Abstract
Leishmaniasis is one of the most important vector borne diseases caused by kinetoplastid protozoa Leishmania sp. Among all forms of Leishmaniasis, Visceral leishmaniasis (VL) or Kala-azar is the severest form of the illness. VL is characterized by fever, hepatosplenomegaly, anaemia, edema, weight loss and invariably fatal if left untreated. Characterization of Leishmania sp. is extremely necessary to understand the epidemiology, taxonomy and population genetics of the parasites which ultimately helps in designing appropriate drug regimen to combat the disease. In this study, we aimed to type the clinical isolates of Leishmania species collected in the period 2006–2010 from patients (n = 9) diagnosed with Kala-azar and post Kala-azar dermal leishmaniasis (PKDL) by RAPD-PCR method using eight selected primers. Genome of the clinical isolates were amplified and electrophoresed in agarose gel. These were compared with the RAPD PCR profiles of WHO reference strains for L. donovani (DD8) and L. tropica (K27) respectively. We calculated the Jaccard’s Similarity Coefficient and found one (study code T5) out of nine isolates as L. tropica while the rest were L. donovani. This pilot study supports the earlier single report claiming that both the species are responsible for Kala-azar in India and it also emphasizes the need for more systematic typing of clinical isolates of Indian Kala-azar.
Keywords: Indian Kala-azar, Leishmania donovani, Leishmania tropica, Random Amplified Polymorphic DNA-PCR
Introduction
Kinetoplast protozoan parasites belonging to the Genus Leishmania cause a spectrum of diseases ranging from simple cutaneous lesions to devastating mucocutaneous disfiguring to fatal systemic form. The parasites are transmitted by the bite of the female sand fly and are prevalent in four continents, being endemic in 88 countries, 72 of which are developing countries (Desjeux 2004). The visceral form of the disease known as Visceral Leishmaniasis (VL) or Kala-azar (KA) occurs in India, Bangladesh, Brazil, Iran, Nepal and Sudan (Sundar 2001). The symptoms of VL include fever, weight loss, anemia and hepatosplenomegaly (Bittencourt and Barral-Netto 1995). Post Kala-azar Dermal Leishmaniasis (PKDL), on the other hand, is a sequel of KA where 10–20% of the patients develop unusual cutaneous lesions in the form of hypopigmented macules, erythema and nodules that appear several months or years after apparent cure from the disease. The epidemiology of the disease is further complicated due to the overlapping of HIV infection with VL (Desjeux and Alvar 2003).
In India, VL dramatically broke out in epidemic proportions in the early nineties after an apparent dormancy with the state of Bihar and adjoining districts of Uttar Pradesh and West Bengal facing the brunt (Thakur 1993; Mallik and Basu 1993; Desjeux 1993). The situation has been aggravated further due to the emergence of over 60% KA cases becoming unresponsive to antimonials, the first-line drug (Croft et al. 2006). The aromatic diamidine pentamidine represents the second line of treatment (Amato et al. 1998) and along with Amphotericin B has high toxicity (Jackson et al. 1990).
Leishmania parasites have similar morphology and sometimes cause similar clinical manifestations and therefore, differentiation between species requires molecular or biochemical techniques. The characterization of Leishmania by the Isoenzyme method is considered as the ‘Gold standard’ even today (WHO 1994) but the procedure is lengthy, complicated and expensive. The Random Amplified Polymorphic DNA (RAPD) technique (Welsh and McClelland 1990; Williams et al. 1990) has extensively been used for molecular characterization as well as phylogenetic relationship studies of parasitic organisms including kinetoplastids (Tibayrenc et al. 1993). The RAPD PCR has been used to discriminate between different species of Leishmania (Gomez et al. 1995; Manna 1998; Hajjaran et al. 2004; Manna et al. 2005; Oliveira et al. 2007; Alimoradi et al. 2009; Hamad et al. 2010). RAPD uses random oligomers to amplify genomic DNA and thus does not necessitate any prior knowledge of the organism’s genome. As the Leishmania genome is GC rich, the randomly selected primers (all decamers but one twelve-mer) were selected with 60–70% GC (Noyes et al. 1996). In the present study, we have described the RAPD PCR profiles of nine recent (2006–2010) clinical isolates of KA and PKDL of which three were from Bangladesh and rest from India.
