Abstract
Glutaredoxin belongs to the oxidoreductase family with cytosolic glutaredoxin 1 (Grx1) and mitochondrial gluraredoxin 2 (Grx2) isoforms. Of the two isozymes, the function of Grx2 is not well understood. This paper studied the effect of Grx2 deletion on cellular function using primary lens epithelial cell cultures isolated from Grx2 gene knockout (KO) and wild type (WT) mice. We found that both cell types showed similar growth patterns and morphology, and comparable mitochondrial glutathione pool and complex I activity. Cells with deleted Grx2 did not show affected Grx1 or thioredoxin (Trx) expression but exhibited high sensitivity to oxidative stress. Under treatment of H2O2, the KO cells showed less viability, higher membrane leakage, enhanced ATP loss and complex I inactivation, and weakened ability to detoxify H2O2 in comparison with that of the WT cells. The KO cells had higher glutathionylation in the mitochondrial proteins, particularly the 75-kDa subunit of complex I. Recombinant Grx2 deglutathionylated complex I, and restored most of its activity. We conclude that Grx2 has a function to protect cells against H2O2-induced injury via its peroxidase and dethiolase activities; particularly, Grx2 prevents complex I inactivation and preserves mitochondrial function.
Keywords: glutaredoxin 2, oxidative stress, complex I, mitochondira, glutathionylation
INTRODUCTION
Oxidative stress is considered as one of the most important risk factors for many diseases in humans, including cancer [1], Alzheimer’s disease, Parkinson’s disease [2], macular degeneration [3] and cataract [4]. Often the oxidative damage resulting from external or internal oxidants such as superoxide, hydrogen peroxide and hydroxyl radicals is found in proteins, lipids and nucleic acids. In proteins, thiol groups are most reactive and easily oxidized to affect enzyme activity or cellular function. Mammalian cells have a battery of oxidation defense systems, including small molecules such as glutathione (GSH) and vitamin E that neutralize the oxidants, and enzymes specialized in oxidant detoxification, such as glutathione peroxidase, glutathione S-transferase and superoxide dismutase. Furthermore, there are oxidation damage repair systems present in almost all mammalian cells, including the glutaredoxin (Grx) and thioredoxin (Trx) systems. Both Grx and Trx belong to the large oxidoreductase family. The former is GSH-dependent and is specialized in dethiolating glutathionylated proteins (or protein-thiol mixed disulfide, PSSG) while the latter is NADPH-dependent and dethiolates protein-protein disulfide (PSSP).
Grx has two isoforms, the cytosolic Grx1 (also known as thioltransferase) and the mitochondrial Grx2 [5, 6]. Grx1 has been studied extensively and a number of key proteins/enzymes, including glyceraldehyde 3-phosphate dehydrogenase [7], protein tyrosine phosphatase 1B [8], HIV protease [9], P65 [10] and caspase-3 [11] have been identified as substrates for Grx1 dethiolase. The recently identified Grx2 also has a vicinyl cysteine (CSYC) at its active site, similar to other oxidoreductases, but less is known about its target proteins. Compared with Grx1, Grx2 has a higher affinity with glutathionylated proteins and is less likely to be inactivated by oxidation [5]. Recently Lillig et al. [12] have characterized Grx2 as an iron–sulfur containing protein, the first of such kind in the oxidoreductase family, and the Fe-S cluster bridges two Grx2 molecules through their two cysteine residues at the N-terminal end to form a stable and inactive dimer under GSH-rich environment. However, in the presence of oxidative stress or high GSSG environment, the dimer dissociates from the Fe-Cu cluster to form an active monomer. Therefore this iron-sulfur cluster can be considered as a redox sensor for Grx 2 [12]. Furthermore, our previous data also showed that Grx2 has GSH-dependent and thioredoxin reductase-dependent peroxidase activities [13], and that Grx2 can protect cells from oxidative stress-induced apoptosis by preserving the activity of complex I [14]. A recent study in another laboratory also reported that Grx2 can reduce selenium compounds like selenite and selenodiglutathione [15].
The eye lens is very sensitive to oxidative stress, and the combination of redox imbalance and protein modification can easily cause structural crystallin proteins to aggregate and deflect light, resulting in loss of transparency, known as cataract. Thus, the lens is a very good model to study oxidation and oxidation-induced protein damage and repair. In view of the importance of Grx in protecting lens proteins/enzymes from oxidative damage, we examined Grx2 function in the lens using primary cell cultures developed from Grx2 knockout (KO) and wild type (WT) mice. We found that although deletion of the Grx2 gene showed no phenotypical changes in the cells, the cells showed a low tolerance to oxidative stress. The KO cells exhibited a weak ability to detoxify oxidants when exposed to H2O2, which resulted in an accelerated loss of viability and membrane integrity, and shrinking pool size for ATP and GSH. Furthermore, the mitochondrial complex I in KO cells was heavily modified by glutathionylation and its activity was suppressed extensively. However an infusion of recombinant Grx2 into the KO cells was able to correct all these changes, suggesting that Grx2 may be an essential protector for the function of mitochondria.
MATERIALS AND METHODS
Materials
DMEM medium, fetal bovine serum (FBS), gentamicin, penicillin and 0.05% trypsin were all purchased from Gibco (Carlsbad, CA, USA). Hydrogen peroxide and all other chemicals were obtained from Sigma Chemical Company (St Louis, MO, USA) unless otherwise stated. Antibodies against αA-crystallin, GAPDH, complex I 75-kDa subunit (NDUFS1) and complex I 30-kDa subunit (NDUFS3) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). A complex I immunocapture kit, complex II immunocapture antibody and Complex II 30 kDa subunit (HDSB) monoclonal antibody were purchased from Mitosciences (Eugene, OR, USA). Anti-Glutathione monoclonal antibody (anti-PSSG antibody) was purchased from Virogen (Watertown, MA, USA). The recombinant protein A/G agarose beads were purchased from Exalpha Biological Inc. (Shirley, MA, USA). Glutathione ethyl ester was purchased from Sigma (St Louis, MO, USA). Sulfo-NHS-biotin and streptavidin-agrose beads were purchased from Thermo Fisher scientific (Fair Lawn, NJ, USA). 10% lauryl maltoside was obtained from Mitoscience (Eugene, OR, USA)
Purification of recombinant Grx2 and its mutants, and antibodies for Grx2
Mouse Grx2 was expressed in Escherichia coli and protein expression was induced with 0.4 mM isopropyl-1-thio-β-D-galactopyranoside in 200 ml of Lysogeny broth (LB) culture media. Cells were harvested by centrifugation at 10,000 g for 10 min and re-suspended in 5 ml of ice-cold protein extraction reagent (Novagen, Gibbstown, NJ, USA) containing 125 units of endonuclease. Cell debris was removed by centrifugation at 16,000 g for 20 min at 4°C. Grx2 protein was isolated by using a His-tagged protein purification kit (Novagen, Gibbstown, NJ, USA), following the manufacturer’s instructions.
