibits a unique expression pattern in the zebrafish liver. It is initially expressed in both hepatocytes and BECs and its expression is subsequently restricted to the BECs. A mutation in this gene disrupts the polarization of hepatocytes and the formation of bile canaliculi. Furthermore, the biliary network in cldn15lb mutant larvae is disorganized due to defects in the remodeling process. The livers of cldn15lb mutant adults also present with abnormal lesions. Our study reveals that tight junction proteins play an important role in remodeling the biliary network during morphogenesis, and the cldn15lb mutant provides a new in vivo model to further investigate mechanisms underlying cholestatic diseases caused by tight junction protein mutations.
Materials and Methods
Animals
AB wild-type, clochem39 (Stainier et al., 1995), and the mutant allele cldn15lbfh290 were maintained as described previously (Westerfield, 2000). The cldn15lbfh290 mutant was crossed with Tg(Tp1bglob:eGFP)um14 (Parsons et al., 2009) and genotyped according to the TILLING center protocol with XbaI (http://labs.fhcrc.org/moens/Tilling_Mutants/cldn15lb/allele_1.html). Tg(fabp10:rasGFP)s942 was constructed using the 2.8kb promoter from fabp10 (Her et al., 2003) cloned into a modified rasGFP-pBS vector. The construct was injected with I-SceI meganuclease (Thermes et al., 2002) to generate the transgenic line.
In situ hybridization
Whole-mount in situ hybridizations were performed as described previously (Alexander et al., 1998). The cldn15lb_tv1 (GenBank ID: NM_001002446) and cldn15lb_tv2 (Supp. Table 1) in situ probes correspond to the first exon of each transcript. The in situ constructs were cloned into pGEMT-easy (Promega) with BamHI/XhoI. Antisense DIG-labeled RNA probes were made by linearizing the construct with XhoI and transcribing with T7 RNA polymerase. For in situ hybridization with immunohistochemistry, in situ hybridization was performed first. The larvae were then sectioned with a vibratome and processed as described below (under immunofluorescence).
Expression vectors and cell transfection
Both cldn15lb transcripts were amplified by RT-PCR from RNA of wildtype larvae at 3 dpf and cloned into the mammalian expression vector pCDNA3 (Invitrogen) with BamHI/NotI. The eGFP construct was cloned into pCDNA3 by NotI/XhoI. HEK293 cells were cultured in DMEM/10% FBS and Cos7 cells were cultured in H21 High Glucose media/10% FBS/Pen-Strep to 95% confluency for transfection. Transient transfections were performed with Lipofectamine2000 (Promega) and Opti-MEM medium (Life Technologies). Transfected cells were fixed with 10% trichloroacetic acid, washed with PBS, and stained as previously described (Horne-Badovinac et al., 2001).
Expression and imaging of Cldn15lb in MDCK cysts
pCDNA3 cldn15lb-eGFP plasmid was transfected into MDCK cells with Lipofectamine 2000 (Promega) and stable GFP expressing cells were selected by fluorescence activated cell sorting and maintained under Geneticin (G418) selection (50ug/ml) (Invitrogen). Cysts were cultured as described in (Bryant et al., 2010). Cysts were allowed to grow for 5 days after plating, at which time they were fixed with 4% paraformaldehyde and probed with primary antibodies against Occludin (1:200, Invitrogen) and ZO-1 (1:200, R40.76; a gift from Bruce Stevenson) followed by Alexa-conjugated secondary anti-rabbit and anti-rat antibodies respectively (1:400, Invitrogen) and Hoechst 33342 (Invitrogen) for nuclear staining. Cysts were imaged using a Zeiss 510 laser scanning confocal microscope.
Generation of peptide antibody
The program AnthePro 6.0 was used to analyze the sequence of Cldn15lb and to identify the peptide sequence. General Biosciences Corporation synthesized the peptide, conjugated it to KLH, and used it to immunize two rabbits (GB9 and GB10). We received antisera from the second and third bleeds, and used Nab™ Protein A Plus Spin Kit (Pierce) for purification. The first eluted fraction was used for immunofluorescence studies.
Immunofluorescence
For staining with the Cldn15lb antiserum, larvae were first fixed in Histochoice™ MB® (Electron Microscopy Sciences) at 4°C overnight, and then embedded in 4% low-melt agarose in PBS. 150μm transverse sections were obtained with a vibratome (Leica, Bannockburn, IL, USA). For whole-mount staining, larvae were fixed in 4% formaldehyde in egg water at 4°C overnight. Larvae were fixed in 4:1 (v/v) methanol:DMSO at room temperature for 2 hours for the cytokeratin staining. The fixed larvae were then washed in PBS, and the skin and yolk were removed to expose the endoderm. Vibratome sections or whole embryos were incubated with primary antibodies overnight at 4°C. The following antibodies were used: rabbit anti-ABCB11/BSEP (Kamiya Biomedical, 1:1000), rabbit anti-PKC (C-20) (Santa Cruz Biotechnology, 1:1000), mouse anti-ZO1 (Invitrogen #33-9100, 1:400), rabbit anti-pan-Cadherin (Sigma, 1:5000), mouse anti-cytokeratin 18 (Maine Biotechnology Services, 1:10), rabbit anti-ZO-1 (Invitrogen #61-7300, 1:10), and chick anti-GFP (Aves Labs, 1:500). Samples were then washed and incubated with Alexa Fluor conjugated secondary antibodies (Molecular Probes, 1:500), Alexa Fluor conjugated phalloidin (Molecular Probes, 1:500) or TO-PRO3 (Invitrogen) for 4 hours at room temperature. After washing, samples were mounted on slides in Vectashield and imaged on a Zeiss LSM510 Pascal confocal microscope. For whole-mount larvae, all images were captured from the area where the extrahepatic duct enters the liver and begins to branch in order to minimize sampling error. Images were further analyzed with LSM software (Zeiss, Thornwood, NY, USA) and Image J Software.
Transmission electron microscopy
The larvae were fixed with 0.8% paraformaldehyde/ 2.5% glutaraldehyde in 0.1M Cacaodylate buffer at 4°C overnight. The samples were processed for TEM by the Pathology & Imaging Core of the Liver Center at the University of California, San Francisco.
Histology
One year old zebrafish were euthanized and their digestive systems dissected and fixed overnight with formalin at 4°C. The samples were embedded in paraffin, cut into 5 micron sections, and stained with hematoxylin and eosin.
PED-6 assay
5 dpf larvae were fed PED-6 in a single plate, and then collected at 30 minutes, 45 minutes, 1 hour, 2 hours, or 6 hours and fixed immediately. Biliary secretion of PED-6 was assessed based on gallbladder fluorescence intensity and categorized as strong, faint/small, or no fluorescence relative to other larvae in that sampling group. Larvae were subsequently genotyped.
Results
Identification of a novel claudin gene
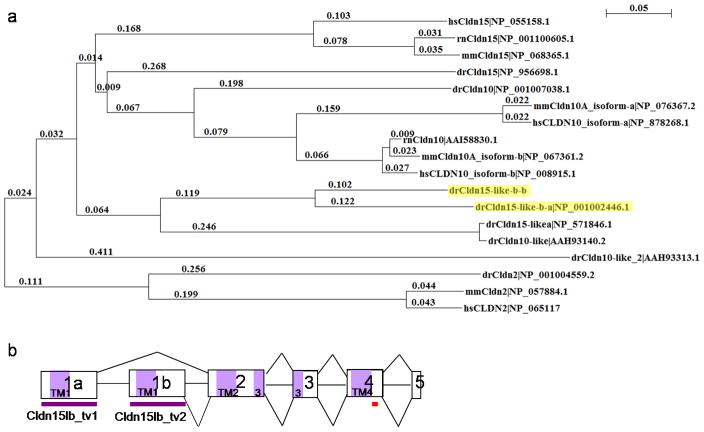
The liver in zebrafish hnf1b mutants is severely underdeveloped (Sun and Hopkins, 2001). To identify genes regulated by Hnf1b, we performed a microarray analysis of hnf1b mutants (Bagnat et al., 2007) and identified a transcript (GenBank ID: NM_001002446) that is 24-fold downregulated in mutants compared to wildtype. It encodes a protein belonging to the large PMP22/EMP/MP20/Claudin superfamily. This protein contains four transmembrane domains, two extracellular loops, and a PDZ-binding domain characteristic of the topology of the Claudin family of proteins. The first extracellular loop also contains the signature WGLWCC motif present in all Claudins. These characteristics suggest that the novel transcript encodes a Claudin. Alignment with other Claudins identified this novel protein to be phylogenetically related to mammalian Claudin-15 (Fig. 1a), which also shares significant homology to mammalian Claudin-10 (Loh et al., 2004). The transcript was subsequently named claudin 15-like b (cldn15lb), in agreement with the Zebrafish Nomenclature Committee.