Materials and methods
Chemicals and reagents
The tissue culture chemicals like medium M199, medium RPMI-1640, sodium bicarbonate, streptomycin, and penicillin and other reagents used were obtained from Sigma chemical (St. Louis, MO, USA). Fetal Bovine Serum (FBS) was purchased from Invitrogen, USA. Random primers were procured from Eurofins Scientific GmbH, Germany.
Clinical isolates and reference strains
We have collected nine clinical isolates of which three were isolates of KA from International Centre for Diarrheal Disease Research, Dacca, Bangladesh. The Indian isolates of KA were taken from patients admitted in the Calcutta National Medical College, Kolkata. The presence of amastigotes in the bone marrow of the patients were confirmed by Giemsa staining as well as transforming into promastigote forms in culture medium M199. The PKDL isolate was collected from Department of Dermatology, School of Tropical Medicine, Kolkata, India. World Health Organization (WHO) reference strains for Leishmania donovani (DD8) and L. tropica (K27) were kindly provided by Dr. L. F. Schnur, Hebrew University-Hadassah Medical School, Jerusalem, Israel.
Preparation of nucleic acid from the clinical isolates of KA and PKDL
Total nucleic acids from all clinical isolates and two WHO reference strains were prepared as per standard protocol (Bhattacharya et al. 1993). Briefly, Leishmania promastigotes were suspended in Phosphate Buffered Saline (PBS, 0.02 M, pH 7.4). The cell pellets from different strains were obtained by centrifugation at 6000 rpm for 10 min at 4°C. Pellets were washed in PBS and resuspended in 400 μl of NET buffer. DNA was prepared from the samples by digestion with 100 μg/ml proteinase K in the presence of 1% SDS overnight at 37°C, followed by phenol chloroform extraction and ethanol precipitation. The DNA was dissolved in Tris EDTA buffer and stored at −20°C till use.
PCR amplification of the DNA of clinical isolates of KA and PKDL with random primers:
DNA amplification using random primers were preformed according Manna et al. (2005) with the following eight primers of which, seven were decamers while Universal Minicircle Sequence (UMS) was twelve-mer. The sequences were given as follows: 5′-TGC CGA GCT G-3′ (OPA 2), 5′-GGG TAA CGC C-3′(OPA 5), 5′-GTG ATC GCA G-3′(OPA 6), 5′-GAA ACG GGT G-3′(OPA 7), 5′-AGT CGG GCT G-3′(OPA 8), 5′-AAT CGG GCT G-3′(OPA 9), 5′-GGT CCC TGA C-3′(OPA 10), 5′-GGG GTT GGT GTA-3′(UMS). As Leishmania genome is GC rich, sequences were selected randomly with the requirement that their GC contents are 60–70% and that they have no self complementary ends.
In brief, the procedure was as follows, reaction mixtures (20 μl) contained 10× PCR buffer, 50 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer, 25 ng total nucleic acid and 1.2 units of Taq DNA polymerase, volume was made up to 20 μl with PCR grade distilled water. Amplification was performed in a thermal cycler with the protocol: 30 total cycles using the program, 5 min at 94°C, 1 min at 94°C, 2 min at (50–60°C), 2 min at 72°C. At the end of the PCR cycle, all tubes were kept for 5 min at 72°C for extension to go to completion. The products after the PCR run were electrophoresed in 1.2% agarose gel in TAE buffer and analyzed.
