Abstract
Pemphigus vulgaris (PV) is an epidermal blistering disorder caused by antibodies directed against the desmosomal cadherin desmoglein-3 (Dsg3). The mechanism by which PV IgG disrupt adhesion is not fully understood. To address this issue, primary human keratinocytes and patient IgG were utilized to define the morphological, biochemical and functional changes triggered by PV IgG. Three phases of desmosome disassembly were distinguished. Analysis of fixed and living keratinocytes demonstrated that PV IgG cause rapid Dsg3 internalization which likely originates from a non-junctional pool of Dsg3. Subsequently, Dsg3 and other desmosomal components rearrange into linear arrays that run perpendicular to cell contacts. Dsg3 complexes localized at the cell surface are transported in a retrograde fashion along these arrays before being released into cytoplasmic vesicular compartments. These changes in Dsg3 distribution are followed by depletion of detergent insoluble Dsg3 pools and by the loss of cell adhesion strength. Importantly, this process of disassembly can be prevented by expressing exogenous Dsg3, thereby driving Dsg3 biosynthesis and desmosome assembly. These data support a model in which PV IgG cause the loss of cell adhesion by altering the dynamics of Dsg3 assembly into desmosomes and the turnover of cell surface pools of Dsg3 through endocytic pathways.
Introduction
Desmosomes are adhesive intercellular junctions that mediate attachment of intermediate filament networks to sites of cell to cell contact (Desai et al., 2009; Green and Simpson, 2007; Holthofer et al., 2007). Desmosomes are prominent in tissues that experience substantial mechanical stress, including the heart and the epidermis (Bazzi and Christiano, 2007). The desmogleins and desmocollins are the major desmosomal adhesion molecules and are members of the cadherin gene superfamily (Garrod and Chidgey, 2008; Wheelock and Johnson, 2003). Adhesive interactions mediated by the desmogleins and desmocollins are coupled to the intermediate filament cytoskeleton by an elaborate arrangement of cytoplasmic proteins. These desmosomal plaque molecules include the armadillo family proteins plakoglobin and the plakophilins (Hatzfeld, 2007), as well as members of the plakin cytolinker family, such as desmoplakin (Green and Simpson, 2007; Sonnenberg and Liem, 2007).
Desmosome function is compromised in a number of inherited (Awad et al., 2008; Bazzi and Christiano, 2007; Lai-Cheong et al., 2007) and autoimmune disorders (Stanley and Amagai, 2006). Pemphigus vulgaris (PV) is an acquired disease in which the desmosomal cadherin Dsg3 is targeted by autoantibodies (Kottke et al., 2006; Waschke, 2008). Patients with this disorder exhibit severe oral erosions as well as epidermal blistering if the disease progresses to include targeting of Dsg1. Importantly, IgG can be isolated from PV patient serum and used in mouse and tissue culture models to investigate the mechanism of disease pathogenesis (Amagai, 2008; Kottke et al., 2006). For example, injection of IgG from pemphigus patients into newborn mice causes epidermal blistering that is indistinguishable from that observed in patients (Stanley and Amagai, 2006). Similarly, PV IgG disrupts desmosomes and compromises adhesion strength when incubated in the media of cultured keratinocytes (Payne et al., 2004). This ability to passively transfer the disease from the patient to various model systems has resulted in significant advances in our understanding of PV pathobiology. Nonetheless, the precise mechanism by which PV IgG causes loss of keratinocyte adhesion is not fully understood.
Evidence from mouse models as well as atomic force microscopy suggests that PV IgG may directly disrupt Dsg3 trans (adhesive) interactions (Heupel et al., 2008; Shimizu et al., 2004). Similarly, mapping of pathogenic epitopes from both patient IgG and experimentally generated monoclonal antibodies indicate that pathogenic antibodies target the amino-terminal region of Dsg3, a cadherin domain critical for adhesive interactions (Tsunoda et al., 2003). These findings offer compelling evidence that disruption of Dsg3 adhesive interactions causes the loss of adhesion and blistering that characterizes the disease in patients. However, other work suggests a more complicated effect of PV IgG on keratinocytes (Muller et al., 2008; Sharma et al., 2007; Waschke, 2008). For example, a number of studies have demonstrated that pharmacological approaches targeting various signaling pathways can prevent loss of adhesion both in vitro and in vivo (Berkowitz et al., 2005; Berkowitz et al., 2006; Williamson et al., 2006). These studies suggest that PV IgG trigger keratinocyte responses and signal transduction cascades that are required for loss of adhesion. Similarly, previous work from our laboratory demonstrated that keratinocyte exposure to PV IgG at 4°C is insufficient to cause loss of adhesion, even though PV IgG bind Dsg3 and decorate cell-cell borders (Calkins et al., 2006). In fact, incubation in PV IgG for several hours is required before detectable changes in cell-cell adhesive strength are measurable. These and other data suggest that PV IgG binding to Dsg3 is necessary but not sufficient to disrupt keratinocyte adhesion (Kitajima, 2002; Mao et al., 2009; Sato et al., 2000; Yamamoto et al., 2007).
In the current study, a combination of biochemical and imaging approaches was used to define the dynamics of the PV-IgG-Dsg3 complex during desmosome disassembly. The results reveal that desmosome disassembly occurs in discrete phases that proceed in a temporally predictable sequence. These phases of disassembly include endocytosis of non-desmosomal Dsg3 as previously demonstrated by Sato and colleagues using immunogold electron microscopy (Sato et al., 2000). Internalization of non-junctional Dsg3 is followed by the rearrangement of desmosomal components into linear structures that appear to function as sites for endocytosis of desmosomal pools of Dsg3. This second phase is followed by depletion of junctional Dsg3 and the loss of cell adhesion. Remarkably, this process can be prevented by expressing exogenous Dsg3 to counteract this disassembly pathway with enhanced Dsg3 biosynthesis. These findings have important implications for understanding the basic cellular mechanisms of desmosome disassembly, and for designing therapeutic strategies to treat PV and related disorders.