Grx2 antibody was prepared as described previously [10]. Briefly, recombinant mouse Grx2 protein was purified and used to immunize a rabbit. The first booster injection was given 4 weeks later, followed by three more booster injections. Antiserum was collected at 2 weeks. The IgG fraction was then isolated using a protein A Sepharose column (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Generation of Grx2 Knockout Mice
A Grx2 global knockout mouse model was established in S-Y Ho’s laboratory, and details of the derivation will be described elsewhere. Briefly, exon 2 was deleted in the mutated Grx2 gene. The expressed Grx2 mRNA found in homozygous knockout mice is likely derived from fusion of exons 1, 3, and 4. This assumption was confirmed by sequencing DNA derived from RT-PCR of mutant Grx2 mRNA isolated from the heart of a homozygous knockout mouse. As a consequence, translation of protein from this mRNA would stop at exon 3, as the codons of exon 3 are not in-frame with those of exon 1. The resulting protein would contain only the mitochondrial translocation signal, which is encoded by exon 1, followed by 8 amino acids translated from the out-of-frame exon 3. This aberrant polypeptide is probably very labile and therefore less capable of preventing mRNA from degradation in cells, leading to a lower level of mutant Grx2 mRNA in tissues of homozygous knockout mice compared to that of wild type mice. Indeed, protein blot study showed an approximately 50% decrease of Grx2 protein in tissues of heterozygous knockout mice compared to those of wild type mice, and no Grx2 protein in the same tissues of homozygous knockout mice (Ho, unpublished results).
Primary mouse lens epithelial cell (LEC) cultures
Primary LEC cultures were established from four 2-week old WT or Grx2 KO mice in 129SV × C57BL/6 mixed backgrounds. Mouse lens capsules with attached epithelial layers were cut into small pieces and treated with 0.05% trypsin at 37°C for 10 mins. The cells loosened from the capsule were placed into a 24-well plate containing 1 ml DMEM with 20% FBS and 50 μg/mL gentamicin per well. The cultures were incubated for 1 week in a humid atmosphere with 5% CO2 at 37°C. Medium was changed every 3 days. After the primary cultures achieved confluence, the cells were subcultured by using 0.05% trypsin.
Mitochondria isolation
Mitochondrial fractions was isolated according to Christian et al. [16]. Briefly, mouse LECs were trypsinized and centrifuged. The cell pellets were suspended in 3 ml ice-cold mitochondria isolation buffer containing 0.2 M sucrose, 10 mM MOPS, 10 mM EGTA and 10 mM Tris-HCl (pH 7.4) and homogenized using a glass homogenizer, and followed by centrifugation at 600 g for 10 min. The supernatant was saved and centrifuged at 7,000 g for 10 min. Pellets were collected and washed with 200 μl of isolation buffer followed by centrifugation at 7,000 g for another 10 min. The final fraction enriched in mitochondria was re-suspended in isolation buffer and immediately used for measurement of Grx2 and complex I activities.
For isolating liver mitochondria, the liver was quickly removed from euthanized mouse, rinsed 3 times using ice-cold mitochondria isolation buffer (same buffer as above), minced into small pieces and homogenized with a glass-to-glass homogenizer. The homogenate was processed to extract mitochondria with successive centrifugation as described above. The large quantity of hepatic mitochondria was used for immunocapture studies for complex I, and complex II.
Western blot analysis
Equal amounts of protein were subjected to SDS-PAGE on a 12% polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Boulder, CO, USA). Detection was done using the ECL Western blotting detection system (Thermo Scientific, Rockford, IL, USA). The immunoblot was analyzed with an imaging system (Versadoc 5000 MP Imaging System, Bio-Rad, Richmond, CA).
Grx2 activity assays
Grx2 activity was assayed according to a previously described method [5]. Briefly, the reaction mixture contained 0.2 mM NADPH, 0.5 mM GSH, 0.1 M potassium phosphate buffer (pH 7.4), 0.4 units of GSSG reductase and an aliquot of mitochondrial fraction in a total volume of 1 ml. The reaction was carried out at 30 °C following a 5-min pre-incubation with 2 mM hydroxyethyldisulfide (HEDS). The decrease in absorbance of NADPH at 340 nm was monitored for 3 min using a Beckman DU 640 Spectrophotometer (Beckman, Fullerton, CA). To determine the Grx2 activity, the slope of the linear portion of the time course for 340-nm absorption loss in a control (Grx2-free) sample was subtracted from the slope of the samples containing Grx2.
Cytotoxicity assays
Cell viability was measured by a colorimetric cell viability kit (PromoKine, Heidelberg, Germany). The kit uses tetrazolium salts WST-8 that are reduced to water-soluble, orange formazan dye by dehydrogenases present in the metabolically active cells. Absorbance of the formazan dye is proportional to the number of viable cells. LECs were deprived of serum gradually by first culturing overnight in DMEM with 2% FBS followed by incubating in serum-free medium for 30 min. Then LECs were then treated with 100 μM of H2O2 for the indicated times. After treatment, the cells were incubated with 10 μl WST-8 solution at 37°C for another 1 hr. Transmission at 450 nm was evaluated using a 96-well microplate reader (Bio-Rad, Richmond, CA).
Lactate dehydrogenase (LDH) release assay
LDH activity was measured by using a cytotoxicity detection kit (Roche, Mannheim, Germany). The integrity of the cell membrane was measured by comparing level of LDH release into the medium with total LDH level. Total activity was defined as the maximum releasable LDH activity in the entire cell lysate. Wild type and Grx2 KO mouse LECs were gradually serum starved before exposure to a bolus of 100 μM H2O2 for 0-6 hrs. Cell lysates were centrifuged and supernatants were analyzed for LDH activity.
Caspase 3/7 assay
A caspase-Glo assay kit (Promega, Madison, USA) was used to detect caspase 3/7 activity. Cells (104) were plated onto a 96-well microplate. After H2O2 treatment, the cells were mixed with 100 μl caspase-Glo reagent, followed by incubation at room temperature for 30 mins. Luminescence was then measured by a microplate reader (BMG labtech).
DCF fluorescence in H2O2-treated wild type and Grx2 knockout mice
Lens epithelial cells from WT and Grx2 KO mice were washed with Ca2+- and Mg2+-free phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, MO), and incubated with PBS containing 50 μM DCFH-DA. After 5 min the cells were again washed twice with PBS before determination of the basal DCF fluorescence level. After a baseline was acquired, 50 μM H2O2 was added to all cell groups and DCF fluorescence levels were determined at given time points. The intensity of fluorescence was detected with a Floustar Optma microplate reader (BMG Labtech, Offenburg, Germany).
GSH and ATP determinations
For GSH measurement, an aliquot of fresh cell lysate was treated with an equal volume of 20% trichloroacetic acid followed by centrifugation. The supernatant was used immediately for GSH analysis using DTNB reagent [17], following the method of Lou et al. [18].
ATP level was measured by an ATP bioluminescence assay kit CLS II (Roche, Mannheim, Germany) according to the manufacturer’s recommendation. Briefly, cells were diluted with PBS to 108 cells/ml cell suspensions, into which 9 volumes of boiling solution containing 100 mM Tris and 4 mM EDTA were added and incubated for 2 minutes at 100°C. The cell suspension was centrifuged at 1,000 g for 1 min, and 50 μl of the sample was mixed with 50 μl luciferase reagent by automated injection. The luminescence intensity was detected by a Floustar Optma microplate reader (BMG labtech, Offenburg, Germany, integrating for 1 to 10 seconds.