Figure 1. Identification of a novel, alternatively spliced, claudin gene.
a, Phylogenetic tree of mammalian and zebrafish Claudin-10, Claudin-15, and their related proteins show that the novel protein, highlighted in yellow, is most similar to mammalian Claudin-15. Claudin-2, also expressed in the liver, is distantly related to Cldn15lb. This tree was constructed using ClustalX with neighbor joining and 1000 bootstrap replicates. Zebrafish (dr), Human (hs), Mouse (mm), rat (rn). b, Schematic of the claudin 15-like b gene. cldn15lb is a 5 exon gene and the first exons, 1a and 1b, are alternatively spliced to generate 2 different transcripts. The first exon encodes the first transmembrane domain and first extracellular loop. Purple boxes represent the coding regions for each transmembrane domain in the protein. Purple bars underline the region recognized by the transcript variant-specific in situ hybridization probes. Red bar underlines the region targeted by the antibody.
Genomic structure analysis identified this transcript to include five exons. Further investigation of the EST database revealed another five-exon transcript from the same gene that resulted from the alternative splicing of the first exon (Fig. 1b, Supp. Table 1). The two transcripts, cldn15lb_transcript variant1 (cldn15lb_tv1) and cldn15lb_transcript variant2 (cldn15lb_tv2), share the last four exons and only differ in the first exon. The first exon encodes the first transmembrane domain and the first extracellular loop (Fig. 1b).
cldn15lb transcripts are differentially expressed
In situ hybridization analysis was performed to study the expression pattern of the two transcripts. The use of transcript specific probes targeting the first exon of each transcript (Fig. 1b, purple bars) revealed that the two transcripts are expressed differentially. cldn15lb_tv1 is not maternally deposited (Fig. 2a). We detected its expression in the liver from 42 hours post fertilization (hpf) onwards (Fig. 2a). In addition, minimal expression was observed in the gut and pancreas starting at 60 and 72 hpf, respectively (Fig. 2a). Closer examination of the liver expression at 72 hpf revealed that cldn15lb_tv1 is expressed in a tree-like pattern (Fig. 2a′). In contrast, the second transcript, cldn15lb_tv2, is maternally deposited and expressed mainly in the head and gut starting at 42 hpf (Fig. 2b). We also detected cldn15lb_tv2 expression in the liver at 72 hpf but it did not appear as the branched pattern exhibited by the first transcript variant (Fig. 2b). The tree-like expression pattern of cldn15lb_tv1 is reminiscent of the intrahepatic biliary and vascular networks. To determine whether cldn15lb_tv1 is expressed in the vascular network, we performed in situ hybridization on Tg(kdrl:eGFP)s843 (Beis et al., 2005) larvae that express eGFP in endothelial cells. A cldn15lb probe that identifies both transcripts did not appear to co-localize with Tg(kdrl:eGFP) expression (Fig. 2c). Furthermore, cldn15lb expression was present in clochem39 mutants which lack most endothelial cells (Fig. 2c) (Stainier et al., 1995), suggesting that cldn15lb is not expressed in the intrahepatic vasculature. Instead, it is likely that cldn15lb is expressed in the intrahepatic biliary network. These data indicate that the two transcript variants of the cldn15lb gene are differentially expressed during zebrafish development.
Figure 2. The two isoforms of Cldn15lb are differentially expressed during development.
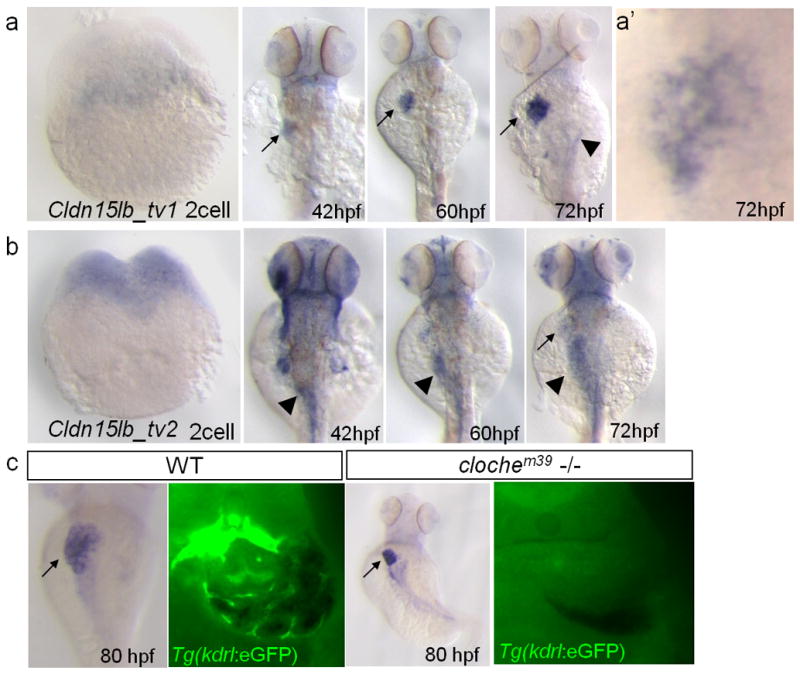
a–b, In situ hybridization with transcript variant-specific probes. a, cldn15lb_tv1 is not expressed at the 2-cell stage. At 42 hpf, it is expressed mainly in the liver (arrow) with some expression in the head. Its expression in the liver intensifies through 60 and 72 hpf. Low level expression is observed in the pancreas at 72 hpf (arrowhead). a′, Detailed analysis at 72 hpf shows that expression in the liver is in a unique, tree-like pattern. b, cldn15lb_tv2 is detected at 2-cell stage indicating that it is maternally deposited. cldn15lb_tv2 transcripts can be found in the gut (arrowhead), head, and pectoral fins at 42 hpf. Minimal expression of cldn15lb_tv2 is found in the liver at 72 hpf (arrow). c, Left, in situ hybridization of cldn15lb in wildtype and clochem39mutant larvae. Probe used was against the 3′UTR of cldn15lb and thus labels both transcripts. Right, vibratome cross section of in situ hybridization with fluorescence microscopy. In situ hybridization signal does not co-localize with GFP-positive endothelial cells and is present in clochem39 mutants, which lack most endothelial cells, indicating that cldn15lb is not expressed in the intrahepatic vasculature.
Cldn15lb is expressed in the hepatocytes and biliary epithelial cells
To characterize the cellular localization of Cldn15lb, we generated a peptide antibody against a sequence in its C-terminal region (Fig. 1b, red bar; Supp. Fig. 1a). To validate the antibody, we transfected HEK293 cells, which do not express endogenous Claudins, with a construct encoding zebrafish Cldn10-like, Cldn15, or Cldn15lb-a. [Cldn15lb-a and -b are the proteins encoded by cldn15lb_tv1 and _tv2, respectively.] The antibody did not label cells transfected with Cldn10-like or Cldn15 but labeled those transfected with Cldn15lb-a (Supp. Fig. 1b). To determine whether the antibody recognized both Cldn15lb isoforms, we generated GFP-tagged versions of Cldn15lb-a and Cldn15lb-b and transfected them into Cos7 cells. Cells transfected with either GFP-tagged construct were labeled by the antibody, while non-transfected cells were not labeled (Supp. Fig. 1c). These results indicate that the antibody recognizes both isoforms of Cldn15lb, as expected from the location of the antigen used to generate it.