Results
Time Course of Desmosome Disassembly in Response to PV IgG
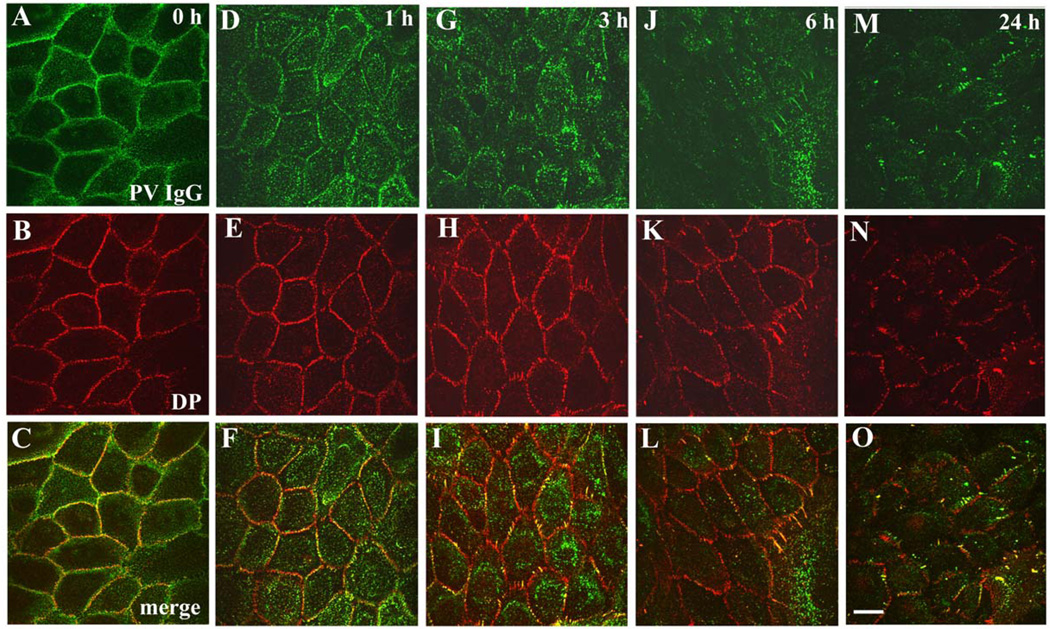
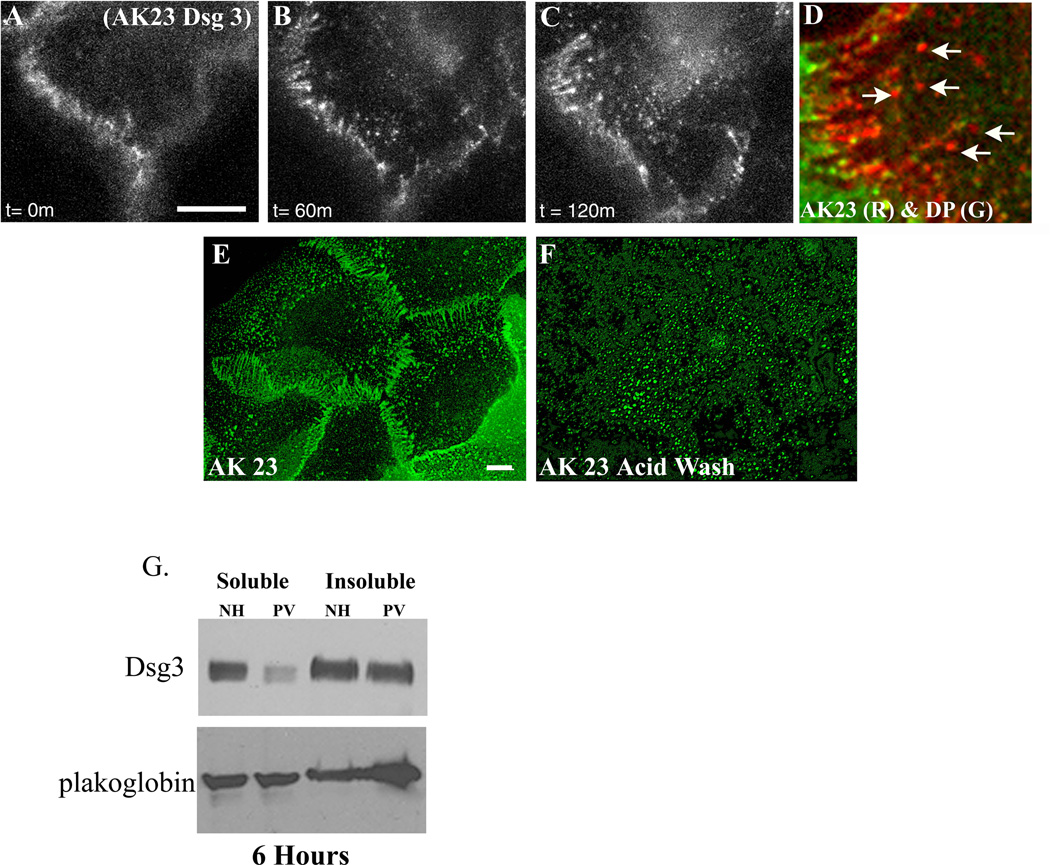
In the current study, a series of biochemical and imaging approaches were used to define the spatial and temporal relationships between desmosomal components during desmosome disassembly in response to PV IgG. As shown in Figure 1, PV IgG bind to keratinocyte intercellular junctions when cells are incubated at 4°C and colocalize with desmoplakin along cell borders (Fig. 1A–C). After 1 hr at 37°C, PV IgG-Dsg3 complexes clustered into punctuate structures that were distal to cell contacts and lacked desmoplakin (Fig. 1D–F). This pool of PV IgG-Dsg3 is likely to represent internalization of Dsg3 from non-desmosomal compartments as previously described by Sato and colleagues (Sato et al., 2000). This change in Dsg3 distribution preceded disruption of desmoplakin organization (Fig. 1E). However, by 3–6 hrs, desmoplakin was markedly disrupted and junctional Dsg3 localization decreased (Fig. 1G–L). By 24 hrs, Dsg3 was largely depleted, desmoplakin localization was highly disorganized (Fig. 1M–O) and cell-cell adhesion strength was significantly compromised (Fig. 8H and (Calkins et al., 2006; Delva et al., 2008). Control experiments using a monoclonal antibody against Dsg3 confirmed that PV IgG colocalized with Dsg3 over the 24 hour time course (Supplemental Fig. 1). To determine if PV IgG-induced desmosome rearrangement leads to depletion of desmosomal components, keratinocytes were incubated with NH or PV IgG for 24 hours. Sequential Triton X-100 detergent extraction and western blot analysis was carried out to determine if membrane associated (Triton soluble) or cytoskeleton associated (Triton insoluble) pools of desmosomal proteins were depleted after 24 hours exposure to PV IgG (Supplemental Fig. 2). The Triton soluble pools of desmoplakin, desmocollin-2 (Dsc2) and plakoglobin in PV IgG treated cells were similar to the NH control, whereas the soluble pool of Dsg3 was dramatically reduced. The Triton insoluble pools of desmoplakin and plakoglobin were not altered by PV IgG. The insoluble pool of Dsc2 exhibited a slight decrease in response to PV IgG, whereas Dsg3 levels were considerably down regulated (see also Supplemental Fig. 4 and (Calkins et al., 2006). These data demonstrate that morphological changes in Dsg3 are followed by selective depletion of Dsg3 protein levels.