Protein glutathionylation detection
Purified WT mouse Grx2 and Grx2-C70S/C73S double mutant (20 μM) were loaded into LECs from Grx2 KO mice by using protein transfection reagent as previously described [10]. The cells were then treated with or without 1 mM H2O2 for 30 min and lysed with RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate and protease inhibitor cocktail). Mitochondrial fractions were isolated by using the method described above. Equal amounts of protein (40 μg) in each group were loaded onto an SDS-PAGE gel under non-reducing conditions and probed with an anti-PSSG antibody.
Complex I activity assays
The enzymatic activity of complex I was assayed using the sensitive spectrophotometric method developed recently [19]. In brief, 50 μg of mitochondrial proteins isolated from mouse LEC was mixed with 960 μl reaction mixture containing 20 mM KH2PO4, 3.5 mg/ml BSA, 60 μM 2,6-dichloroindophenol (DCIP), 70 μM decylubiquinone (prepared in dimethyl sulfoxide, DMSO), and 1 μM antimycin-A at 30°C. After 3 min, 20μl of 10 mM NADH was added and the absorbance was measured at 30 sec intervals for 4 min at 37°C. After that, 1.0 μl rotenone (1 mM, in DMSO) was immediately added and the absorbance was measured again at 30 sec intervals for additional 4 min to confirm it is the rotenone-sensitive complex I activity. The enzyme activity was expressed as nmol DCIP reduced per min/mg protein. The substrate DCIP, which is a water-soluble final electron acceptor, is specific for complex I since DCIP receives electrons only from complex I, and not from other non-mitochondrial NADH dehydrogenases. BSA is a necessary component in the assay mixture since it acts as a solubilization agent for rotenone and decylubiquinone.
Immunocapture of Complex I and complex II
In order to detect the glutathionylation of complex I or II, a commercial immunocapture kit was used to pull down target proteins according to the manufacturer’s instructions. The recommended starting amount was 5 mg mitochondrial protein. Such large quantity of protein is extremely difficult to obtain from primary lens epithelial cells, and thus liver mitochondrial proteins were used for this study. Hepatic mitochondrial proteins from a Grx2 KO mouse were pre-incubated with 20 μM recombinant mouse Grx2 proteins and 5 mM GSH for 10 min. After that, mitochondrial proteins from WT or Grx2 KO mice were stimulated with or without 1 mM H2O2 for 30 min, followed by washing twice with cold mitochondria isolation buffer. The immunocapture kit for complex I or II was then added to the samples, and immunocaptured products were separated under non-reducing conditions by SDS-PAGE and probed with a specific anti-PSSG antibody.
Detecting Glutathionylated Complex I by Biotinylated GSH ethylester (BioGEE)
Biotinylated GSH ethylester (BioGEE) was made by incubating 25 mM of GEE (Sigma, St Louis, MO, USA) with 25 mM sulfo-NHS-biotin (Thermo fisher scientific, Fair Lawn, NJ, USA) in 50 mM NaHCO3 (pH 8.5) at room temperature for 1 hr following the previously published method [20]. The biotinylating reaction was terminated by adding 1.25 mM NH4HCO3 (pH 8.5) followed by a 1 hr incubation at room temperature to quench the un-used biotinylation reagents. LECs (80% confluent) either from WT or Grx2 KO mice were cultured overnight in DMEM with 2% FBS followed by incubating in serum-free medium for 30 min before exposing to 250 μM BioGEE for 1 hr. The cells were then treated with or without 100 μM H2O2 for 6 hrs, and washed with cold PBS containing 50 mM N-ethylmalemide (NEM) to scavenge remaining BioGEE. The mitochondrial fraction of BioGEE-treated cells was purified by using mitochondria isolation buffer containing 0.2 M sucrose, 10 mM MOPS, 10 mM EGTA, 10 mM Tris-HCl (pH 7.4) and 50 mM iodoacetamide (IAM). Equal amounts of mitochondrial protein was treated with 1% lauryl maltoside for 30 mins followed by centrifugation at 16,000 g for 30 mins to pre-clear the sample. The supernatant was collected and proteins covalently bound to biotin were captured by binding to streptavidin-agrose beads (Thermo fisher scientific, Fair Lawn, NJ, USA) at 4°C for 1 hr, and the beads were washed 3 times with 1 ml washing buffer (20 mM Tris-HCl pH 7.5, 137 mM NaCl, 2 mM EDTA and 10% glycerol). Glutathionylated mitochondrial proteins in the BioGEE-treated samples were released from the beads by incubating with 50 μl of non-denaturing lysis buffer (20 mM Tris-HCl, pH 8, 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA and 10 mM DTT) for 15 mins with gentle shaking at 4°C. Samples released from streptavidin-Agrose beads were separated by SDS-PAGE and the glutathionylated 75-kDa subunit of complex I was detected by immunoblotting with polyclonal anti- NDUFS1 (75-kDa subunit of complex I) antibody (Santa Cruz, CA, USA).
Statistics
Each experiment was performed at least three times and statistical analyses were performed using one-way ANOVA followed by Tukey’s test as a post hoc test with the SPSS software. The number of experimental samples used in each group was presented in the figure legends. All data were expressed as means ± S.D. and differences were considered significant at P < 0.05.
RESULTS
Characterization of wild type (WT) and Grx2 knockout (Grx2 KO) primary mouse lens epithelial cells (LECs)
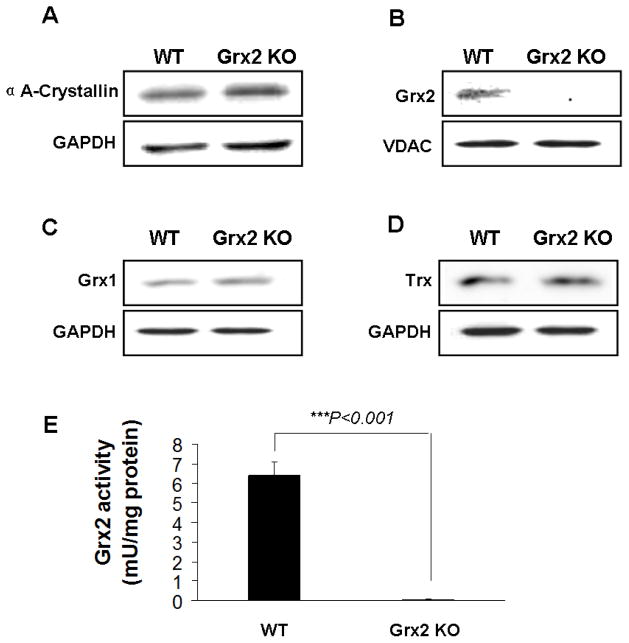
Primary cultures of WT and Grx2 KO lens epithelial cells were used to study the physiological role of Grx2. The cells were confirmed to be lens specific by the presence of αA-crystallin protein (Fig. 1A). Immunoblotting also showed that Grx2 was expressed in the mitochondrial fraction of WT but absent in that of the KO cells (Fig. 1B). Cytosolic glutaredoxin (Grx1 or thioltransferase) and thioredoxin (Trx) expressions were equally detectable in both types of cells (Figs. 1C, 1D), indicating that this KO model is specifically targeted to Grx2 and does not affect expression of other thioldisulfide oxidoreductases. Grx2 activity was also examined in both cell types. As shown in Figure 1E, Grx2 enzyme activity in the mitochondrial fraction from WT cells was 6.4±0.4 mM/mg protein while no activity could be detected in Grx2 KO cells.