Results
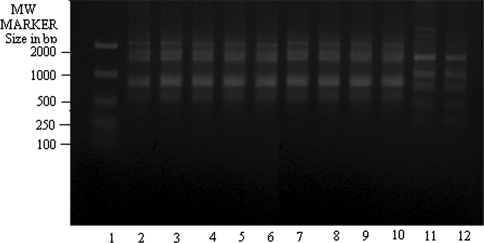
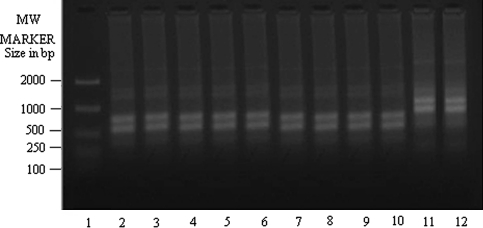
We amplified all clinical isolates with eight random decamers (sequences stated in “Materials and methods” section). Information regarding the patients’ sex, age, country and year of collection of the isolates from KA and PKDL patients had been described in Table 1. We checked for each primer whether they could amplify genomic DNA of reference strains of different species in a species specific manner and if they did so, we used these primers for further amplification experiments taking the DNA isolated from all clinical isolates of KA and PKDL. The random primer OPA 2 (Fig. 1) has been found out to discriminate between DD8 (L. donovani) in lane 2 and K27 (L. tropica) in lane 12 faithfully and can be used for identification and characterization of the clinical isolates of Leishmania sp. This is true for all the random primers stated here but OPA 7 i.e. out of eight random primers, OPA 7 could not score well to amplify the Leishmania genome characteristically.
Table 1.
List of different clinical isolates of KA and PKDL used for present study
| Study codea | Reference Codeb | Agec | Sexd | Countrye/Datef |
|---|---|---|---|---|
| T2 | MHOM/IN/10/IICB-AKS | 2 years 3 months | Male | INDIA/2010 |
| T3 | MHOM/IN/10/IICB-MS | 11 years | Male | INDIA/2010 |
| T4 | MHOM/IN/10/IICB-LH | 56 years | Male | INDIA/2010 |
| T5 | MHOM/IN/10/IICB-LB | 12 years | Male | INDIA/2010 |
| T7 | MHOM/IN/10/IICB-SIB | 17 years | Female | INDIA/2010 |
| P1g | MHOM/IN/08/PG-MA | 35 years | Male | INDIA/2008 |
| PG2 | MHOM/BD/06/MJ | 39 years | Male | BANGLADESH/2006 |
| PG3 | MHOM/BD/07/MM | 17 years | Male | BANGLADESH/2007 |
| PG4 | MHOM/BD/07/SA | 8 years | Male | BANGLADESH/2007 |
| DD8h | MHOM/IN/80/DD8 | WHO reference strain for Leishmania donovani | ||
| K27h | MHOM/SU/74/K27 | WHO reference strain for L. tropica | ||
aStudy codes given to the collected clinical isolates of KA and PKDL for the present study
bReference codes were given to the isolates according to the WHO nomenclature rules followed for Leishmania sp. [Evans DA (1989) World Health Organization, Geneva, pp 15–40]
cAge of the patients of KA or PKDL
dSex of the patients of KA or PKDL
eCountry of the patients
fYear of collection of the isolates from patients
gPKDL isolate
hDD8 and K27, two WHO reference strains for Leishmania donovani and L. tropica
Fig. 1.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of Kala-azar along with DD8 and K27 using primer OPA2. Lane 1 MW marker (100–2000 size in bp); lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4; lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
Amplification of genomic DNA of all clinical isolates along with WHO reference stains, DD8 (L. donovani) and K27 (L. tropica) with random primer OPA 2 showed (Fig. 1) that T5 (lane 11) was matching exactly with K27(lane 12) while banding patterns present in lanes 3–10 (other KA and PKDL isolates) were identical with DD8(lane 2). Bangladeshi KA isolates, PG2 (lane 6), PG3 (lane 7) and PG4 (lane 8) were all very similar to DD8 and different from K27. The same was true for PKDL isolate, P1 in lane 4. The other KA clinical isolates collected from Calcutta National Medical College, Kolkata, T2 (lane 3), T3 (lane 5), T4 (lane 9) and T7 (lane 10) respectively showed similar banding profiles to that of DD8 and they were different from K27 in terms of the mobilities of the amplified DNA.