Figure 1. Time course of desmosome disassembly in response to PV IgG.
Keratinocytes were exposed to PV IgG at 4°C for 20 minutes and subsequently shifted to 37°C for 1, 3, 6, and 24 hours. The localization of human IgG and desmoplakin (DP) was monitored by immunofluorescence microscopy. In cells incubated at 4°C (A–C), PV IgG labels cell borders and desmoplakin staining is predominantly in punctate linear patterns at cell-cell junctions. After 1 hour treatment with PV IgG (D–F), the PV-IgG-Dsg3 molecules accumulate in puncta that are distal to cell-cell borders, while desmoplakin (E) staining is unchanged. Keratinocytes treated with PV IgG for 3 and 6 hours exhibit a rearrangement of desmoplakin into linear arrays emanating from cell borders which contain both Dsg3 and desmoplakin. Following treatment with PV IgG for 24 hours (M–O) both Dsg3 and desmoplakin are noticeably mislocalized and/or absent from cell-cell junctions. Bar, 10µm.
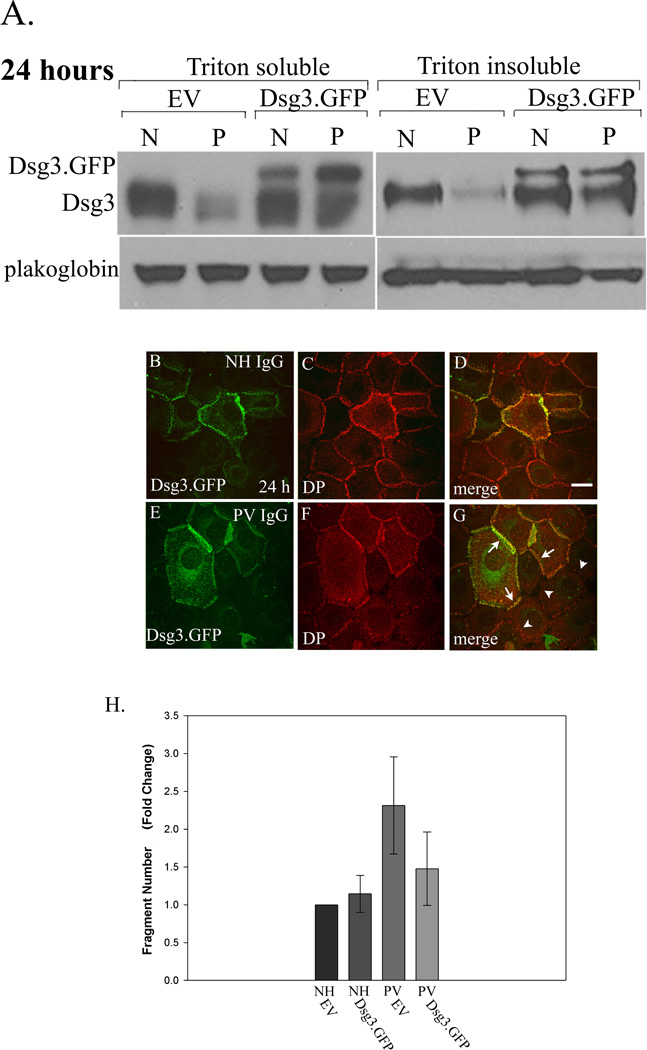
Figure 8. Expression of exogenous Dsg3.GFP prevents desmoplakin mislocalization and loss of cell adhesion in PV IgG treated keratinocytes.
Dsg3.GFP was expressed in keratinocytes using an adenoviral delivery system 24 hours prior to treatment with NH or PV IgG. After an additional 24 hrs, cells were processed for sequential detergent extraction and western blot (Panel A), immunofluorescence (Panels B–G), or subjected to mechanical stress to measure cell adhesion strength (Panel H). Note that expression of exogenous Dsg3.GFP prevented the down-regulation of Dsg3 in both the Triton X-100 soluble and insoluble pools in PV IgG (P) treated cells compared to cells treated with NH IgG (N) (Panel A). Note that Dsg3.GFP colocalized with desmoplakin in NH IgG treated cells (B–D) and prevented mislocalization of desmoplakin in PV IgG treated cells (E–G). Note also the robust desmoplakin staining in cells that are expressing Dsg3.GFP (arrows, G) in sharp contrast to adjacent cells lacking Dsg3.GFP (arrowheads, G). To determine if Dsg3.GFP expression could prevent loss of cell adhesion strength in PV IgG treated cells, NH IgG or PV IgG treated keratinocytes were released from the substrate using the enzyme dispase and subjected to mechanical stress. The number of resulting monolayer fragments was quantified. Note that exogenous Dsg3.GFP rendered PV IgG treated keratinocytes resistant to mechanical stress. The graph represents an average of three separate experiments quantified by a blinded observer. PV IgG treated keratinocytes expressing Dsg3.GFP exhibited significantly less fragments than those expressing empty adenovirus as determined using ANOVA with post hoc least significant difference analysis (**p < 0.01). Error bars represent the standard deviation. Bar, 10µm.