Fig. 1. Characterization of wild type (WT) and Grx2 knockout Grx2 KO) primary mouse lens epithelial cells (LEC)s.
Primary mouse LECs were isolated from WT and Grx2 KO lens epithelial layers. (A) Expression of αA-crystallin in wild type and Grx2 knockout LECs. Whole cell lysates (40 μg) were obtained from wild type or Grx2 knockout mouse LECs and analyzed by immunoblot by using anti-αA- crystallin antibody. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Detection of Grx2 in WT and Grx2 KO mouse LECs. 40 μg of mitochondrial isolated from WT and Grx2 KO cells were analyzed by Western blot with anti-Grx2 antibody. Voltage-dependent anion channels (VDAC) was used as both a mitochondrial marker and loading control. (C) Expression of glutaredoxin 1 (Grx1) in WT and Grx2 KO LECs. 40 μg of whole cell lysates obtained from WT or Grx2 KO cells were analyzed by immunoblot by using anti-glutaredoxin antibody. GAPDH was used as a loading control. (D) Expression of thioredoxin (Trx) in WT or Grx2 KO LECs. 40 μg of whole cell lysates obtained from wild type or Grx2 knockout cells were analyzed by immunoblot by using anti-thioredoxin antibody. GAPDH was used as a loading control. (E) Grx2 activity in wild type and Grx2 knockout LECs. Mitochondrial fraction isolated from WT or Grx2 KO cells were used to detect Grx2 activity as described under “Materials and methods”. Error bars indicate S.D., n=3, *P<0.05 vs. WT.
Effect of Grx2 deletion on H2O2-induced LEC injury
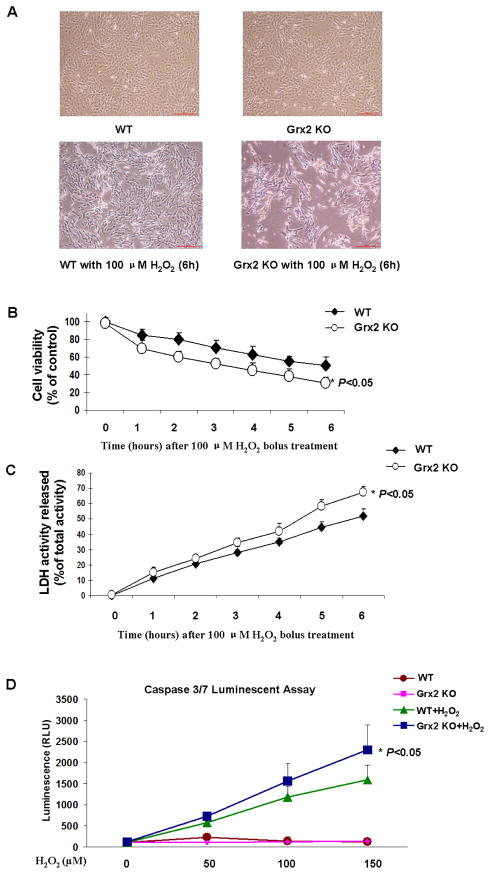
To test if deletion of Grx2 increases LEC sensitivity toward oxidative stress, we treated the cells with a bolus of H2O2 and cell morphology was observed under phase-contrast microscopy. Based on our preliminary studies with various doses of H2O2 (50 μM to 500 μM) for the treatment, we observed that 100 μM H2O2 for 6 hrs provided the best results (data not shown) and thus this concentration was used to induce LEC injury. As shown in Fig. 2A, WT and Grx2 KO cells showed no obvious differences when cultured under normal culture conditions. Both types of cells displayed typical morphological characteristics of epithelial cells, including tight packing and cuboidal shape. Following exposure to 100 μM H2O2 for 6 hrs, Grx2 KO cells showed a marked decrease in number with shrinkage in size and loss of adhesion as compared with H2O2-treated WT cells, indicating that Grx2 depletion in LECs results in more severe damage from oxidative stress.
Fig. 2. Effect of H2O2 in wild type (WT) and Grx2 knockout (Grx2 KO) lens epithelial cells.
(A) Comparison of the cell density and morphology after H2O2 treatment. WT and Grx2 Grx2 KO primary mouse LECs were treated with or without 100 μM H2O2 for 6 hrs. After treatment, cell morphology was observed under phase contrast microscopy. (B) Effect of H2O2 on cell viability in WT and Grx2 KO cells. WT and Grx2 KO primary mouse LECs were plated in 96-well plate (all normalized to 10,000 cells/well) and treated with or without 100 μM H2O2 at for indicated times. Cell viability in each time point was measured by adding WST-8 reagent and then the transmission was evaluated under 450 nm using a 96-well microplate reader. Error bars indicate S.D., n=3, *P<0.05 vs. WT treated with H2O2. (C) Effect of H2O2 on LDH release in WT and Grx2 KO cells. Wild type and Grx2 knockout primary mouse LECs were treated with or without 100 μM H2O2 at different time point from 0 to 6 hours. LDH activity was measured as described as under “Materials and methods”. Error bars indicate S.D., n=3, *P<0.05 vs. WT treated with H2O2. (D) Effect of H2O2 on caspase 3/7 activation in WT and Grx2 KO cells. Wild type and Grx2 knockout primary mouse LECs were plated in 96-well plate (all normalized to 10,000 cells/well) and treated with 0, 50, 100 and 150 μM H2O2, respectively for 6 hrs. Caspase 3/7 activity was measured by using luminogenic substrate containing the DEVD sequence. Error bars indicate S.D., n=3, *P<0.05 vs. WT treated with H2O2.
To further evaluate the functional effect of Grx2 deletion on oxidatively stressed LECs, cell viability was determined by WST-8 assay. As shown in Fig. 2B, the cell viability initially showed no difference between the WT and Grx2 KO cells, but 100 μM H2O2 treatment (0, 1, 2, 3, 4, 5 and 6 hrs) caused a more extensive viability loss in KO cells than that of the WT cells. At the end of a 6-hr H2O2 treatment, 50.3±9.8% of WT cells survived but only 30.4±7.0% of KO cells was viable.
H2O2-induced cell damage due to Grx2 gene deletion was also examined by quantifying leakage of lactate dehydrogenase (LDH) to assess the integrity of cell membrane. As shown in Figure 2C, upon H2O2 stress, both cell types showed a time-dependent LDH leakage, which was more severe in the KO cells than that of the WT control cells.
Malfunction of mitochondria is known to lead to cell apoptosis [21]. Oxidative stress can induce apoptosis via caspase 3/7 activation [22]. To examine if Grx2 deletion can accelerate this apoptotic pathway, we examined the WT and KO cells with and without H2O2 treatment. As shown in Figure 2D, untreated WT and Grx2 KO cells both have no caspase3/7 activation; however, when the cells were treated with an increment dosage of H2O2 (50, 100 and 150 μM) for 6 hrs, both cell types showed a dose-dependent activation of caspase 3/7, which was higher in Grx2 KO cells than WT cells. These data further indicate that Grx2 deletion can lead to increased apoptosis in stressed LECs.