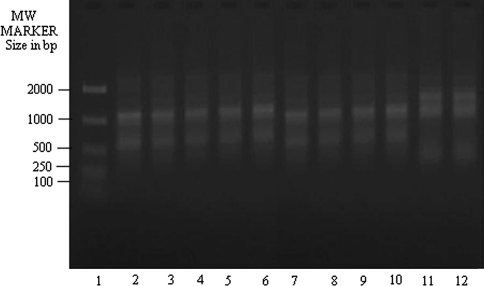
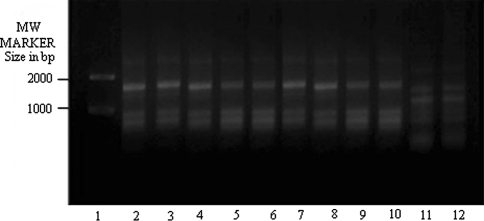
The amplification of genomic DNA with OPA 5 (Fig. 2) showed three Bangladeshi isolates (lanes 6–8 respectively) having similar banding pattern to DD8 and they were not like K27. The clinical isolate, T5 (lane 11), on the other hand, again showed matching banding profile with K27 but not with DD8.
Fig. 2.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of KA along with DD8 and K27 using primer OPA5. Lane 1 MW marker (100–2000 size in bp), lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4; lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
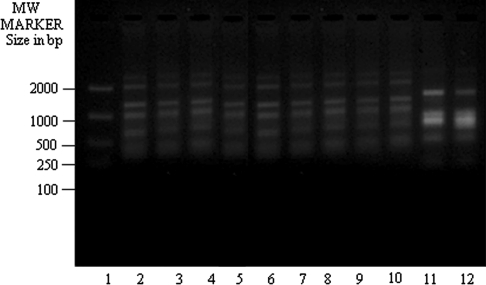
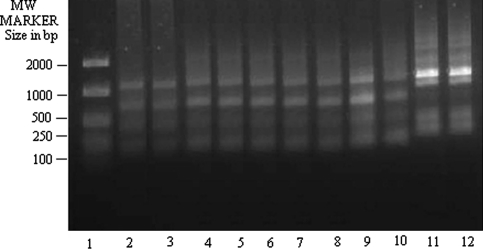
These findings were true for all other primers studied here i.e. OPA 6 (Fig. 3), OPA 8 (Fig. 4), OPA 9 (Fig. 5) and UMS (Fig. 6). T5 (lane 11 for each figure) showed banding profile very much similar to L. tropica (lane 12 in each figure) but no similarity with L. donovani (lane 2 in each figure). On contrary, other clinical isolates of KA from both India and Bangladesh and one PKDL isolate studied here showed congruence with DD8 in term of genome amplification with the primers mentioned in the present study but they showed dissimilarity with K27. These observations hold true also for OPA 10 (data not shown).
Fig. 3.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of KA along with DD8 and K27 using primer OPA6. Lane 1 MW marker (100–2000 size in bp), lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4, lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
Fig. 4.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of KA along with DD8 and K27 using primer OPA8. Lane 1 MW marker (100–2000 size in bp), lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4, lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
Fig. 5.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of KA along with DD8 and K27 using primer OPA9. Lane 1 MW marker (100–2000 size in bp), lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4, lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
Fig. 6.
RAPD-PCR amplification of 25 ng of total nucleic acid of different clinical isolates of KA along with DD8 and K27 using primer Universal Minicircle Sequence (UMS). Lane 1 MW marker (100–2000 size in bp), lane 2 DD8, lane 3 T2, lane 4 P1, lane 5 T3, lane 6 PG2, lane 7 PG3, lane 8 PG4, lane 9 T4, lane 10 T7, lane 11 T5, lane 12 K27. DD8 and K27 are WHO reference strains for Leishmania donovani and Leishmania tropica respectively, T2, T3, T4, T7 and T5 were clinical isolates of Kala-azar from India; P1 was PKDL isolate; PG2, PG3 and PG4 were clinical isolate of Kala-azar from Bangladesh
We calculated the relatedness among different clinical isolates studied here (Table 2) by Jaccard’s Similarity Coefficient method (Sneath and Sokal 1973) using our RAPD PCR data set. In short, the calculation for Jaccard’s Coefficient was done with numbers of total bands and with numbers of primers used in the study and the Similarity Coefficient, Sj was defined as
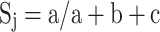 |
where a = number of bands (DNA) in common, b = bands unique to the first or reference stock, c = bands unique to the second stock whose relatedness to the first stock needs to be ascertained. In simpler cases, where only relatedness or similarity between two stocks or clinical isolates were to be determined, the mean of all the Sj values for different arbitrary primers were calculated.