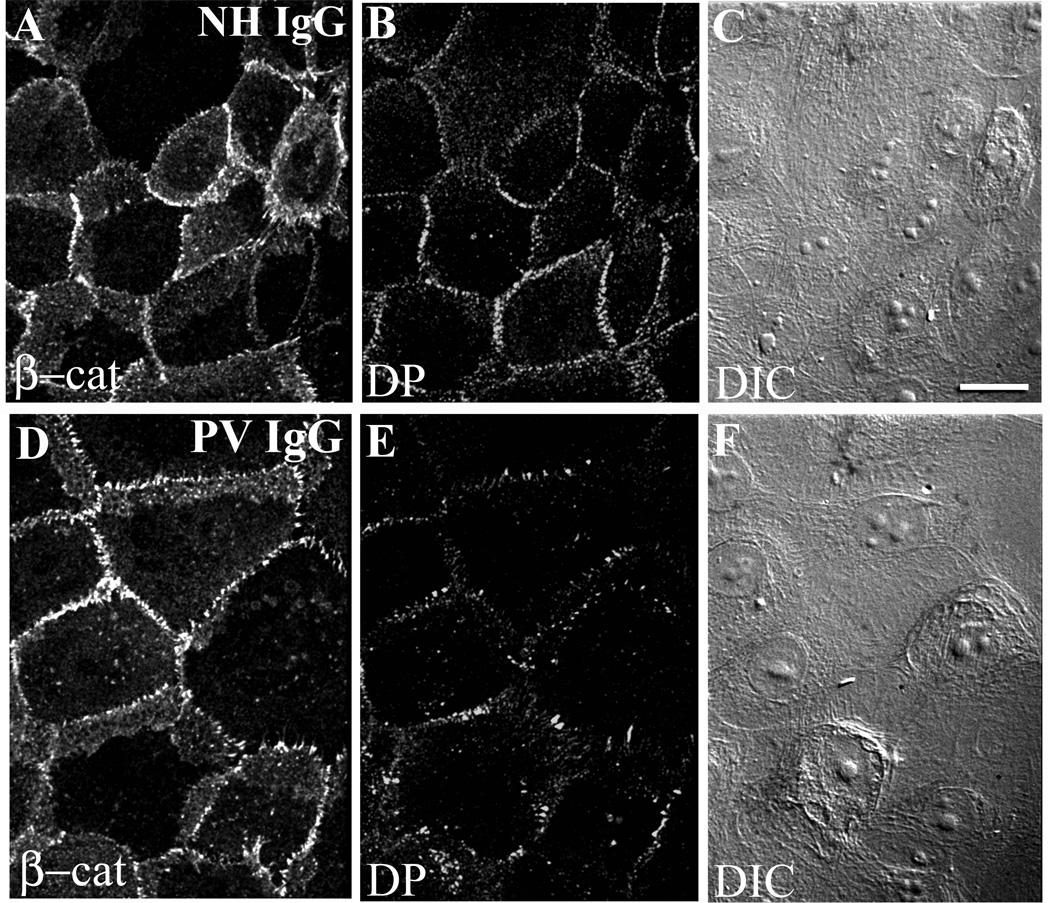
To further assess changes in the distribution of junction components and overall cell shape, keratinocytes were treated with either NH IgG (Fig. 2A–C) or PV IgG (Fig. 2D–F) and the localization of the adherens junction protein β-catenin was used to outline cell-cell contacts. After 6 hrs in PV IgG, alterations in desmoplakin localization were observed, but only minimal changes in β-catenin localization were detected (Fig. 2D, E). Furthermore, differential interference contrast microscopy demonstrated that PV IgG treatment did not cause a dramatic alteration in cell shape (Fig. 2F). These data are consistent with previous findings (de Bruin et al., 2007) and indicate that changes in desmosomal protein distribution precede alterations in adherens junctions and changes in cell shape.
Figure 2. Desmosomes are disrupted by PV IgG but β-catenin is minimally affected and cells remain in close apposition.
Keratinocytes were treated with NH IgG (A–C) or PV IgG (D–F) for 6 hours and subsequently processed for immunofluorescence. Note robust β-catenin and desmoplakin (DP) staining (A, B) in cells treated with NH IgG. The PV IgG treated cells exhibit markedly disrupted desmoplakin staining at borders (E), whereas β-catenin remains largely unaltered (D). Differential interference contrast imaging shows that cells do not undergo major changes in cell shape (F). Bar, 10µm.
Non junctional Dsg3 is internalized and degraded before desmosomes are disrupted by PV IgG
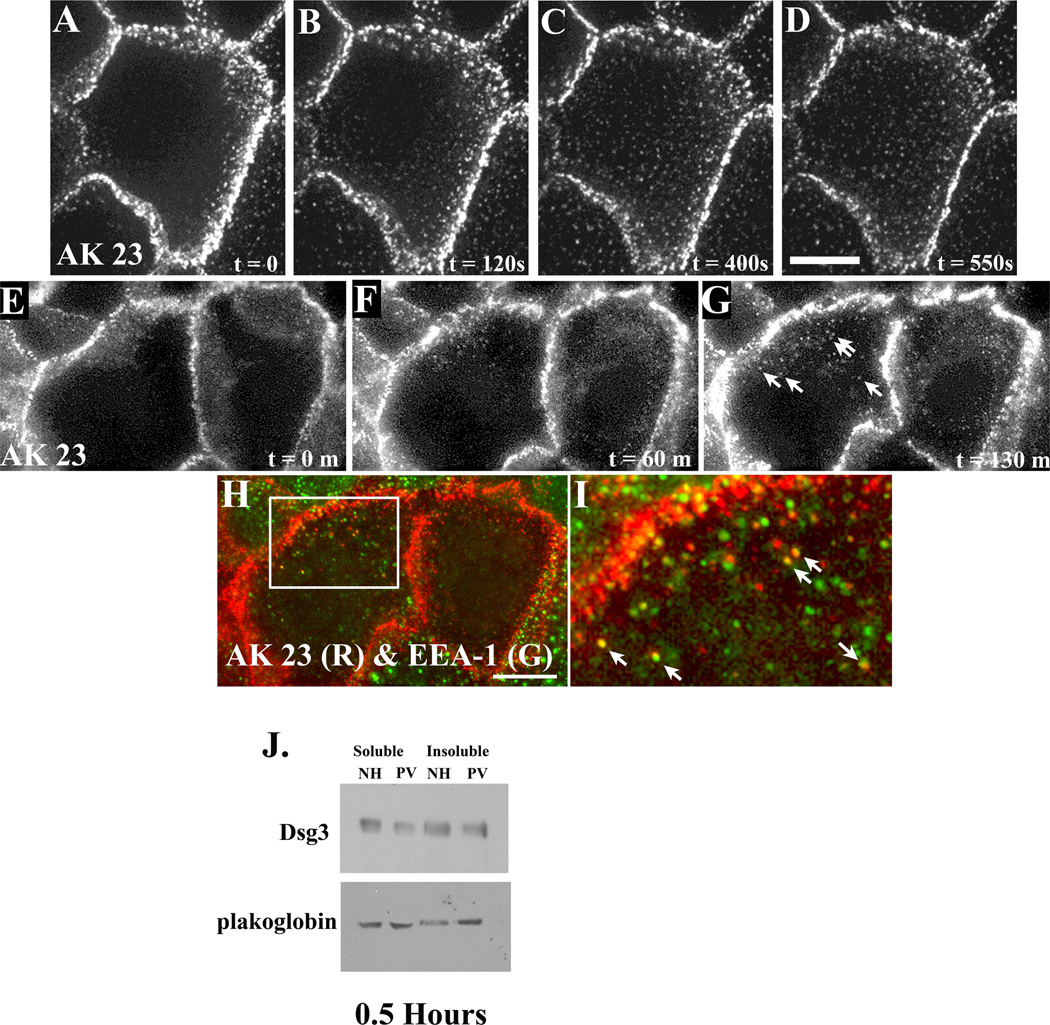
The results shown in Figure 1 suggest that the distribution of desmosomal components is altered in a sequential manner after exposure of keratinocytes to PV IgG. To monitor sequential changes in Dsg3 localization after addition of PV IgG, time lapse fluorescent imaging experiments were conducted using fluorescently tagged Dsg3 monoclonal antibodies (AK15 or AK23) (Tsunoda et al., 2003) to label cell surface pools of Dsg3. To verify that these tracer antibodies did not cause significant alterations in cell surface Dsg3 distribution, AK23 was allowed to bind to keratinocytes at 4°C. Excess fluorescently tagged AK23 was then removed from the medium and keratinocytes treated with either NH IgG or PV IgG for 6 hrs at 37°C (Supplemental Fig. 3). Note that very little change in AK23 distribution was observed after 6 hr exposure to NH IgG, whereas the distribution of the AK23 complex was dramatically altered by addition of PV IgG. Similar results were obtained using AK15 (not shown). Therefore, a series of experiments were conducted using fluorescently tagged AK15 or AK23 monoclonal antibodies to monitor Dsg3 distribution after exposure to PV IgG using four dimensional time lapse fluorescence microscopy of living keratinocytes. Over a time course of 4 hrs, time lapse imaging demonstrated that Dsg3 is first clustered by PV IgG into puncta, followed by