Effect of Grx2 depletion on H2O2 detoxification in mLECs
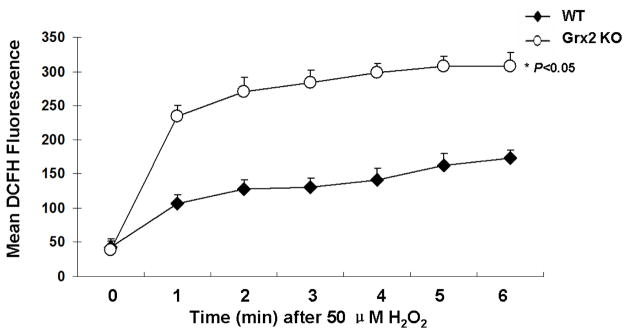
Since Grx2 is known to possess peroxidase activity [13], we examined the rate of detoxification of H2O2 in Grx2 KO and WT LECs. To ensure that the dose of H2O2 would not cause a fluorescence overload, we chose a moderate concentration of 50 μM for this study. As shown in Figure 3, the intensity of the basal DCF fluorescence (a marker for intracellular ROS) is equally low in both WT and Grx2 KO cells at zero time. Upon H2O2 treatment, fluorescence gradually increased in both, but a much higher elevation was seen for the Grx2 KO cells during the course of 6 min of study. These data indicate that Grx2 deletion decreased the LECs’ ability to detoxify H2O2.
Figure 3. H2O2 detoxification in wild type and Grx2 knockout lens epithelail cells.
Wild type (WT) and Grx2 knockout (Grx2 KO) primary mouse LECs were plated in 96-well plate (all normalized to 10,000 cells/well) and treated with PBS containing 50 μM DCFH-DA. After a baseline was acquired, 50 μM H2O2 was added to the cells and DCF fluorescence levels were determined at given time points. Error bars indicate S.D., n=6, *P<0.05 vs. WT.
Effect of Grx2 deletion on the levels of free GSH and bound GSH (PSSG) in LECs under oxidative stress conditions
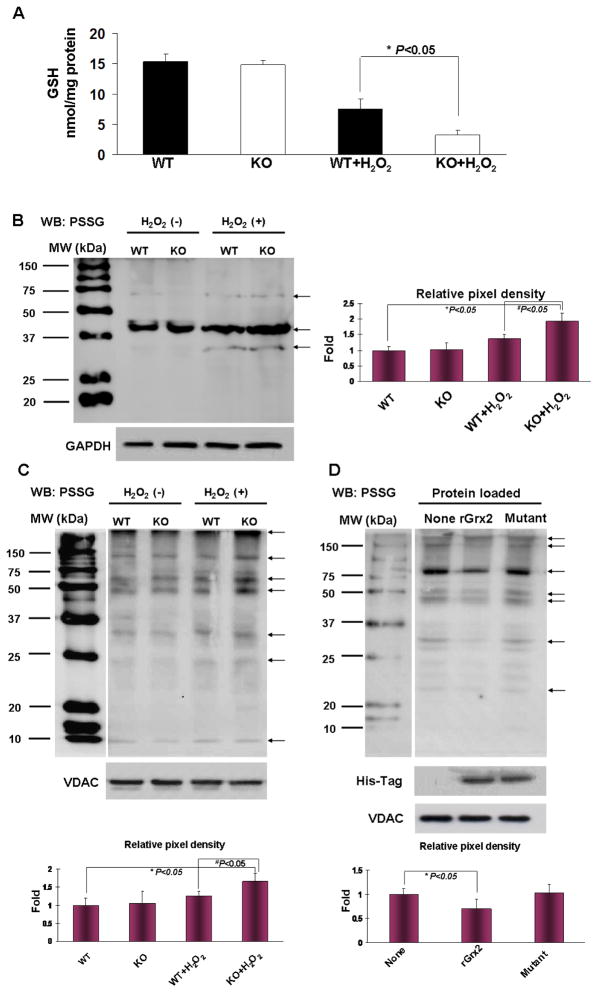
Since GSH is an important antioxidant in the lens [4] and also a cofactor for Grx2 dethiolase activity [5], we examined levels of free and protein-bound GSH in LECs before and after H2O2 treatment. Free GSH levels were similar in both WT and KO cells (15.3±1.2 nmol/mg protein WT, 14.8±0.8 nmol/mg protein KO) under stress-free conditions (Fig. 4A). However, when cells were treated with H2O2 (100 M for 6 hrs), the GSH concentration of Grx2 KO cells was depleted nearly 80% to 3.2±0.7 nmol/mg protein while the WT cells were able to maintain half of GSH at a level of 7.5±1.7 nmol/mg protein.
Figure 4. Free GSH and glutathionylated proteins (PSSG) in wild type (WT) and Grx2 knockout (KO) lens epithelial cells after H2O2 treatment.
(A) GSH level in WT and Grx2 KO lens epithelial cells (LECs). WT and Grx2 KO primary mouse LECs were treated with or without 100 μM H2O2 for 6 hrs. Cells were lysed in lysis buffer and GSH concentrations in cell lysates were determined using Ellman reagent described under “Material and methods”. Error bars indicate S.D., n=6, *P<0.05 vs. WT treated with H2O2. (B) PSSG levels in whole cell lysates in WT and Grx2 KO LECs. Wild type and Grx2 knockout were treated with or without 1 mM H2O2 for 30 min. Then the cells were lysed in lysis buffer and the total cell lysates were immunoblotted with anti-GSH (PSSG) antibody under non-reducing conditions. GAPDH was used as a loading control. The right panel depicts the relative pixel density of all the PSSG bands (see arrows) over GAPDH (with the untreated WT normalized to 1.0). Data presented are a typical representation of triplicate experiments. *P<0.05, vs. WT LECs. #P<0.05, vs WT LECs treated with 1 mM H2O2 for 30 min. (C) PSSG levels in mitochondrial fraction of WT and Grx2 KO cells. Cells in (B) were used for mitochondrial isolation and the mitochondrial extracts were used for Western blot (WB) detection for PSSG. Mitochondrial protein VDAC was used as a loading control. The bottom panel depicts the relative pixel density of all the PSSG bands (see arrows) over VDAC (with the untreated WT normalized to 1.0). Data presented are a typical representation of triplicate experiments. *P<0.05, vs. WT LECs. #P<0.05, vs WT LECs treated with 1 mM H2O2 for 30 min. (D) Deglutathionylation of mitochondrial PSSG with imported Grx2. Grx2 KO cells were imported with 20 μM purified wild type Grx2 recombinant protein (rGrx2), Grx2-C70S/C73S double mutated protein (Mutant). After 4 hours of incubation, the cells were treated with 1 mM H2O2 for 30 min. After treatment, the mitochondrial fraction of each group was analyzed for PSSG. His-Tag immunoblotting was done to verify the delivery of recombinant Grx2 protein. The bottom panel depicts the relative pixel density of all the PSSG bands (see arrows) over VDAC (with the KO LECs without import normalized to 1.0). Data presented are a typical representation of triplicate experiments. *P<0.05, vs. KO LECs treated with 1 mM H2O2 for 30 min.