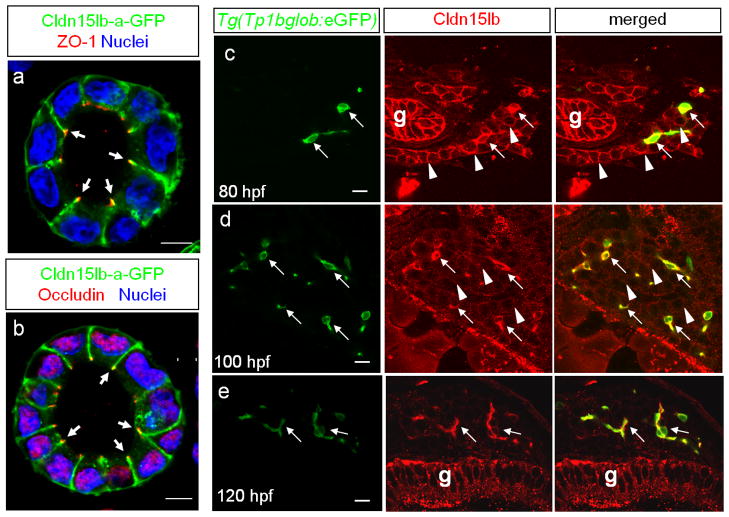
To determine the sub-cellular localization of Cldn15lb, we generated stable MDCK lines expressing Cldn15lb-a-GFP and found that it co-localizes with the tight junction markers ZO-1 and Occludin (Fig. 3a, b), further indicating that this novel protein is a tight junction protein. It must be noted that Cldn15lb-a-GFP is also expressed along the lateral membranes and at lower levels in the basal membrane (Fig. 3a, b). Expression of a tight junction protein along lateral membranes has been reported previously (Bryant et al., 2010). Next, we performed immunofluorescence analysis to determine the cellular localization of Cldn15lb in zebrafish larvae. At 80 hpf, Cldn15lb immunostaining co-localizes with cells expressing the Notch-responsive fluorescent reporter Tg(Tp1bglob:eGFP)um14 (Parsons et al., 2009) suggesting that Cldn15lb is expressed in BECs (Fig. 3c, arrow) (Lorent et al., 2010). The antibody also labels the surrounding hepatocytes at slightly lower levels (Fig. 3c, arrowhead in middle panel). Cldn15lb immunostaining becomes markedly enhanced in the BECs by 100 hpf while the immunostaining in the hepatocytes becomes punctated (Fig. 3d). By 120 hpf, Cldn15lb immunostaining appears only in the BECs (Fig. 3e). Since the antibody recognizes both isoforms of Cldn15lb, the signal in the gut epithelial cells is likely due to Cldn15lb-b as predicted by the in situ hybridization data. The antibody also labels neuromasts of the lateral line (data not shown) which express other Claudins such as Claudin-b (Haas and Gilmour, 2006), suggesting that it may cross-react with other members of the Claudin family not tested in vitro. In summary, Cldn15lb is initially expressed in both hepatocytes and BECs and its localization becomes restricted to the intrahepatic biliary network by 120 hpf.
Figure 3. Cldn15lb is dynamically expressed in hepatocytes and biliary epithelial cells during development.
a–b, Single confocal plane of MDCK cysts stably expressing Cldn15lb-GFP immunostained with the tight junction markers ZO-1(a) or Occludin (b). Cldn15lb-GFP co-localizes with the tight junction markers (arrows). c–e, 150 μm transverse sections through the liver at 80 (c), 100 (d), and 120 (e) hpf. c, At 80 hpf, Cldn15lb immunostaining is observed in biliary epithelial cells (BECs) (arrows) as marked by Tg(Tp1bglob:eGFP) expression as well as the surrounding hepatocytes (arrowheads). d, At 100 hpf, Cldn15lb immunostaining in BECs (arrows) increases while that in hepatocytes (arrowheads) becomes more punctate. e, At 120 hpf, Cldn15lb immunostaining is only observed in BECs (arrows). Green: Tg(Tp1bglob:eGFP); red: Cldn15lb antibody. The antibody also labels cells in the gut (g). Scale bars are 10um.
Cldn15lb plays a role in hepatocyte polarization and bile canaliculus development
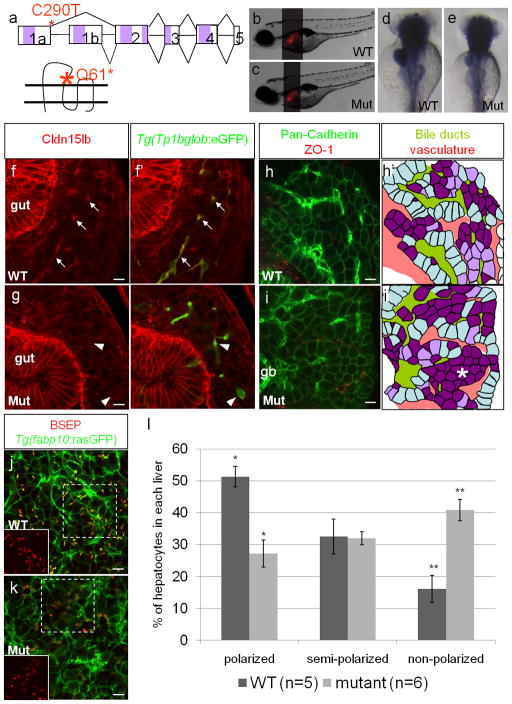
To determine the role of Cldn15lb in the development of the intrahepatic biliary network in zebrafish, we identified a mutation in the corresponding gene by TILLING. The nonsense point mutation, C290T, is located at the 3′ end of exon1a, and thus, is specific to cldn15lb_tv1 (Fig. 4a); it changes glutamine at amino acid 61 to a stop codon, resulting in a protein truncated at the end of the first extracellular loop (Fig. 4a). cldn15lbfh290 mutant larvae exhibit a normally sized liver and do not present with any discernible body phenotype (Fig. 4b–c). They survive to become normal looking adults. Expression of foxa3 and ceruloplasmin in the mutant larvae at 3 days post fertilization (dpf) appears unaffected indicating that endodermal and hepatic development, respectively, are normal (data not shown). In situ hybridization analysis shows that cldn15lb_tv1 transcripts in the liver and pancreas are expressed at a lower level in cldn15lbfh290 mutants while cldn15l_tv2 expression in the gut is not affected (Fig. 4d–e). Cellular analysis of the pancreas did not reveal any abnormalities in the cldn15lbfh290 mutants (data not shown). 100 hpf mutant livers stained with the Cldn15lb antibody did not show immunofluorescence, suggesting that the full-length, functional Cldn15lb-a is absent in the mutant livers (Fig. 4f–g). However, the antibody still recognizes Cldn15lb-b in wildtype and mutant guts. These data further confirm that the antigen detected by the Cldn15lb antibody in the liver is Cldn15lb-a and not Cldn15lb-b.