rearrangement of junctional pools of Dsg3 (Supplemental movie 1). Additional time lapse imaging focusing on earlier time points revealed that Dsg3 formed clusters on the dorsal surface of keratinocytes (Fig. 3A–D and Supplemental Movie 2). This clustering occurred rapidly and preceded any obvious changes in Dsg3 at cell-cell borders. To determine the fate and composition of these Dsg3 complexes, longer time lapse imaging experiments were conducted. In addition, the cells were fixed at the end of the time lapse series and processed for retrospective immunofluorescence microscopy using antibodies directed against early endosome antigen-1 (EEA-1). This approach demonstrated that clusters of Dsg3 formed within the first two hours of PV IgG incubation at 37°C (Fig. 3E–G). Furthermore, retrospective analysis revealed that many of the Dsg3 particles observed colocalize with EEA-1 (Fig. 3H–I), demonstrating that Dsg3 is internalized. Immunoblot analysis of Triton X-100 extracts of keratinocytes indicated that the soluble pool of Dsg3 is only slightly diminished after incubation with PV IgG at these early time points, and no consistent changes were observed in the insoluble pool of Dsg3 (Fig. 3J). Taken together, these results suggest that the internalization of non-junctional pools of Dsg3 occurs rapidly upon exposure of keratinocytes to pathogenic PV IgG and that these changes precede alterations in the junctional pools of Dsg3.
Figure 3. Non-junctional pools of Dsg3 are rapidly internalized after exposure to PV IgG.
The Dsg3 antibody AK23 was fluorescently tagged with Alexa Flour 555 and incubated with human keratinocytes for 30 minutes at 4°C. Unbound antibody was removed and media containing PV IgG was added. Living cells were imaged using time lapse fluorescence microscopy (A–D and E–G) to follow the dynamics of the Dsg3 complex in PV IgG treated cells. Note the appearance of AK23-Dsg3 clusters and vesicular structures that form rapidly after exposure to PV IgG, while junctional Dsg3 remains largely unaltered (Supplemental movie 2). To determine the composition of the AK23-Dsg3 puncta, cells shown in panel G were rapidly fixed and processed for indirect immunofluorescence using antibodies directed against EEA-1. Note co-localization between the AK23-Dsg3 complex and EEA-1 (H, I), indicating that the AK23-Dsg3 complex undergoes internalization to endosomes. Each panel in the series represents a single projection image of 7 (A–D) or 8 (E–G) z-axis images collected at each time point. Sequential detergent extraction and western blot analysis was carried out to determine if membrane associated (Triton X-100 soluble) or cytoskeleton associated (Triton X-100 insoluble) pools of Dsg3 were depleted. A decrease in the soluble pool of Dsg3 was detected within 30 minutes of incubation with PV IgG (J). Results are representative of at least three independently conducted experiments. Bar, 10µm.