Next we examine the effect of H2O2 stress on protein glutathionylation in LEC cells. As the protein glutathionylation process is relatively fast, we chose to treat the cells with a higher concentration of H2O2 (1 mM) for 30 min to compare the oxidation-induced PSSG formation in WT and KO lens epithelial cells in the following studies. PSSG content was measured in cell lysates by immunoblotting with anti-PSSG antibody. As shown in Figure 4B, untreated WT and KO LECs showed no difference in the basal levels of PSSG; however, cells treated with H2O2 (1 mM, 30 min) displayed an enhanced PSSG formation, especially in the KO cells, indicating that Grx2 gene knockout can also affect non-mitochondrial proteins.
Because Grx2 is located in the mitochondria, we also examined PSSG formation in the mitochondrial fraction of both cell types. As shown in Figure 4C, levels of PSSG in WT and KO cells were equally low under un-stressed conditions; however, H2O2 stress (1 mM, 30 min) raised PSSG levels extensively in Grx2 KO cells over that of WT. These data strongly suggest that some of the target proteins of Grx2 are located in the mitochondria.
Next, we used recombinant protein of Grx2 (rGrx2) or a double mutation of Grx2 at its active site (mutant) to examine if oxidation-induced PSSG elevation in the mitochondrial fraction of LECs can be corrected. The recombinant proteins were delivered using a BioPorter system (Gene Therapy Systems, San Diego, CA), following the previous procedure [23]. Figure 4D clearly shows that imported rGrx2, but not its imported mutant, can substantially reduce PSSG levels in the mitochondria of stressed LECs, indicating the specificity of this dethiolating function of Grx2. The successful delivery of rGrx2 or mutant Grx2 into the mitochondrial fraction was confirmed by observing the presence of His-Tag on the recombinant proteins, which was positively reacted to Hig-Tag antibody (Fig. 4D). The respective cytosolic and mitochondrial proteins, GAPDH and VDAC, were used for Western blotting to ensure loading of equal amounts of protein (Fig. 4).
Effect of Grx2 deletion on ATP level and complex I activity in LECs
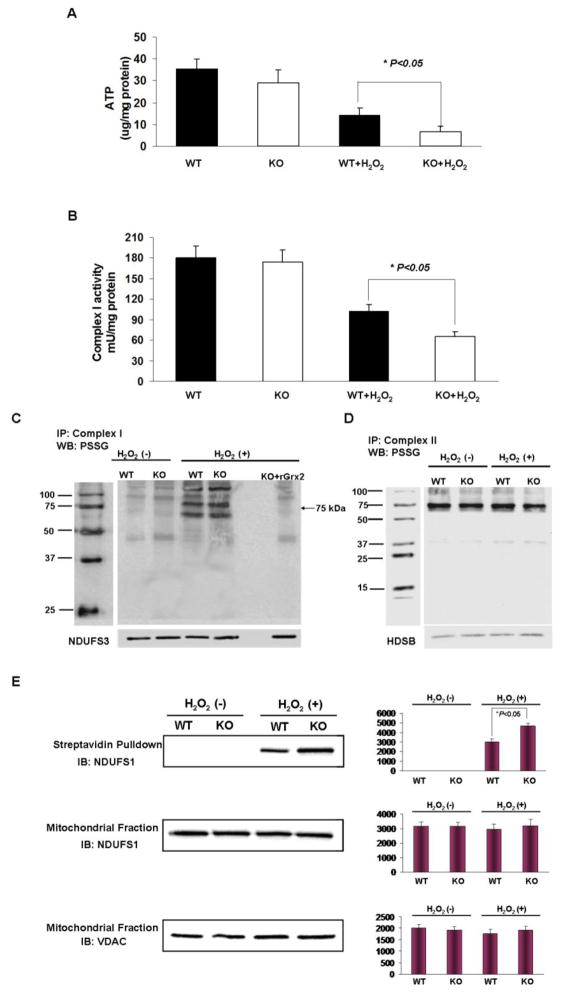
The major function of mitochondria is to carry out molecular oxygen reduction coupled with ATP production through the electron transport system (ETS). Therefore, ATP production is a very important index of mitochondrial function. To test whether Grx2 deletion affects mitochondrial energy homeostasis in LECs, we compared ATP levels in WT and Grx2 KO cells before and after H2O2 challenge (100 μM, 6 hrs). The results in Figure 5A indicates that there was no significant difference in the basal ATP levels of the two types of unstressed cells, but H2O2 treatment suppressed 60% of ATP production in the WT cells (from 35.4±4.7 μg/mg protein to 14.2±3.3 μg/mg protein), and over 80% in the KO cells (from 29.2±5.9 μg/mg protein to 6.7±2.7 μg/mg protein).
Figure 5. Protective effect of Grx2 against complex I dysfunction induced by H2O2.
(A) ATP level in wild type and Grx2 knockout LECs. Wild type (WT) and Grx2 knockout (Grx2 KO) primary mouse LECs were treated with or without 100 μM H2O2 for 6 hrs. ATP level in each group was determined using ATP assay kit. The data are expressed as mean±S.D., n=6, *P<0.05 vs. WT treated with H2O2. (B) Complex I activity in wild type and Grx2 knockout LECs. The mitochondrial lysate from (A) was used to detect complex I activity. The data are expressed as mean±S.D., n=6, *P<0.05 vs. WT treated with H2O2. (C) Glutathionylation and de-glutathionylation of complex I in wild type and Grx2 knockout mouse. Liver mitochondrial lysates (5 mg) isolated from WT or Grx2 KO mouse were incubated with or without 20 μM purified mouse Grx2 recombinant protein (rGrx2) and 5 mM GSH for 10 min. Then, mitochondrial lysates were treated with or without 1 mM H2O2 for 30 min. After treatment, mitochondrial lysates were washed twice with cold mitochondria isolation buffer. Agarose beads irreversibly cross-linked to complex I monoclonal antibodies were added. Immunocaptured complex I in each group was used for Western blot (WB) detection of PSSG. A specific antibody to 30 kDa subunit of complex I (NDUFS3) was used as a loading control. The position of 75 kDa subunit of complex 1 is indicated by an arrow. (D) Glutathionylation of complex II in wild type and Grx2 knockout mouse. Mitochondrial lysates were treated with or without 1 mM H2O2 for 30 min and complex II was pulled down by using complex II antibody. Immunocaptured complex II from each group was used for Western blot (WB) detection of PSSG. HDSB was used as a loading control. IP: mmunoprecipitation; MW: molecular weight (E) Wild type (WT) and Grx2 knockout (KO) primary mouse LECs were preloaded with Biogee for 1 hour. Then the cells were treated with or without 100 μM H2O2 for another 6 hrs. Streptavidin extracts from the mitochondrial proteins were resolved by SDS-PAGE and hybridized with NDUFS1, a specific antibody against 75 kDa subunit (upper panel). NDUFS1 protein level was detected in the mitochondria fraction (middile panel). VDAC was used as a loading control (lower panel). The bar graph with average pixel density for each western blot is shown. IB: Immunoblotting.