Table 2.
Jaccard similarity coefficient of different clinical isolates of Leishmania sp. calculated from RAPD-PCR study
| L. donovani (DD8) | T2 | T3 | T4 | T7 | P1 | PG2 | PG3 | PG4 | T5 | L. tropica (K27) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L. donovani (DD8) | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.24 | 0.24 |
| T2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.24 | 0.24 | |
| T3 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0.24 | 0.24 | ||
| T4 | 0 | 1 | 1 | 1 | 1 | 1 | 0.24 | 0.24 | |||
| T7 | 0 | 1 | 1 | 1 | 1 | 0.24 | 0.24 | ||||
| P1 | 0 | 1 | 1 | 1 | 0.24 | 0.24 | |||||
| PG2 | 0 | 1 | 1 | 0.24 | 0.24 | ||||||
| PG3 | 0 | 1 | 0.24 | 0.24 | |||||||
| PG4 | 0 | 0.24 | 0.24 | ||||||||
| T5 | 0 | 1 | |||||||||
| L. tropica (K27) | 0 |
The Jaccard Coefficient has values between 1, for identical Operational Taxonomic Unit (OTU) and 0 for OTUs with no similarity at all. For an average analysis with ten primers, if the coefficient falls between any values, 0.75–1, one arbitrarily assumes the strains to belong to the same species or species complex (Tibayrenc et al. 1993)
The Jaccard’s Similarity Coefficient has values between 1, for identical Operational Taxonomic Unit (OTU) and 0 for OTUs with no similarity at all. For an average analysis with ten primers, if the coefficient falls between any values, 0.75–1, one arbitrarily assumes the strains to belong to the same species or species complex (Tibayrenc et al. 1993). In our study, we calculated (Table 2) the Coefficient of Similarity for all clinical isolates with L. donovani and L. tropica and found that besides T5, all were within the L. donovani species range while T5 showed 24% homology with DD8 and with other isolates including the PKDL (P1) and three isolates from Bangladesh (PG2, PG3 & PG4 respectively). Interestingly, it showed 100% homology with K27, the WHO reference strain for L. tropica.
Discussion
Our study with limited number of isolates (nine) showed eight of them were L. donovani while remaining one (T5) possessed RAPD PCR profiles similar to L. tropica reference strain, K27. This observation is lending credence to the previous findings by other group in the past that L. tropica causes Visceral Leishmaniasis in India (Sacks et al. 1995). Historically, it is known that Leishmania donovani is the aetiological agent for Indian Kala-azar. The species is endemic in the eastern part of the country (Salotra and Singh 2006) whereas L. tropica causing cutaneous disease in India is restricted to the western part (Kumar et al. 2007). The causal agent for PKDL is found out to be L. donovani (Salotra et al. 2001) and the incidence of PKDL might have importance in the transmission of VL in India, as PKDL provides the only known reservoir of the parasites (Thakur and Kumar 1992). The population structure of Leishmania donovani causing Indian Kala-azar was repeatedly reported to be homogeneous (El Tai et al. 2000; Chatterjee et al. 1995; Manna et al. 2005). Same type of observation was made in adjacent country, Bangladesh (Shamsuzzaman et al. 2000) which shares same border with eastern part of India and occasional human migration is common. Alam et al. (2009) claimed that the circulation of a single homogeneous population of L. donovani is responsible for the epidemic outbreak in India [Bihar], Bangladesh and Nepal and there exists a grade of responsiveness of the parasites to the antimonial drugs.