Figure 4. Hepatocyte polarization and bile canaliculi development are affected in cldn15lbfh290 mutant livers.
a, Schematic of the TILLING mutation. The nonsense mutation is situated at the end of exon1a making this mutation specific to cldn15lb_tv1. The mutation is situated in the sequence encoding the extracellular region immediately before the second transmembrane domain. b–c, Brightfield pictures of wildtype (b) and mutant (c) larvae at 5 dpf. The overall body morphology and liver size, as assessed by Tg(fabp10:RFP)gz12 expression, appear unaffected. d–e, in situ hybridization analysis of wildtype (d) and mutant (e) larvae at 80 hpf with a probe that recognizes both transcript variants. The mutant larvae exhibit decreased expression of cldn15lb in the liver and pancreas while gut expression appears unaffected. f–g, 150 μm transverse sections through the liver at 100 hpf. Green: Tg(Tp1bglob:eGFP); red: Cldn15lb antibody. Wildtype larvae (f) express Cldn15lb in BECs (arrows). Cldn15lb expression in mutant larvae (g) is severely reduced (arrowhead). h–i, Whole-mount analysis of 80 hpf larvae stained for pan-cadherin (green) and ZO-1(mouse) (red). Anti-pan-cadherin outlines hepatocytes at lower intensity while it marks BECs at higher intensity. Anti-mouse ZO-1 outlines the intrahepatic vasculature. h′–i′, Red: vasculature; Yellowish-green: biliary network. Hepatocytes are color-coded by their cell shape and location relative to the biliary and vascular networks. Hepatocytes that are sandwiched between the networks and columnar in shape are labeled in light blue. These cells appear to be fully polarized. Hepatocytes that are either columnar but not sandwiched between the two networks or are cuboidal and between the networks are labeled in light purple. Hepatocytes that are not polarized are non-uniformly shaped and not lined by the networks. These cells are labeled in dark purple. Most hepatocytes in mutant larvae do not appear to be polarized and are found in rosettes (* in i′) while a higher number of hepatocytes in wildtype larvae are polarized. Gallbladder (gb) j–k, Whole-mount analysis of 100 hpf Tg(fabp10:rasGFP) s942 larvae stained for BSEP (red) which marks canaliculi. Canaliculi in mutant livers are shorter and wider than those in wildtype livers. Insets highlight the boxed areas (red channel only). l, Percentage of hepatocytes that are polarized (light blue), semi-polarized (light purple), and non-polarized (dark purple). Wildtype livers have significantly more polarized cells while having almost three times fewer non-polarized cells (* p = 0.0018, ** p = 0.0012). Error bars represent SEM. All scale bars are 10um.
Since Claudins are one of the core components of tight junctions and Cldn15lb is expressed in both hepatocytes and BECs, we examined the three types of tight junctions in wildtype and mutant livers. We observed “kiss-point” contacts between membranes of hepatocytes characteristic of tight junctions in electron micrographs of 4 and 5 dpf mutant livers (Supp. Fig. 2a–b) indicating that junctions are formed properly. Furthermore, the adherens junction protein atypical Protein Kinase C (aPKC) and tight junction protein Zona Occludens-1 (ZO-1) appear to be properly localized in the apical membranes of mutant hepatocytes (data not shown). Localization of ZO-1 between BECs is also comparable between wildtype and mutant larvae at 100 hpf, indicating that those junctions are also not affected by the cldn15lb mutation (Supp. Fig. 2c). Tight junctions between hepatocytes and BECs also do not appear to be affected as cytokeratin-18 labeling of the canals of Hering in mutant larvae is similar to wildtype (Supp. Fig. 2d). These data suggest that tight junction formation is not affected in cldn15lbfh290 mutants.
Using a cadherin antibody that outlines hepatocytes and, with higher intensity, bile ducts as well as a ZO-1 antibody to mark the vasculature, we found that the patterning of the intrahepatic vascular and biliary networks is disrupted in cldn15lbfh290 mutants (Fig. 4h–i). In wildtype livers, the vascular and biliary networks are arranged in an alternating pattern such that a single row of hepatocytes separates them (Fig. 4h, light blue cells in h′) (Sakaguchi et al., 2008). However, cldn15lbfh290 mutant livers do not exhibit this pattern and hepatocytes are often found in clusters of cells two to three layers thick or in rosettes (Fig. 4i, dark purple cells in i′). During development, the patterning of the intrahepatic vascular and biliary networks occurs in conjunction with hepatocyte polarization. Between 50 and 80 hpf, hepatocytes receive signals from invading endothelial cells and undergo polarization. The side of the hepatocyte lined by endothelial cells becomes the basal domain. As the hepatocytes polarize, apical proteins, including those involved in bile secretion, become restricted to the membrane facing the biliary network (Sakaguchi et al., 2008). Since a defect in the patterning of the intrahepatic vascular and biliary networks was observed in cldn15lbfh290 mutants and Claudins are core components of tight junctions that have been implicated in the maintenance of apicobasal polarity (Shin et al., 2006), we analyzed hepatocyte polarization. In 80 hpf wildtype livers, most hepatocytes are sandwiched between the biliary network and the vasculature, and are columnar in shape indicating that they are fully polarized (Fig. 4h, light blue cells in h′). There is also a small population of cells that are undergoing polarization. These cells are either cuboidal-shaped and located between the two tubular networks, or columnar-shaped but not yet clearly sandwiched between the networks (Fig. 4h, light purple cells in h′). In contrast, only a small number of columnar cells were found in cldn15lbfh290 mutant livers. The majority of the mutant hepatocytes are non-uniformly shaped suggesting that they are not polarized (Fig. 4i, dark purple cells in i′). Often, these non-polarized cells are clustered together and exclude the tubular networks (Fig. 4i, region of purple cells highlighted by *). To quantify this phenotype, we categorized each cell in confocal stacks of the liver; optical sections were taken every 3 um, and 1452 wildtype and 2292 mutant hepatocytes were analyzed. We found that mutant livers have significantly fewer polarized hepatocytes (27.2% vs. 52.1%) and almost three times as many non-polarized hepatocytes (40.8% vs. 16.2%) (Fig. 4l).
Establishing apicobasal polarity not only ensures the proper establishment of liver architecture, it is also important for later stages of liver development. Apical pumps and transporters such as Bile Salt Export Pump (BSEP) and Multidrug Resistance Protein 2 (MRP2) are directed to the canalicular membrane on the apical region of hepatocytes where they facilitate bile acid secretion (Alrefai and Gill, 2007). Since hepatocyte polarization is affected in cldn15lbfh290 mutants, we wanted to assess canalicular formation. We immunostained Tg(fabp10:rasGFP)s942 larvae, whose hepatocytes are outlined by rasGFP with enhanced fluorescence on their basal membranes, with an antibody against BSEP (Gerloff et al., 1998), and found that canaliculi in 100 hpf wildtype larvae appear as long indentations into the apical domain of hepatocytes (Fig. 3j). However, in the mutant larvae, the canaliculi appear shorter and wider suggesting that their formation is affected (Fig. 3k). By 6 dpf, two-thirds of the mutant larvae have recovered and exhibit wildtype-looking canaliculi (data not shown). Since the canaliculi are initially malformed, we tested whether bile secretion and lipid metabolism might be disrupted using the fluorescently labeled fatty acid reporter PED-6 (Farber et al., 2001). Since PED-6 accumulates in the gallbladder after biliary secretion, we categorized the intensity of gallbladder fluorescence to assess secretion efficiency and found that fatty acids could be secreted by cldn15lbfh290 mutants, but at a slower rate. After 45 minutes of feeding, most of the mutant larvae had not processed, or had only processed a small amount of, PED-6 while most of the wildtype larvae had processed it completely. However, after 6 hours of feeding, almost all of the mutant larvae, similar to wildtype, had processed the PED-6 reporter (Supp. Fig. 3). Thus, our data show that loss of Cldn15lb-a disrupts bile canaliculi formation and delays bile secretion but that the mutant larvae appear to recover by 6dpf.
Cldn15lb is important for the organization of the intrahepatic biliary network
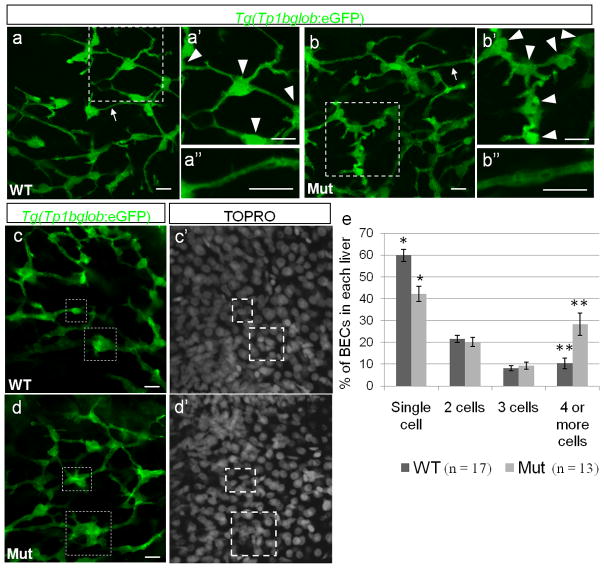
Given that Cldn15lb becomes localized to the intrahepatic biliary network after 100 hpf and that defects in hepatocyte polarization have been shown to affect biliary duct development (Cullinane et al., 2010b), we decided to investigate the role of Cldn15lb in the development of the intrahepatic biliary network. We used the Tg(Tp1bglob:eGFP) um14 line which labels cell bodies and cytoplasmic extensions of BECs (Parsons et al., 2009; Lorent et al., 2010). The cellular extensions connect to each other to form the tubular network of the biliary system. We found that at 100 hpf, the biliary network in the mutant livers is disorganized (Fig. 5a–b). In wildtype livers, most cell bodies of BECs (arrowheads) are well-separated from each other (Fig. 5a′). Furthermore, BECs interconnect through long cytoplasmic extensions that form thin tubules (Fig. 5a″). However, in the mutant livers, cell bodies are closer to each other (Fig. 5b′) and the tubules appear dilated (Fig. 5b″).