PV IgG cause rearrangement of cell surface Dsg3 into linear arrays and subsequent internalization of Dsg3 from cell-cell junctions
The data shown above indicate that internalization of non-junctional Dsg3 precedes alterations in junctional pools of Dsg3 and desmoplakin. Therefore, additional time lapse microscopy was carried out using fluorescently tagged AK23 at later time points after addition of PV IgG. As shown in Figure 4, over a time course of 2 hours, Dsg3 reorganizes from the typical desmosomal pattern into linear arrays that extend perpendicularly from cell borders (Fig. 4A–C and Supplemental Movie 3). Interestingly, the AK23-Dsg3 complex appears to be released from the tips of these linear arrays into vesicular structures that are transported in a retrograde fashion away from cell-cell junctions. Retrospective immunolocalization was used to define the composition of these linear arrays and vesicular structures. AK23-Dsg3 complexes in linear arrays colocalized with desmoplakin, whereas vesicular Dsg3 that is released from the linear arrays did not (Fig. 4D, see also Fig. 5). These findings suggest that the Dsg3-desmoplakin complex is disrupted upon Dsg3 endocytosis from the cell surface. To test this possibility, cell surface Dsg3 was labeled with AK23 and keratinocytes were treated with PV IgG for 3 hrs. The cells were then transferred to 4°C and either fixed immediately (Fig. 4E) or washed in a low pH buffer before fixation to remove AK23 bound to the cell surface (Fig. 4F) (Delva et al., 2008). Importantly, the low pH wash removed AK23 that localized to linear arrays, but the punctate AK23-Dsg3 particles remained after the acid wash, indicating that this pool of Dsg3 had been internalized. At this time point, soluble levels of Dsg3 are notably reduced, whereas the Triton-X 100 insoluble pool of Dsg3 exhibits little or no change (Fig. 4G). These data suggest that the Dsg3 localized to linear arrays is on the cell surface, whereas the vesicular structures released from the linear arrays represent Dsg3 that has entered an endocytic compartment.
Figure 4. The PV IgG-Dsg3 complex reorganizes into linear arrays that exhibit retrograde movement before entering vesicular compartments.
Keratinocytes were incubated at 4°C with Alexa Flour labeled AK23 monoclonal antibody for 30 minutes. Unbound antibody was removed followed by the addition of keratinocyte media containing PV IgG. Cells were then imaged every 5 minutes for two hours at 37°C. Notice the formation of linear arrays of AK23-Dsg3, followed by apparent budding of vesicular structures from the tips of the arrays (B, C). (Supplemental movie 3). Retrospective immunofluorescence analysis (D) indicates that Dsg3 that has entered this vesicular compartment is no longer associated with desmoplakin (arrows, panel D). To determine if Dsg3 in the linear arrays was present on the cell surface, living cells were incubated with the pathogenic monoclonal AK23 for 3 hrs in the presence of PV IgG. Cells were then incubated on ice and either fixed immediately (E) or incubated in a low pH acid wash before fixation (F) to remove cell surface bound AK23 IgG. Note that the linear arrays are not observed in cells exposed to the low pH wash, demonstrating that Dsg3 in these structures is present at the cell surface and that the punctate vesicular Dsg3 observed in panel F is intracellular. Western blot analysis of keratinocyte lysates treated for 6 hrs in PV IgG reveals a decrease in the non-junctional Triton X-100 soluble pool of Dsg3, whereas the Triton insoluble pool of Dsg3 exhibited little or no change. No changes in plakoglobin levels were observed (G). Results are representative of at least three independently conducted experiments. Bar, 10µm.
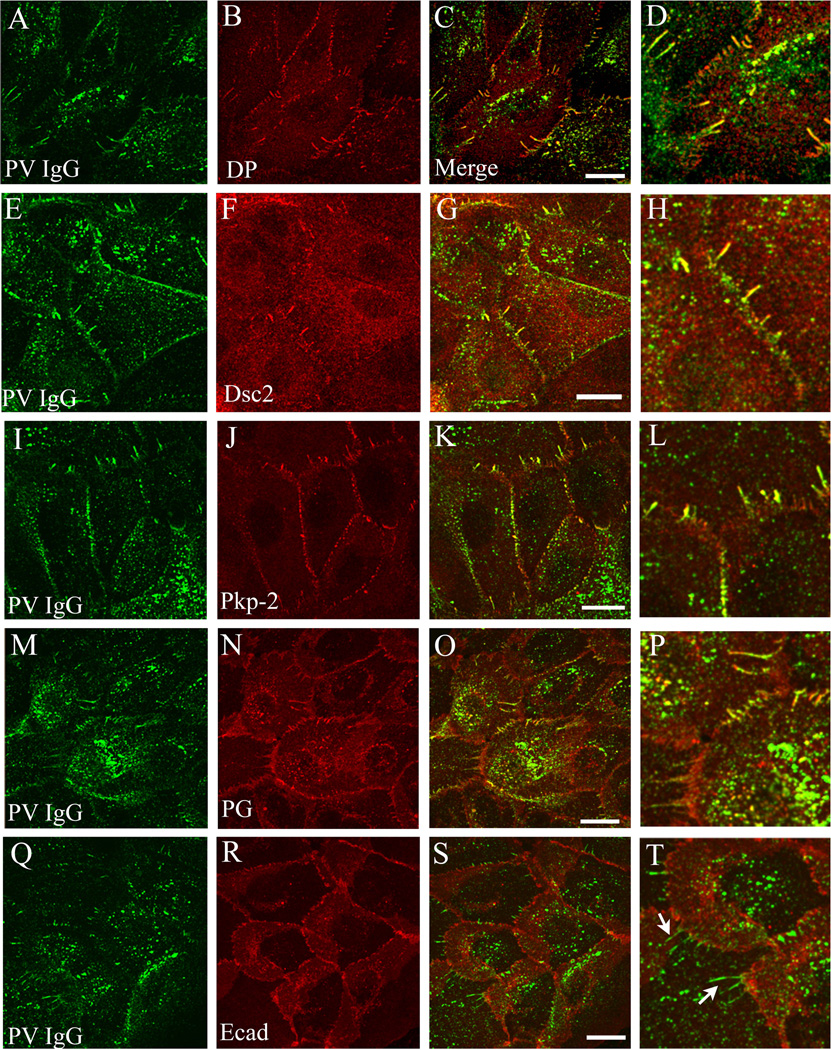
Figure 5. Linear arrays contain desmosomal but not adherens junction components.
Primary uman keratinocytes were incubated with PV IgG for 6 hours and then processed for confocal immunofluorescence microscopy for PV IgG and either desmoplakin (DP, A–D), desmocollin-2 (Dsc2, E–H), plakophilin-2 (Pkp-2, I–L), plakoglobin (M–P), or E-cadherin (Ecad, Q–T). Note that the PV IgG complex colocalizes with each of the desmosomal components in linear arrays, but does not colocalize with the adherens junction marker E-cadherin (arrows panel T). Bar, 10µm.