There was no report of co endemicity of both L. donovani and L. tropica in eastern part India until 1995 when Sacks et al. (1995) claimed that L. tropica causes nearly 25% of current Kala-azar cases in India. The author also suggested that this has some bearing with the unresponsiveness to pentavalent antimonials in the field. This relationship between unresponsiveness to antimonials and L. tropica infection manifested in visceral Leishmaniasis in Kenya was first reported by Mebrahtu et al. (1989). Visceral infection caused by L. tropica has also been reported from the veterans of ‘Operation Desert Storm’ (Magill et al. 1993) though the clinical features were not same with the classical KA. Hajjaran et al. (2004) reported the association of L. tropica with canine visceral leishmaniasis in Iran which was in concordance with the observation made by Sacks et al. (1995) for Indian Kala-azar. In recent time, Sharma et al. (2005) reported the occurrence of both L. donovani and L. tropica in the Localized Cutaneous Leishmaniasis (LCL) or ‘Oriental sore’ patients from Himachal Pradesh. LCL is endemic in the deserts of Rajasthan and is known to be caused by L. tropica (Kumar et al. 2007). In Sri Lanka, similar type of observation had been documented by Karunaweera et al. (2003), where they claimed the cases of Cutaneous Leishmaniasis caused by L. donovani. Concomittant natural infection of L. major with L. donovani had been seen in Iraq (Al-Diwany et al. 1995). There are occasional reports of co endemicity of other species of Leishmania with L. donovani or vice versa. In India, this situation has become complicated further as, in somewhat startling report Srivastava et al. (2010) showed the association of Leptomonas sp. parasites with Kala-azar patients.
All the above observations along with our finding strongly suggested that the issue of systematic typing of the clinical isolates of Indian Kala-azar should be taken into serious consideration which is still lacking. In Latin America where different Leishmania sp. are endemic (Reithinger and Dujardin 2007), characterization and identification of the clinical isolates has been given utmost importance. We are employing other molecular characterization techniques like amplification of Internal Transcribed Spacer (ITS) of ribosomal operon of the Leishmania parasites, Restriction Fragment Length Polymorphism analysis (RFLP) of ITS by various restriction enzymes and sequencing (unpublished data) to ascertain the extent of genetic diversity present among the clinical isolates of Leishmania sp. in the eastern endemic zone of India. This kind of attempt is extremely necessary to understand the population dynamics of the causal agents of Indian Kala-azar.
Acknowledgements
The authors sincerely acknowledge the financial support from University Grant Commission, India. For the infrastructure help, the co operations of Director of Public Instructions, Govt. of West Bengal, India and Officer in Charge, Bethune College, Kolkata are duly acknowledged. Supriya Khanra is the project fellow in the UGC Major Project, F. No. 35-57/2008 (SR) dated 20.3.2009.
Conflict of interest None.
References
- Alam MZ, Kuhls K, Schweynoch C, Sundar S, Rijal S, Shamsuzzaman AK, Raju BV, Salotra P, Dujardin JC, Schonian G. Multilocus microsatellite typing (MLMT) reveals genetic homogeneity strains in India subcontinent. Infect Genet Evol. 2009;9:24–31. doi: 10.1016/j.meegid.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Al-Diwany LJ, Al-Awkati NA, Atia M, Rassam MB. Concomitant natural infection with L. donovani and L. major: a case report from Iraq. Soz Praventivemed. 1995;40(4):234–238. doi: 10.1007/BF01354478. [DOI] [PubMed] [Google Scholar]
- Alimoradi S, Hajjaran H, Mohebali M, Mansouri F. Molecular identification of Leishmania species isolated from human cutaneous leishmaniasis by RAPD-PCR. Iran J Public Health. 2009;38(2):44–50. [Google Scholar]