Figure 5. The intrahepatic biliary network is disorganized in cldn15lbfh290 mutant livers.
a–d, Whole-mount confocal projections of Tg(Tp1bglob:eGFP) um14 wildtype and mutant livers at 100 hpf. a–b, The biliary network in the mutant larvae appears to be disorganized. a′–b′, Magnification of the boxed regions. In the mutant livers, the cell bodies of the BECs (arrowheads) are closer together, and thus the ducts are shorter. a″–b″, Magnification of the duct labeled by the arrow. In the mutant livers, the ducts appear dilated. c–d, At 100 hpf, a majority of the BECs present as individual cells in wildtype livers. In the mutant livers, multiple BECs are clustered together at this stage (boxed regions). e, Percentage of BECs that present as single cells, doubles, triples, or in clusters of four or more cells. Wildtype livers have almost double the number of single cells and almost 3 times less cells in clusters of 4 or more cells (* p = 0.00056, ** p = 0.006). Error bars represent SEM. All scale bars are 10um.
Lorent et al. (2010) described that the intrahepatic biliary network undergoes population expansion via proliferation between 80 and 100 hpf and extensive remodeling between 3 and 4 dpf to give rise to a highly elaborate network. During the remodeling process, the BECs segregate from each other and the connection between them narrows. To determine whether the disorganized appearance of the biliary network in the cldn15lbfh290mutants was due to a defect in cell proliferation or in remodeling, we carried out various analyses at several stages from 50 to 120 hpf. Using an antibody against phospho-histone3 (PH3) to label proliferating cells (Ke et al., 2008), we found that wildtype and mutant larvae have similar numbers of PH3-positive cells (data not shown). In addition, the total number of BECs is similar in wildtype and mutant larvae (Supp. Fig. 4). These data suggest that Cldn15lb does not play a role in BEC proliferation and that the disorganized appearance is not due to the presence of more cells, but rather to a difference in BEC distribution. To determine whether the distribution defect was due to abnormalities in remodeling, we analyzed the organization of BECs and found that the network structures are comparable in wildtype and mutant larvae at 50, 64, 74, and 80 hpf (data not shown). However, at 100 hpf, we began to observe a difference in the distribution of the cell bodies (Fig. 5c–d). In wildtype larvae, most cell bodies of the BECs have undergone remodeling and are clearly separated from each other. They present as individual cells connected to each other by thin cytoplasmic bridges (Fig. 5c). In contrast, most of the cell bodies of the BECs in the mutant livers are clustered together in groups (Fig. 5d). To quantify this phenotype, we categorized each BEC as presenting as a single cell, two cells, three cells, or in a cluster of 4 or more cells, and calculated the respective percentages (total BECs counted: 898 for wildtype, 818 for mutants). In the cldn15lbfh290 mutant larvae, we observed a significant decrease in the percentage of BECs present as single cells (59.8% to 42.2%) and a significant increase in the percentage of BECs in clusters of four or more cells (10.4% to 28.3%) (Fig. 5e). These data suggest that a defect in the remodeling process leads to the disorganized biliary network observed in cldn15lbfh290 mutant larvae.
cldn15lbfh290 mutants survive to adulthood and appear grossly normal but microscopic examination of 1-year-old fish (4 wildtype and 4 mutants) revealed hepatic abnormalities in cldn15lbfh290 mutants. Scattered well-circumscribed lesions composed of multiple layers of epithelioid cells surrounding eosinophilic material, golden-brown pigment granules, and nuclear debris were identified in all mutant livers (Supp. Fig. 5b,d). Although similar lesions were noted in one of the four wildtype fish, mutant livers contained significantly more lesions and the lesions in the mutant livers were significantly larger in size (Supp. Fig. 5a,c,e,g). We measured the largest distance that runs through the center of each lesion to determine their diameter, and found that the average diameter was at least twice as large in mutants as in wildtype. Some lesions in the mutant livers, especially that of the most severely affected (Supp. Fig. 5e,f), had dystrophic calcifications. Focal lymphocytic inflammation was found around a few of the lesions, but most of the lesions were associated with minimal adjacent inflammatory reaction. The hepatic parenchyma in mutant livers was relatively unremarkable in some areas, and in other areas showed varying degrees of disorganization and loss of normal architecture. In the most severely affected mutant (Supp. Fig. 5e, f), the liver had a spongy appearance suggesting hepatocyte apoptosis or necrosis, and increased fibrous tissue surrounded some of the lesions. Overall, the phenotypes observed were more severe in the mutant livers than in the wildtype livers suggesting that cldn15lb-a deficiency facilitates the formation and/or progression of hepatic abnormalities in adult livers.
Discussion
The intrahepatic biliary system is a network of tubes within the liver that transports bile away from hepatocytes. Formation of this system requires specification of BECs and subsequent remodeling of the BECs to form a functional three-dimensional tubular network. Defective formation of the network or its canalicular system disrupts bile flow and causes cholestatic disease. The mechanisms that underlie cholestatic disease due to mutations in genes encoding tight junction proteins have not been identified (Carlton et al., 2004). Here, we present evidence that a tight junction protein, Cldn15lb-a, plays a role in the remodeling of the biliary network. In zebrafish cldn15lbfh290 mutants, BECs do not remodel properly and they remain clustered or in close proximity to each other, resulting in a malformed network with larger tubules. In addition, hepatocytes are not properly polarized and bile canaliculus formation is disrupted. This and previous studies, taken together, identify components of tight junctions to be important to establish a functional biliary network for effective bile flow (Baala et al., 2002; Hadj-Rabia et al., 2004; Son et al., 2009; Yeh et al., 2010).
cldn15lb is a novel claudin gene that is alternatively spliced to generate two isoforms expressed in a tissue- and cell-specific manner. The two isoforms differ in their first extracellular loop whose amino acid composition determines barrier properties (Van Itallie and Anderson, 2006). As such, the two variants may convey different barrier properties and functions. Even though Cldn15lb is phylogenetically similar to mammalian Cldn15, based on amino acid sequence, it is also closely related to mammalian Cldn10. Both Cldn15lb-a and human Cldn10 are expressed in hepatocytes and BECs (Nemeth et al., 2009). Mouse Cldn10 is weakly expressed in tight junctions of hepatocytes (Van Itallie et al., 2006). On the other hand, expression of Cldn15lb-b in the gut is more similar to that of mammalian Cldn15. Interestingly, mammalian Cldn10 has also been described to be alternatively spliced in the first exon similarly to cldn15lb (Van Itallie et al., 2006; Gunzel et al., 2009). Mammalian Cldn10 has six transcript variants that are differentially expressed in various tissues and exhibit different functions (Gunzel et al., 2009). Three of these six transcripts also lack the fourth exon and the resulting proteins localize intracellularly suggesting that they may play a novel role within the cell (Gunzel et al., 2009). cldn15lb transcripts lacking the fourth exon are not present in the zebrafish EST database at this time.