Since complex I is the main entry point of ETS, we next examined whether the ATP loss was associated with loss in complex I activity, and whether Grx2 deletion causes a differential complex I inactivation in WT and Grx2 KO cells. As shown in Figure 5B, basal complex I activity is similar in WT and Grx2 KO LECs (180.0±14.2 mU/mg protein verses 174.2±19.8 mU/mg protein). After H2O2 treatment, complex I activity lost 40% in WT cells (102.1±14.2 mU/mg protein), and more than 60% in Grx2 KO cells (66.4±8.7 mU/mg protein).
Grx2 deletion enhances accumulation of glutathionylated complex I in mouse LECs and liver mitochondria under oxidative stress
We speculate that the oxidation-induced complex I inactivation shown above may associated with glutathionylation (or PSSG formation) at active sites of one or more of its key subunits that are sensitive to oxidation. Therefore, we examined the status of complex I glutathionylation in KO and WT mouse mitochondrial models before and after oxidation (100 μM H2O2 for 6 hrs). PSSG in complex I was examined using two methods. One was an in vitro study with isolated mitochondria in which the complex I protein complex was isolated by immunocapture pull-down (Mitosciences, Eugene, OR, USA), followed by immunoblotting with anti-PSSG antibody. Since the pull-down experiment requires a large quantity of mitochondria, we used liver mitochondria for this study. The other method was an in vivo amplification of oxidation-induced PSSG formation in Grx2 KO and WT LECs by delivering GSH through biotinated glutathione ethyl ester (BioGEE). Figure 5C shows that under un-stressed conditions, complex I glutathionylation is equally low in both liver mitochondrial preparations (WT and Grx2 KO), however, after oxidative stress, the level of PSSG in complex I is much higher in the liver sample from Grx2 KO mice than that of the WT mice. Our data also showed in Figure 5C that although the complex I subunit at 75-kDa does not have detectable PSSG in both liver samples under non-stressed conditions, a heavy band appeared at the 75-kDa position (see arrow) in both groups after H2O2 stimulation, and the intensity was much higher in the Grx2 KO sample. The PSSG in Grx2 KO could be eliminated when the mitochondrial protein fraction was pretreated with recombinant Grx2 (right column in Fig. 5C). This result suggests that the observed elevated glutathionylation at 75-kDa was a direct result of Grx2 deletion.
To ensure that oxidation-induced glutathionylation in mouse liver mitochondria occurred specifically in complex I, we used complex II pull-down for comparison. As shown in Figure 5D, complex II glutathionylation was seen but was not affected by oxidative stress, and the absence of Grx2 did not increase the level of PSSG in the complex II-associated proteins.
To confirm that oxidation-induced PSSG formation in complex I also occurs in the LECs, we used a BioGEE approach. Biotin-conjugated GSH ester can be delivered easily into WT or Grx2 KO LECs and subsequently hydrolyzed into biotinylated GSH intracellularly. An oxidation-induced biotinylated PSSG complex can be pulled down by streptavidin-agrose beads for immunoblot analysis by NDUFS1, a specific antibody for the 75-kDa protein. As shown in Figure 5E, no obvious glutathionylation in the 75-kDa of complex I could be seen under normal culture conditions. However, after exposing to H2O2 (1 mM, 30 min), KO cells showed much more elevated PSSG in the subunit of complex 1 than WT cells.
DISCUSSION
This study with cultured primary lens epithelial cells demonstrated that Grx2 is critical for cell survival when the cells are subjected to oxidative stress. This conclusion is based on the findings that although LECs from Grx2 KO mice showed comparable morphological and biochemical properties as that of the WT cells under unstressed conditions, cells with Grx2 deletion had difficulty coping with oxidative stress (H2O2). The stressed KO cells displayed weakened viability with signs of apoptosis and disrupted mitochondrial function, such as lowered mitochondrial ATP and GSH pools, enhanced complex I inactivation and apoptotic caspase 3/7 activation. Furthermore, Grx2 absence also impaired the cells’ ability to detoxify H2O2, resulting in a marked increase in the level of glutathionylated proteins in mitochondria. These deviations from WT cells are likely a direct result of a lack of both the dethiolase and peroxidase functions of Grx2, since importing recombinant Grx2 into Grx2 KO cells rescued and corrected many of the above abnormalities. The current findings are in agreement with our earlier report, in which Grx2 over-expression in a human lens epithelial B3 cell line protected the cells against H2O2-induced apoptosis, while suppression of Grx2 expression with siRNA showed an opposite effect [23].
It is interesting to note that although Grx 2 is an isozyme of Grx1, the phenotypic change associated with Grx 2 deletion was relatively minor in comparison with lens primary epithelial cell cultures isolated from Grx1 knockout mice. Under unstressed conditions, and compared to cells from WT mice with a same background, cells with Grx2 deletion were essentially the same. Cells with Grx1 deletion on the other hand showed a phenotype in both morphological and biochemical alterations; the cells were slower in growth, showed a resistance to spread and contained less free GSH but more protein-bound GSH [24]. However, both mutant cells showed an elevated sensitivity to oxidative stress. This discrepancy between the two Grxs may be explained by the fact that Grx1 has many cellular functions, including regulation of NF-kB, actin polymerization and apoptosis. Thus, deletion of Grx1 would affect cellular physiology on many fronts. On the other hand, Grx2 phenotypical change observed only under oxidative stress The reason for this difference is unclear, but likely associated with the unique property of Grx2, which in a normal, reduced environment, it forms an inactive dimeric complex with a [Fe-S] cluster while the oxidation-sensitive Fe-S cluster would dissociate Grx2 from the complex and convert it into an active monomer to exert protective function [25].
What causes cells to be sensitive to oxidative stress is an intriguing question. Many proteins/enzymes lose activities or biological functions when critical cysteine moieties on the structure are reversibly oxidized, resulting in the formation of protein-thiol mixed disulfides, for example, protein glutathionylation (PSSG), thus affecting cellular functions in general. The dethiolation properties of Grx1 and Grx2 are clearly essential for the rescue of such cellular injury. In a previous report, we showed that importing recombinant Grx1 into LECs made it possible to rescue H2O2-induced activity loss for the ATP-producing enzyme glyceraldehyde-3 phosphodehydrogenase (GAPDH), and assisted the mutant cells in detoxifying H2O2 after a bolus addition of this oxidant [26]. The current study showed that Grx2 could prevent H2O2-induced cell apoptosis, and that rescuing the activity of H2O2-inactivivated complex I in the mitochondria with imported Grx2 was an essential event. Furthermore, only nascent Grx2 protein, and not C70S or C73S mutant protein, was effective [23].
Based on the current results, it is likely that glutathionylation and inactivation of complex I or other key enzymes in the mitochondria may play a major role regarding the pathological state of the cells. Impaired complex I can trigger ROS generation, which can cause further damage to mitochondrial structure, affecting its function and triggering activation of the apoptotic pathway, resulting in apoptosis. Our data clearly showed that oxidation induced more PSSG in total proteins as seen by Western blot analysis (Fig. 4B), and results from immunocaptured glutathionylated-mitochondrial proteins were enhanced in H2O2-treated cells, particularly in H2O2-treated Grx2 KO cells (Fig. 4C). Therefore, deletion of Grx2 gene appears to affect more than just the mitochondrial proteins. The glutathionylated protein bands were validated by their removal with nascent Grx2 protein, but not with Grx2 mutant protein (Figure 4C, D). It is worth noting that the pattern of PSSG bands in the mitochondrial fraction of H2O2-treated KO cells in Figure 4C (far right lane) and in Figure 4D (far left lane) showed differential intensity, likely due to the difference in the quality of isolated mitochondria in separate preparation. Regardless, this difference does not diminish the impact of Grx2 gene deletion in elevated glutathionylated proteins found in the mitochondria.