- Amato V, Amato J, Nicodemo A, Uip D, Amato-Neto V, Duarte M. Treatment of mucocutaneous leishmaniasis with pentamidine isothionate. Ann Dermatol Venereol. 1998;125(8):492–495. [PubMed] [Google Scholar]
- Bhattacharyya R, Singh R, Hazra TK, Majumder HK. Application of polymerase chain reaction with specific and arbitrary primers to identification and differentiation of Leishmania parasites. FEMS Microbiol Lett. 1993;114(1):99–104. doi: 10.1111/j.1574-6968.1993.tb06557.x. [DOI] [PubMed] [Google Scholar]
- Bittencourt A, Barral-Netto M (1995) Leishmaniasis. In: Tropical pathology vol 8, 2nd edn. Springer, Berlin, pp 597–651
- Chatterjee M, Manna M, Bhaduri AN, Sarkar D. Recent Kala-azar cases in India: isozyme profiles of Leishmania parasites. Ind J Med Res. 1995;102:165–172. [PubMed] [Google Scholar]
- Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. J Clin Microbiol Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjeux P (1993) WHO/CID internal document, the leishmaniasis, meeting of interested parties on management and financing of the control of tropical diseases other than malaria, CTD/MIP/WP/93.8, Geneva, 15 Sep
- Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Desjeux P, Alvar J. Leishmania/HIV co-infections: epidemiology in Europe. Ann Trop Med Parasitol. 2003;97(suppl):3–15. doi: 10.1179/000349803225002499. [DOI] [PubMed] [Google Scholar]
- El Tai NO, Osman OF, El Fari M, Presber W, Schonian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Tran R Soc Trop Med Hyg. 2000;94:1–5. doi: 10.1016/S0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- Evans DA. Handbook on isolation characterization and cryopreservation of Leishmania. Geneva: World Health Organization; 1989. pp. 15–40. [Google Scholar]
- Gomez RF, Macedo AM, Pena SDJ, Melo MN. Leishmania (Vianna) braziliensis: Genatic relationships between strains isolated from different areas of Brazil as revealed by DNA fingerprinting and RAPD. Exp Parasitol. 1995;80:681–687. doi: 10.1006/expr.1995.1084. [DOI] [PubMed] [Google Scholar]
- Hajjaran H, Mohebali M, Razavi MR, Rezai S, Kazemi B, Edrissian GH H, Mojtabavi J, Hooshmand B. Identification of Leishmania species isolated from human cutaneous leishmaniasis, using Random Amplified polymorphic DNA(RAPD-PCR) Iran J Public Health. 2004;33(4):8–15. [Google Scholar]
- Hamad SH, Khalil EAG, Musa AM, Ibrahim ME, Younis BM, Elfaki MEE, El-Hassan AM. Leishmania donovani: Genetic diversity of isolates from Sudan characterized by PCR-based RAPD. Exp Parasitol. 2010;125:389–393. doi: 10.1016/j.exppara.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Jackson JE, Tally JD, Ellis WY, Mebrahtu YB, Lawyer PG, Were JB, Reed JG, Panisko DM, Limmer BL. Quantitative in vitro drug potency and drug susceptibility evaluation of Leishmania sp. from patients unresponsive to pentavalent antimony therapy. Am J Trop Med Hyg. 1990;43:464–480. doi: 10.4269/ajtmh.1990.43.464. [DOI] [PubMed] [Google Scholar]
- Karunaweera ND, Pratlong F, Siriwardane HV, Ihalamulla RL, Dedet JP. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme MON-37. Trans R Soc Trop Med Hyg. 2003;97:380–381. doi: 10.1016/S0035-9203(03)90061-7. [DOI] [PubMed] [Google Scholar]
- Kumar R, Bumb RA, Ansari NA, Mehta RD, Salotra P. Cutaneous Leishmaniasis caused by Leishmania tropica in Bikaner, India: Parasite identification and characterization using molecular and immunologic tools. Am J Trop Med Hyg. 2007;76(5):896–901. [PubMed] [Google Scholar]
- Magill AJ, Grogl M, Gasser RA, Sun W, Oster CN. Visceral infection caused by Leishmania tropica in veterans of ‘Operation Desert Storm’. New Engl J Med. 1993;328:1383–1387. doi: 10.1056/NEJM199305133281904. [DOI] [PubMed] [Google Scholar]
- Mallik KK, Basu D (1993) Recent advances in the management of Indian Kala-azar. In: Bhaduri AN, Basu MK, Sen AK, Kumar S (eds) Current trends in leishmania research. New Delhi Publication of Information Directorate (CSIR) 263