Expression of this novel gene in cultured MDCK cells showed that it is expressed in tight junctions. However, we could not identify defects in any of the three types of tight junctions in the liver based on immunostaining. Furthermore, tight junctions between hepatocytes observed in electron micrographs also appear to be normal. Unfortunately, we were not able to identify tight junctions between BECs or between BECs and hepatocytes in the electron micrographs. Such analyses may reveal tight junction abnormalities that cannot be detected by immunostaining. These tight junctions may indeed have functional defects that are too subtle or minor to detect but could account for the observed delay in PED-6 processing in the mutants. Alternatively, it is possible that tight junctions are not significantly affected in the mutants due to redundancy within the Claudin family. Cldn-1, -2, -3, -4, -7, -8, and -10 are all expressed in the mammalian liver (Nemeth et al., 2009), and the other isoform of Cldn15lb is expressed at low levels in the liver starting at 72 hpf. Furthermore, Cldn2 was shown to be upregulated in patients with Claudin-1 mutations (Hadj-Rabia et al., 2004). Thus, other members of the Claudin family could compensate for the lack of Cldn15lb-a function.
Since we observed GFP-tagged Cldn15lb in the lateral and basal membranes of MDCK cells, Cldn15lb might have yet other functions that could account for the mutant phenotypes. Our immunostaining in zebrafish also localizes Cldn15lb to the entire membrane of hepatocytes and BECs. Cldn15lb is initially expressed in both hepatocytes and BECs and is subsequently restricted to BECs. This expression profile is reminiscent of that of the adhesion molecules Cadherin and Alcam (Sakaguchi et al., 2008; Curado et al., 2010; data not shown). Since Claudins were first identified as calcium-independent adhesion molecules (Kubota et al., 1999) and members of the JAM family of tight junction proteins have been shown to co-localize with ZO-1 at adherens-like cell-cell contacts in fibroblasts (Morris et al., 2006), it is possible that Cldn15lb-a functions as an adhesion molecule during liver development.
It is also possible that Cldn15lb functions to maintain hepatocyte polarity and that the absence of Cldn15lb-a function gives rise to defective bile canaliculi. Similar to what is observed in cldn15lbfh290 mutant larvae, bile canaliculi in Cldn2-deficient mice and Cldn2-deficient hepatic cell lines display abnormal morphology (Son et al., 2009; Yeh et al., 2010). During the development of bile canaliculi, apical pumps and transporters involved in bile secretion, such as BSEP and MRP2, are delivered to each canaliculus via a process called apical membrane biogenesis. In hepatocytes where Claudin expression is disrupted, the apical domain is not well-defined, and thus, targeted delivery of apical proteins and apical membrane biogenesis is likely to be affected. In support of this hypothesis, disruption of Rab11a-mediated protein trafficking causes bile canaliculi malformation (Wakabayashi et al., 2005; Cullinane et al., 2010a). In cldn15lbfh290 mutants where hepatocyte polarity is disrupted, trafficking of BSEP and other apical proteins is also likely to be affected.
Polarization of hepatocytes has been associated with biliary duct development and biliary physiology (Sakaguchi et al., 2008; Cullinane et al., 2010a). Patients with arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome have mutations in genes encoding proteins involved in membrane trafficking used to establish hepatocyte polarity. Disruption of these proteins in mouse and zebrafish results in reduced levels of hepatic E-cadherin, abnormal duct development, and disrupted bile flow (Matthews et al., 2005; Cullinane et al., 2010a). Our study provides additional support for the hypothesis that hepatocyte polarity and biliary duct remodeling are linked, and raise the possibility that Claudins and other tight junction proteins influence this remodeling process. First, the remodeling defect in cldn15lbfh290 mutants appears to occur between 80 and 100 hpf when Cldn15lb-a is still expressed in hepatocytes. Second, apical lumen remodeling was shown to be defective in hepatocytes with defective adherens junctions (Theard et al., 2007). Since Claudin-containing tight junctions in hepatocytes are important to define their apical domain, one hypothesis is that BECs receive signals from a polarized epithelium to direct the remodeling process. In the absence of a polarized hepatic epithelium, BECs would not receive such signals and thus, the remodeling of the biliary network would be affected.
Although we were not able to fully characterize the hepatic lesions we observed in adult cldn15lbfh290 mutants, it is interesting to speculate on the etiology of these lesions, particularly given the hepatobiliary abnormalities we observed during development. The central necrosis and pigment deposition raise the possibility that the lesions may represent granulomas, which can be caused by infections, including mycobacterial or fungal infections, or non-infectious entities including stress-induced sarcoidosis, primary biliary cirrhosis, or foreign body reaction (Lamps, 2008; Swaim et al., 2006). In this case, it is possible that loss of Cldn15lb-a results in damage to the hepatobiliary system that renders the mutants more susceptible to infection or other cause of granuloma formation. Alternatively, it is possible that the epithelioid cells at the periphery of at least some of the lesions represent squamous metaplasia of the biliary epithelium. In this case, the lesions may represent a response to abnormal bile flow and/or bile accumulation, which could be caused by leakage of bile through an abnormal biliary epithelium, by accumulation of bile due to partial obstruction of an abnormally-developed biliary network, or by some other biliary abnormality present in the cldn15lbfh290 mutants. Regardless of the identity of these lesions, it is important to note that lesions in the mutant livers are significantly larger than those found in only one of the wildtype livers. Clearly, loss of Cldn15lb-a is associated with hepatic abnormalities in adult fish which may be due to a chronic, possibly subtle, defect with effects that accumulate over time resulting in more abundant and larger lesions. Additional experiments will be necessary to determine whether the adult phenotype is a direct or indirect result of the developmental defects, and how the loss of Cldn15lb-a might cause the exacerbated adult phenotype.
Targeted deletion of Hnf1b has been shown to cause a loss of organization of biliary ductal structures (Coffinier et al., 2002), and our microarray analysis suggests that Hnf1b may do so in part by regulating the expression of cldn15lb to mediate biliary remodeling during morphogenesis. Our study of the liver defects in the cldn15lbfh290 mutants raises the intriguing possibility that cholestasis in patients with mutations in genes encoding tight junction proteins may arise in response to abnormal bile canaliculi and/or biliary duct architecture due to defects in remodeling. Therefore, understanding the cellular mechanisms by which Claudins and other tight junction proteins regulate the development of the intrahepatic biliary network should also bring novel insights into our understanding of cholestasis.
Supplementary Material
Generation and validation of Cldn15lb peptide antibody.
a, Sequence alignment between zebrafish Cldn10, Cldn15, Cldn15lb-a, Cldn15lb-b, and Cldn10l. [Cldn15lb-a and –b are the proteins encoded by cldn15lb_tv1 and cldn15lb_tv2, respectively.] Purple shading boxes indicate transmembrane regions. Red box highlights the epitope targeted by the peptide antibody. b, HEK293 cells were transiently transfected with constructs expressing Cldn10l, Cldn15, or Cldn15lb-a. Immunofluorescence with peptide antibody generated against Cldn15lb recognized Cldn15lb-a only. c, Cos7 cells were transiently transfected with untagged or GFP-tagged version of either transcript variant. The antiserum recognized cells that were transfected with either untagged transcript variant and it co-localized with the GFP-tagged version. Thus, the polyclonal antibody in the serum recognizes Cldn15lb but does not distinguish between the 2 isoforms.
Junctions are present in cldn15lbfh290 mutant livers.
Electron microscopy at 4 (a) and 5 (b) dpf. Insets are magnification of the arrowed areas showing junctional complexes that lie between hepatocytes and line bile canaliculi (c). Junctional complexes can be found in both wildtype and mutant livers. Scale bars are 0.4um (inset: 0.1um). (c) Whole-mount confocal projection of 110 hpf larvae immunostained with 2F11 and ZO-1(rabbit) to mark BECs and tight junctions between BECs, respectively. Tight junctions appear to be properly localized between BECs in the mutant livers. (d) Whole-mount confocal projection of 100 hpf Tg(Tp1bglob:eGFP) um14 larvae immunostained for cytokeratin-18 to mark the canals of Hering. These canals, which line the biliary ducts, appear normal in mutant livers. Cytokeratin-18 immunostaining also labels the intrahepatic vasculature. Scale bars in c and d are 10um.