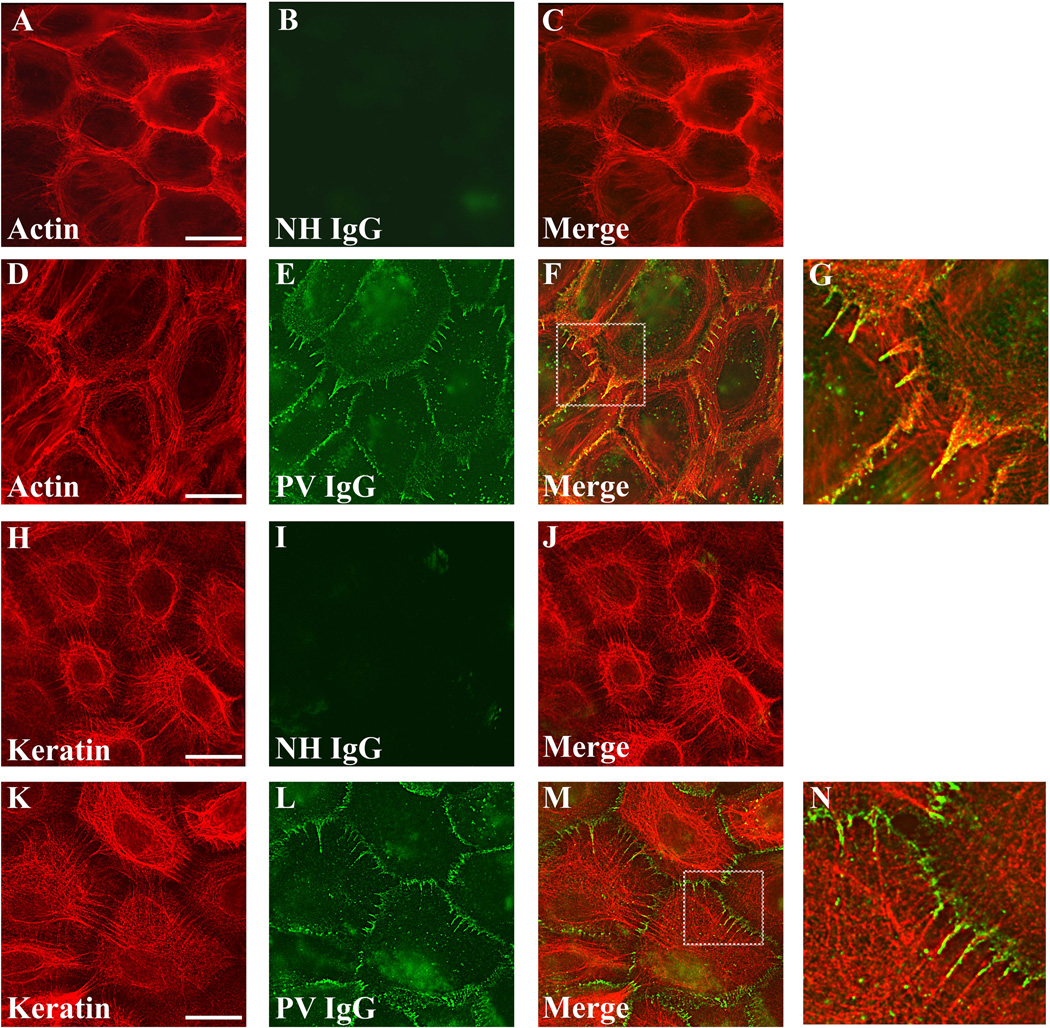
To define the molecular composition of the Dsg3 linear arrays, immunofluorescence microscopy was conducted on keratinocytes treated with PV IgG for 6 hrs. Interestingly, the PV IgG-Dsg3 complex in the linear arrays co-localized with all desmosomal components tested, including desmoplakin (Fig. 5A–D), Dsc2 (Fig. 5 E–H), plakophilin-2 (Fig. 5I–L), and plakoglobin (Fig. 5M–P). However, PV IgG in these linear arrays did not colocalize with the adherens junction component E-cadherin (Fig. 5Q–T). The composition of these structures was analyzed further to determine if cytoskeletal elements were present in the linear arrays. In keratinocytes exposed to NH IgG, actin bundles were prominent at cell-cell contacts (Fig. 6A). As reported previously (Berkowitz et al., 2005), exposure to PV IgG resulted in retraction of actin bundles away from cell borders (Fig. 6D). In addition, actin filaments colocalized with the PV IgG-Dsg3 complex that had reorganized into linear arrays (Fig. 6D–G). Keratin filaments also aligned with PV IgG in the linear arrays (Fig. 6K–N). Together, these data indicate that PV IgG causes reorganization of desmosomal components into linear arrays that contain all of the major desmosomal components, as well as actin and keratin filaments.
Figure 6. PV IgG-Dsg3 in linear arrays colocalize actin and align with keratin filaments.
Keratinocytes were treated with either NH IgG or PV IgG for 6 hrs and processed for immunofluorescence localization of NH and PV IgG with actin (A–G) or NH and PV IgG and cytokeratin (H–N). Note colocalization of PV IgG in linear arrays with actin, which emanate away from cell cell borders in alignment with keratin filaments. Images shown are single z-plane representations that were captured using wide field microscopy followed by deconvolution. Bar, 20µm.