- Manna M (1998) Molecular taxonomic analysis of clinical isolates of Indian Kala-azar patients. PhD Thesis, Jadavpur University, Kolkata
- Manna M, Majumder H, Sunder S, Bhaduri A. Molecular characterizations of the clinical isolates of Kala-azar patients of India suggest Leishmania donovani is the only causal agent of Indian Kala-azar. Med Sci Monit. 2005;11(7):220–227. [PubMed] [Google Scholar]
- Mebrahtu Y, Lawyer P, Githure J. Visceral Leishmaniasis unresponsive to pentostam caused Leishmania tropica in Kenya. Am J Trop Med Hyg. 1989;41(3):289–294. doi: 10.4269/ajtmh.1989.41.289. [DOI] [PubMed] [Google Scholar]
- Noyes HA, Belli AA, Maingoon R. Apprisal of various random amplified polymorphic DNA-polymerase chain reaction primers for Leishmania identification. Am J Trop Med Hyg. 1996;55:98–105. doi: 10.4269/ajtmh.1996.55.98. [DOI] [PubMed] [Google Scholar]
- Oliveira JPC, Fernandes F, Cruz AK, Trombela V, Monteiro E, Camargo AA, Barral A, Oliveira CI. Genetic diversity of Leishmania amazonensis strains isolated in northeastern Brazil as revealed by DNA sequencing, PCR-based analyses and molecular karyotyping. Kinetoplast Biol Dis. 2007;6:1–8. doi: 10.1186/1475-9292-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithinger R, Dujardin JC. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol. 2007;45(1):21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DL, Kenny RT, et al. Indian Kala-azar caused by L. tropica. Lancet. 1995;345:959–961. doi: 10.1016/S0140-6736(95)90703-3. [DOI] [PubMed] [Google Scholar]
- Salotra P, Singh R. Challenges in the diagnosis of post Kala-azar dermal Leishmaniasis. Indian J Med Res. 2006;123:295–310. [PubMed] [Google Scholar]
- Salotra P, Sreenivas G, Pogue GP, Lee N, Nakhasi HL, Ramesh V, et al. Development of a species- specific PCR assay for detection of Leishmania donovani in clinical samples from patients with Kala-azar and post-Kala-azar dermal leishmaniasis. J Clin Microbiol. 2001;39:849–854. doi: 10.1128/JCM.39.3.849-854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamuzzaman SM, Furuya M, Shamsuzzaman Choudhury AKM. Characterization of Bangladeshi Leishmania isolated from Kala-azar patients by isoenzyme electrophoresis. Parasitol Int. 2000;49:139–145. doi: 10.1016/S1383-5769(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Sharma NL, Mahajan VK, Kanga A, Sood A, Katoch VM, Mauricio I, Singh CD, Parwan UC, Sharma VK, Sharma RC. Localized Cutaneous Leishmaniasis due to Leishmania donovani and Leishmania tropica: preliminary findings of the study of 161 new cases from a new endemic focus in Himachal Pradesh, India. Am J Trop Med Hyg. 2005;72(6):819–824. [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco, USA: Freeman; 1973. [Google Scholar]
- Srivastava P, Prajapati VK, Vanaerschot M, Auwera G, Dujardin JC, Sundar S. Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect Genet Evol. 2010;10:1145–1150. doi: 10.1016/j.meegid.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S. Drug resistances in Indian visceral leishmaniasis. Trop Med Int Health. 2001;6:849–854. doi: 10.1046/j.1365-3156.2001.00778.x. [DOI] [PubMed] [Google Scholar]
- Thakur CP (1993) Diminishing effectiveness of currently used drugs in treatment of Kala-azar and Ampphotericine B in antimony and pentamidine resistant Kala-azar. In: Bhaduri AN, Basu MK, Sen AK, Kumar S (eds) Current trends in Leishmania research. Publication and Information Directorate (CSIR) 254, New Delhi
- Thakur CP, Kumar K. Post Kala-azar dermal Leishmaniasis: a neglected aspect of Kala-azar control programme. Ann Trop Med Parasitol. 1992;86:355–359. doi: 10.1080/00034983.1992.11812678. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Neubauer K, Barnabe C, Guerrini F, Skarecky D, Ayala FJ. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (1994) Report on the consultative meeting on Leishmania/HIV co-infection. Co-sponsored by the Instituto Superiore Di Sanita and The Division of the Control of Tropical Diseases, WHO, Rome, pp 4–8