Time course of PED-6 biliary secretion. At 30 minutes, there was no correlation between fluorescence intensity and genotype. However, at 45 minutes, all of the wildtype larvae either had strong or some fluorescence while 50% and 40% of the mutant larvae had no or some gallbladder fluorescence, respectively. This correlation was no longer observed after 6 hours of feeding where almost all larvae, regardless of genotype, displayed PED-6 fluorescence in the gallbladder. All 6 dpf larvae that were fed PED-6 overnight displayed gallbladder fluorescence (data not shown).
Graph showing the number of BECs in each liver from 46 to 120 hpf. At each stage, the number of Tg(Tp1bglob:eGFP) positive cells was counted through a confocal stack 11.5 um thick. The number of BECs in wildtype and mutant larvae is comparable. At least 3 or more larvae were counted for each data point. Error bars represent SEM.
Pathology in adult livers. Low magnification views of wildtype (a) and mutant (c, e) livers stained with hematoxylin and eosin. Scale bars are 200um. Lesions (arrows) are present in 1 of 4 wildtype and 4 of 4 mutant livers analyzed (g). Each liver is outlined by dashed lines. Magnified views of the same livers (b, wildtype; d, f, mutants). Scale bars are 40um. a, Small lesions (70–160 microns in diameter) are scattered throughout the liver parenchyma in one of four wildtype livers. b, An example of one of the lesions found in the wildtype liver. They appear to be well-circumscribed lesions composed of approximately 1–3 layers of flattened epithelioid cells (inset) surrounding eosinophilic material containing occasional golden-brown pigment granules and fragments of nuclear debris. Minimal inflammatory cells are present around the lesions and the hepatic parenchyma appears normal. c, An example of the mutant livers which show abundant and larger lesions (arrows and arrowhead). d, Magnified view of the nodule marked by the arrowhead in c. These lesions have a more exuberant epithelioid proliferation at the periphery, often more than 10 cell layers thick, and contain degenerating cellular debris, golden-brown pigment (arrows), and scattered calcifications. Focal inflammation surrounds a few lesions but they are, for the most part, associated with minimal adjacent inflammatory reaction. The hepatic parenchyma of the mutants is unremarkable in some areas but in other areas shows varying degrees of disorganization. e, Mutant #3 shows abundant and occasionally coalescing lesions (arrowhead). f, A magnified view of the coalesced lesion shows calcified material in the center (*). The hepatic parenchyma in mutant #3 shows hepatocyte dropout and increased fibrous tissue surrounding several of the lesions. g, Plot of the diameter of each lesion found in each liver. The bar indicates the average lesion diameter. Three of the four wildtype livers did not present with lesions. The plot shows that the severity of the phenotype varies in mutants but that on average, the lesions are almost twice to three times the size of those in wildtype #3. Mutant #3, the most severely affected, presented with 74 lesions of varying diameters.
Nucleotide sequence of cldn15lb_tv2
Research Highlights.
A novel gene, Claudin15-like b, is identified in zebrafish and has 2 isoforms.
Cldn15lb is dynamically expressed in hepatocytes and biliary epithelial cells.
Disruption of Cldn15lb affects hepatocyte polarization and canaliculi formation.
Biliary ducts in Cldn15lb mutants do not remodel and form a disorganized network.
Cldn15lb regulates cell behaviors during biliary duct morphogenesis.
Acknowledgments
We thank Sandra Huling for technical help with the EM experiments, Kathryn Helde for developing the genotyping protocol, Yvonne Bradford for her help with the nomenclature, Ana Ayala and Milagritos Alva for fish care, Laura Bull, Silvia Curado and Chunyue Yin for critical reading of the manuscript and helpful discussions, Jackie Maher and JP Grenert for histology advice, and Takashi Mikawa and other Stainier lab members for discussions and technical help. I.D.C. was supported by the ARCS Foundation. M.B. was supported by NIH grant DP2-3034656. K.E. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-#109-10). TILLING of cldn15lbfh290 was supported by NIH grant HG002995 to C.B.M. C.B.M. is an investigator with the Howard Hughes Medical Institute. This work was supported in part by the Pathology & Imaging Core of the UCSF Liver Center (P30 DK026743), as well as grants from the NIH (DK060322) and the Packard Foundation to D.Y.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm Res. 2007;24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, Jacquemin P, Pierreux CE, Clotman F, Lemaigre FP. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Hadj-Rabia S, Hamel-Teillac D, Hadchouel M, Prost C, Leal SM, Jacquemin E, Sefiani A, De Prost Y, Courtois G, Munnich A, Lyonnet S, Vabres P. Homozygosity mapping of a locus for a novel syndromic ichthyosis to chromosome 3q27-q28. J Invest Dermatol. 2002;119:70–76. doi: 10.1046/j.1523-1747.2002.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis D, Bartman T, Jin SW, Scott IC, D’Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nature cell biology. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, Morton DH, Bull LN. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34:91–96. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- Carlton VE, Pawlikowska L, Bull LN. Molecular basis of intrahepatic cholestasis. Ann Med. 2004;36:606–617. doi: 10.1080/07853890410018916. [DOI] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Gresh L, Fiette L, Tronche F, Schutz G, Babinet C, Pontoglio M, Yaniv M, Barra J. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- Cui S, Capecci LM, Matthews RP. Disruption of planar cell polarity activity leads to developmental biliary defects. Dev Biol. 2011;351:229–241. doi: 10.1016/j.ydbio.2010.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane AR, Straatman-Iwanowska A, Zaucker A, Wakabayashi Y, Bruce CK, Luo G, Rahman F, Gurakan F, Utine E, Ozkan TB, Denecke J, Vukovic J, Di Rocco M, Mandel H, Cangul H, Matthews RP, Thomas SG, Rappoport JZ, Arias IM, Wolburg H, Knisely AS, Kelly DA, Muller F, Maher ER, Gissen P. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat Genet. 2010a;42:303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane AR, Straatman-Iwanowska A, Zaucker A, Wakabayashi Y, Bruce CK, Luo G, Rahman F, Gurakan F, Utine E, Ozkan TB, Denecke J, Vukovic J, Di Rocco M, Mandel H, Cangul H, Matthews RP, Thomas SG, Rappoport JZ, Arias IM, Wolburg H, Knisely AS, Kelly DA, Muller F, Maher ER, Gissen P. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nature genetics. 2010b;42:303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S, Ober EA, Walsh S, Cortes-Hernandez P, Verkade H, Koehler CM, Stainier DY. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Disease models & mechanisms. 2010;3:486–495. doi: 10.1242/dmm.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- Gatmaitan ZC, Arias IM. ATP-dependent transport systems in the canalicular membrane of the hepatocyte. Physiol Rev. 1995;75:261–275. doi: 10.1152/physrev.1995.75.2.261. [DOI] [PubMed] [Google Scholar]
- Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- Gunzel D, Stuiver M, Kausalya PJ, Haisch L, Krug SM, Rosenthal R, Meij IC, Hunziker W, Fromm M, Muller D. Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci. 2009;122:1507–1517. doi: 10.1242/jcs.040113. [DOI] [PubMed] [Google Scholar]
- Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10:673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, De Prost Y, Munnich A, Hadchouel M, Smahi A. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 2004;127:1386–1390. doi: 10.1053/j.gastro.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio) FEBS letters. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- Ke Z, Kondrichin I, Gong Z, Korzh V. Combined activity of the two Gli2 genes of zebrafish play a major role in Hedgehog signaling during zebrafish neurodevelopment. Molecular and cellular neurosciences. 2008;37:388–401. doi: 10.1016/j.mcn.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, Tsukita S. Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Current biology : CB. 1999;9:1035–1038. doi: 10.1016/s0960-9822(99)80452-7. [DOI] [PubMed] [Google Scholar]
- Lamps LW. Hepatic granulomas, with an emphasis on infectious causes. Advances in anatomic pathology. 2008;15:309–318. doi: 10.1097/PAP.0b013e31818a6477. [DOI] [PubMed] [Google Scholar]