We further demonstrated in this study that H2O2 not only induced complex I activity loss, but also enhanced glutathionylation at the 75-kDa subunit, the key unit of complex I (arrow in Fig. 5C, and Fig. 5E). This strongly indicates that glutathionylation of complex I in Grx2 KO cells is due to the cells’ inability to dethiolate the conjugates and restore complex I to its reduced state. It is certainly possible that other mitochondrial enzymes/proteins, besides complex I, may be affected by this process in the same way, and collectively contribute to cellular injury. Indeed, other glutathionylated protein bands can be seen in Figure 4C and Figure 5C. Work is underway to characterize these modified target proteins.
Glutathionylation of complex I subunits 75-kDa and 51 kDa is known to occur. As reported by Murphy and associates [27] oxidized glutathione (GSSG)-induced glutathionylation in isolated complex I and led to extensive loss in activity, while recombinant Grx2 could restore this activity loss. Chen et al. [28] also demonstrated the same type of result. Our results for the glutathionylated 75-kDa protein in complex I, using both an immunocapturing technique and a biotin-GEE approach, certainly agree with their findings.
It is also worth noting that Grx2 function appears to be specific for complex I since an enhanced glutathionylated band for complex I showed a differential effect for Grx2 deletion. However, the same analysis used for complex II showed that the glutathionylation level of complex II was not changed under oxidative stress conditions, and there was no obvious difference in glutathionylation level between Grx2 knockout and WT cells (Fig. 5D). This result is consistent with our previous data in which Grx2 was found to bind with complex I, but not with complex II [23]. Thus, our current findings strongly suggest that Grx2 can modulate the mitochondrial complex I by reversible glutathionylation.
In summary, we have provided strong evidence that Grx2 is critical for protecting LECs against oxidative damage. The potential protective effect of Grx2 is associated with its ability to prevent complex I glutathionylation induced by oxidative stress, and thus regulate energy homeostasis and maintain normal mitochondrial biological function. Similar protective properties of Grx2 may also be true in cells other than the lens epithelial cells.
Highlights.
The cellular function of glutaredoxin 2 (Grx2) was studied using primary lens epithelial cell culture from Grx2 knockout mouse. In comparison with the wild type cells, Grx2 knockout results in no obvious phenotype under non-stressed condition. With H2O2 treatment, Grx2 knockout cells show a lower tolerance to oxidative stress. Grx2 deletion induces more glutathionylation in mitochondrial proteins and less ATP production. Imported Grx2 prevents complex I inactivation and preserves mitochondrial function.
Footnotes
Part of the work was presented at the Annual meeting for the Association of Research in Vision and Ophthalmology, May 2-7, 2010, Fort Lauderdale, FL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 2.Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, Floyd RA, Markesbery WR. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci U S A. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 4.Lou MF. Redox regulation in the lens. Prog Retin Eye Res. 2003;22:657–682. doi: 10.1016/s1350-9462(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 5.Gladyshev VN, Liu A, Novoselov SV, Krysan K, Sun QA, Kryukov VM, Kryukov GV, Lou MF. Identification and characterization of a new mammalian glutaredoxin (thioltransferase), Grx2. J Biol Chem. 2001;276:30374–30380. doi: 10.1074/jbc.M100020200. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–26275. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 7.Xing K, Lou MF. The possible physiological function of thioltransferase in cells. FASEB J. 2003;17:2088–2090. doi: 10.1096/fj.02-1164fje. [DOI] [PubMed] [Google Scholar]
- 8.Barrett WC, DeGnore JP, Konig S, Fales HM, Keng YF, Zhang ZY, Yim MB, Chock PB. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 9.Davis DA, Newcomb FM, Starke DW, Ott DE, Mieyal JJ, Yarchoan R. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J Biol Chem. 1997;272:25935–25940. doi: 10.1074/jbc.272.41.25935. [DOI] [PubMed] [Google Scholar]
- 10.Liao BC, Hsieh CW, Lin YC, Wung BS. The glutaredoxin/glutathione system modulates NF-kappaB activity by glutathionylation of p65 in cinnamaldehyde-treated endothelial cells. Toxicol Sci. 2010;116:151–163. doi: 10.1093/toxsci/kfq098. [DOI] [PubMed] [Google Scholar]
- 11.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 12.Lillig CH, Berndt C, Vergnolle O, Lonn ME, Hudemann C, Bill E, Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci U S A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernando MR, Lechner JM, Lofgren S, Gladyshev VN, Lou MF. Mitochondrial thioltransferase (glutaredoxin 2) has GSH-dependent and thioredoxin reductase-dependent peroxidase activities in vitro and in lens epithelial cells. FASEB J. 2006;20:2645–2647. doi: 10.1096/fj.06-5919fje. [DOI] [PubMed] [Google Scholar]
- 14.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallenberg M, Olm E, Hebert C, Bjornstedt M, Fernandes AP. Selenium compounds are substrates for glutaredoxins: a novel pathway for selenium metabolism and a potential mechanism for selenium-mediated cytotoxicity. Biochem J. 2010;429:85–93. doi: 10.1042/BJ20100368. [DOI] [PubMed] [Google Scholar]
- 16.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 17.Ellman GL. A colorimetric method for determining low concentrations of mercaptans. Arch Biochem Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- 18.Lou MF, Dickerson JE, Jr, Garadi R, York BM., Jr Glutathione depletion in the lens of galactosemic and diabetic rats. Exp Eye Res. 1988;46:517–530. doi: 10.1016/s0014-4835(88)80009-5. [DOI] [PubMed] [Google Scholar]
- 19.Janssen AJ, Trijbels FJ, Sengers RC, Smeitink JA, van den Heuvel LP, Wintjes LT, Stoltenborg-Hogenkamp BJ, Rodenburg RJ. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem. 2007;53:729–734. doi: 10.1373/clinchem.2006.078873. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan DM, Wehr NB, Fergusson MM, Levine RL, Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Xing K, Lou MF. Glutaredoxin 2 prevents H(2)O(2)-induced cell apoptosis by protecting complex I activity in the mitochondria. Biochim Biophys Acta. 2010;1797:1705–1715. doi: 10.1016/j.bbabio.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer LM, Lofgren S, Ho YS, Lou M, Wegener A, Holz F, Soderberg P. Absence of glutaredoxin1 increases lens susceptibility to oxidative stress induced by UVR-B. Exp Eye Res. 2009;89:833–839. doi: 10.1016/j.exer.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 26.Lofgren S, Fernando MR, Xing KY, Wang Y, Kuszynski CA, Ho YS, Lou MF. Effect of thioltransferase (glutaredoxin) deletion on cellular sensitivity to oxidative stress and cell proliferation in lens epithelial cells of thioltransferase knockout mouse. Invest Ophthalmol Vis Sci. 2008;49:4497–4505. doi: 10.1167/iovs.07-1404. [DOI] [PubMed] [Google Scholar]
- 27.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J Biol Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 28.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46:5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]