- Lee NP, Luk JM. Hepatic tight junctions: from viral entry to cancer metastasis. World J Gastroenterol. 2010;16:289–295. doi: 10.3748/wjg.v16.i3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaigre FP. Molecular mechanisms of biliary development. Prog Mol Biol Transl Sci. 2010;97:103–126. doi: 10.1016/B978-0-12-385233-5.00004-0. [DOI] [PubMed] [Google Scholar]
- Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res. 2004;14:1248–1257. doi: 10.1101/gr.2400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Plumb-Rudewiez N, Lorent K, Gissen P, Johnson CA, Lemaigre F, Pack M. Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development. 2005;132:5295–5306. doi: 10.1242/dev.02140. [DOI] [PubMed] [Google Scholar]
- Montesano R, Friend DS, Perrelet A, Orci L. In vivo assembly of tight junctions in fetal rat liver. J Cell Biol. 1975;67:310–319. doi: 10.1083/jcb.67.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Tawil A, Berkova Z, Wible L, Smith CW, Cunningham SA. Junctional Adhesion Molecules (JAMs) are differentially expressed in fibroblasts and co-localize with ZO-1 to adherens-like junctions. Cell communication & adhesion. 2006;13:233–247. doi: 10.1080/15419060600877978. [DOI] [PubMed] [Google Scholar]
- Nemeth Z, Szasz AM, Tatrai P, Nemeth J, Gyorffy H, Somoracz A, Szijarto A, Kupcsulik P, Kiss A, Schaff Z. Claudin-1, -2, -3, -4, -7, -8, and -10 protein expression in biliary tract cancers. J Histochem Cytochem. 2009;57:113–121. doi: 10.1369/jhc.2008.952291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of development. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nature genetics. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Current biology : CB. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Theise N. Canals of Hering: recent insights and current knowledge. Seminars in liver disease. 2004;24:43–48. doi: 10.1055/s-2004-823100. [DOI] [PubMed] [Google Scholar]
- Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Son S, Kojima T, Decaens C, Yamaguchi H, Ito T, Imamura M, Murata M, Tanaka S, Chiba H, Hirata K, Sawada N. Knockdown of tight junction protein claudin-2 prevents bile canalicular formation in WIF-B9 cells. Histochem Cell Biol. 2009;131:411–424. doi: 10.1007/s00418-008-0546-0. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim LE, Connolly LE, Volkman HE, Humbert O, Bom DE, Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108–17. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theard D, Steiner M, Kalicharan D, Hoekstra D, van Ijzendoorn SC. Cell polarity development and protein trafficking in hepatocytes lacking E-cadherin/beta-catenin-based adherens junctions. Mol Biol Cell. 2007;18:2313–2321. doi: 10.1091/mbc.E06-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mechanisms of development. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, Anderson JM. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol. 2006;291:F1288–1299. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- Vroman B, LaRusso NF. Development and characterization of polarized primary cultures of rat intrahepatic bile duct epithelial cells. Lab Invest. 1996;74:303–313. [PubMed] [Google Scholar]
- Wakabayashi Y, Dutt P, Lippincott-Schwartz J, Arias IM. Rab11a and myosin Vb are required for bile canalicular formation in WIF-B9 cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15087–15092. doi: 10.1073/pnas.0503702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book a guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. 4. ZFIN; Eugene, Or: 2000. [Google Scholar]
- Yeh TH, Krauland L, Singh V, Zou B, Devaraj P, Stolz DB, Franks J, Monga SP, Sasatomi E, Behari J. Liver-specific beta-catenin knockout mice have bile canalicular abnormalities, bile secretory defect, and intrahepatic cholestasis. Hepatology. 2010;52:1410–1419. doi: 10.1002/hep.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation and validation of Cldn15lb peptide antibody.
a, Sequence alignment between zebrafish Cldn10, Cldn15, Cldn15lb-a, Cldn15lb-b, and Cldn10l. [Cldn15lb-a and –b are the proteins encoded by cldn15lb_tv1 and cldn15lb_tv2, respectively.] Purple shading boxes indicate transmembrane regions. Red box highlights the epitope targeted by the peptide antibody. b, HEK293 cells were transiently transfected with constructs expressing Cldn10l, Cldn15, or Cldn15lb-a. Immunofluorescence with peptide antibody generated against Cldn15lb recognized Cldn15lb-a only. c, Cos7 cells were transiently transfected with untagged or GFP-tagged version of either transcript variant. The antiserum recognized cells that were transfected with either untagged transcript variant and it co-localized with the GFP-tagged version. Thus, the polyclonal antibody in the serum recognizes Cldn15lb but does not distinguish between the 2 isoforms.
Junctions are present in cldn15lbfh290 mutant livers.
Electron microscopy at 4 (a) and 5 (b) dpf. Insets are magnification of the arrowed areas showing junctional complexes that lie between hepatocytes and line bile canaliculi (c). Junctional complexes can be found in both wildtype and mutant livers. Scale bars are 0.4um (inset: 0.1um). (c) Whole-mount confocal projection of 110 hpf larvae immunostained with 2F11 and ZO-1(rabbit) to mark BECs and tight junctions between BECs, respectively. Tight junctions appear to be properly localized between BECs in the mutant livers. (d) Whole-mount confocal projection of 100 hpf Tg(Tp1bglob:eGFP) um14 larvae immunostained for cytokeratin-18 to mark the canals of Hering. These canals, which line the biliary ducts, appear normal in mutant livers. Cytokeratin-18 immunostaining also labels the intrahepatic vasculature. Scale bars in c and d are 10um.
Time course of PED-6 biliary secretion. At 30 minutes, there was no correlation between fluorescence intensity and genotype. However, at 45 minutes, all of the wildtype larvae either had strong or some fluorescence while 50% and 40% of the mutant larvae had no or some gallbladder fluorescence, respectively. This correlation was no longer observed after 6 hours of feeding where almost all larvae, regardless of genotype, displayed PED-6 fluorescence in the gallbladder. All 6 dpf larvae that were fed PED-6 overnight displayed gallbladder fluorescence (data not shown).
Graph showing the number of BECs in each liver from 46 to 120 hpf. At each stage, the number of Tg(Tp1bglob:eGFP) positive cells was counted through a confocal stack 11.5 um thick. The number of BECs in wildtype and mutant larvae is comparable. At least 3 or more larvae were counted for each data point. Error bars represent SEM.
Pathology in adult livers. Low magnification views of wildtype (a) and mutant (c, e) livers stained with hematoxylin and eosin. Scale bars are 200um. Lesions (arrows) are present in 1 of 4 wildtype and 4 of 4 mutant livers analyzed (g). Each liver is outlined by dashed lines. Magnified views of the same livers (b, wildtype; d, f, mutants). Scale bars are 40um. a, Small lesions (70–160 microns in diameter) are scattered throughout the liver parenchyma in one of four wildtype livers. b, An example of one of the lesions found in the wildtype liver. They appear to be well-circumscribed lesions composed of approximately 1–3 layers of flattened epithelioid cells (inset) surrounding eosinophilic material containing occasional golden-brown pigment granules and fragments of nuclear debris. Minimal inflammatory cells are present around the lesions and the hepatic parenchyma appears normal. c, An example of the mutant livers which show abundant and larger lesions (arrows and arrowhead). d, Magnified view of the nodule marked by the arrowhead in c. These lesions have a more exuberant epithelioid proliferation at the periphery, often more than 10 cell layers thick, and contain degenerating cellular debris, golden-brown pigment (arrows), and scattered calcifications. Focal inflammation surrounds a few lesions but they are, for the most part, associated with minimal adjacent inflammatory reaction. The hepatic parenchyma of the mutants is unremarkable in some areas but in other areas shows varying degrees of disorganization. e, Mutant #3 shows abundant and occasionally coalescing lesions (arrowhead). f, A magnified view of the coalesced lesion shows calcified material in the center (*). The hepatic parenchyma in mutant #3 shows hepatocyte dropout and increased fibrous tissue surrounding several of the lesions. g, Plot of the diameter of each lesion found in each liver. The bar indicates the average lesion diameter. Three of the four wildtype livers did not present with lesions. The plot shows that the severity of the phenotype varies in mutants but that on average, the lesions are almost twice to three times the size of those in wildtype #3. Mutant #3, the most severely affected, presented with 74 lesions of varying diameters.
Nucleotide sequence of cldn15lb_tv2