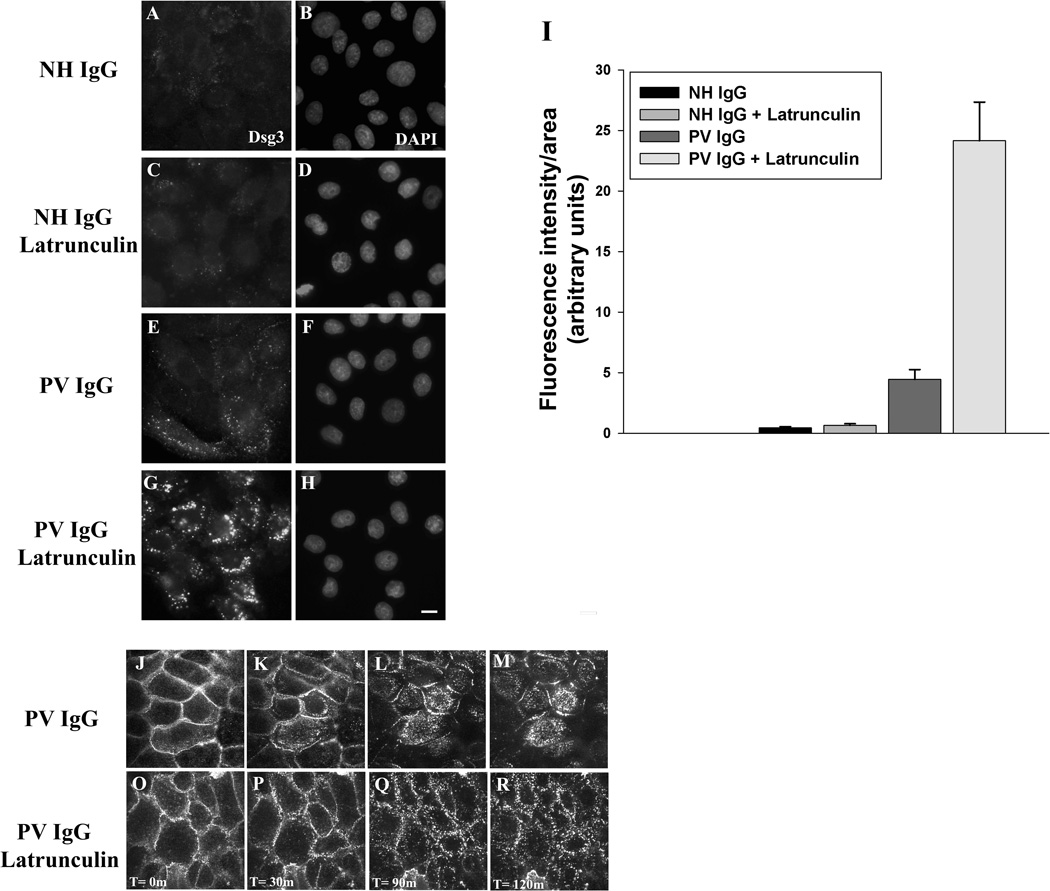
A number of studies have implicated actin and actin associated proteins in the regulation of desmosome assembly (Godsel et al., 2005; Hatzfeld, 2007). The reorganization of actin in response to pathogenic PV antibodies suggests that the actin network may also be playing a role in Dsg3 endocytosis and desmosome disassembly in PV IgG treated cells (Berkowitz et al., 2005). To test this possibility, keratinocytes were treated with latrunculin A to disrupt actin polymerization. Interestingly, latrunculin A treatment resulted in dramatically increased endocytosis of Dsg3 in PV IgG treated cells (Fig. 7). Furthermore, time lapse imaging revealed that latrunculin A treatment eliminated the formation of the typical linear arrays that form in PV IgG treated cells (compare Fig. 7J–M, O–R, and supplemental movies 4–5). Instead, the PV IgG-Dsg3 complex was internalized in large aggregates directly from cell-cell contacts in latrunculin A treated cells. To investigate the role of actin in PV IgG-induced Dsg3 down regulation, western blots were performed on keratinocytes incubated for 24 hours with NH or PV IgG, with or without latrunculin A to inhibit actin polymerization (Supplemental Fig. 4). No changes in desmosomal protein levels were observed in cells treated with NH IgG and latrunculin A for 24 hours. However, both the soluble and insoluble pools of Dsg3 were reduced by PV IgG in the absence and presence of latrunculin A. Interestingly, in cells treated with both PV IgG and latrunculin A the Triton-X soluble pools of Dsc2 and plakoglobin were modestly decreased. Furthermore, the insoluble pools of desmoplakin, Dsc2 and plakoglobin were reduced similarly to Dsg3. Together these data indicate that PV IgG and actin depolymerization act synergistically to decrease steady state desmosomal protein levels.
Figure 7. Actin depolymerization increases PV IgG-induced Dsg3 internalization.
Keratinocytes were untreated (A–B and E–F) or treated with latrunculin A (250 nM) (C–D and G–H) at 37°C for 1 hour. Cells were then incubated at 4°C for 30 min with AK23 IgG to label cell surface pools of Dsg3. Excess AK23 antibody was removed from the medium, NH IgG or PV IgG were added, and the cells were shifted to 37°C for 3 hours. To visualize only the intracellular pool of Dsg3, cells were washed in a low pH buffer at 4°C to remove surface bound antibody prior to processing for immunofluorescence. Bar, 5µm. The amount of internalized AK23-Dsg3 was then quantified (I). Error bars represent the standard error of the mean, where n = 15 fields of view. In parallel experiments, time lapse microscopy was used to visualize Dsg3 dynamics in PV IgG (J–M) or PV IgG + Latrunculin A (O–R) treated keratinocytes (Supplemental movies 4 and 5).
Expression of exogenous Dsg3 prevents desmosome disassembly and loss of adhesion
The rearrangement of Dsg3 into linear arrays followed by release of Dsg3 into vesicular structures precedes the dramatic loss of Dsg3 and desmoplakin from cell-cell borders that are observed by 24 hrs (Fig. 1M–O). By 24 hrs, both the Triton soluble and insoluble pools of Dsg3 are depleted (Fig. 8A, Supplemental Fig. 2 and 4). Recently, we reported that Dsg3 is internalized through a clathrin and dynamin independent mechanism in PV IgG treated keratinocytes (Delva et al., 2008). Therefore, we hypothesized that the disruption of desmosomes and the loss of adhesion in response to PV IgG is caused by the internalization of both non-junctional and junctional pools of Dsg3, leading to depletion of cell surface Dsg3 and the loss of adhesion. Therefore, we reasoned that the loss of adhesion in PV IgG treated cells might be prevented by enhancing Dsg3 biosynthesis rates to counter the increase in Dsg3 endocytosis and turnover. To test this possibility, a GFP tagged Dsg3 fusion protein (Dsg3.GFP) was expressed exogenously using an adenoviral delivery system. As shown in Figure 8A, PV IgG treatment for 24 hours results in decreased Dsg3 levels in both the Triton soluble and insoluble pools. Importantly, expression of exogenous Dsg3.GFP prevented the down-regulation of Dsg3 levels in PV IgG treated keratinocytes. To determine if expression of exogenous Dsg3.GFP could also prevent alterations in desmoplakin localization in PV IgG treated cells, Dsg3.GFP was expressed in NH IgG (Fig. 8B–D) and PV IgG (Fig. 8E–G) treated keratinocytes. Interestingly, expression of Dsg3.GFP prevented the disruption of desmoplakin organization in PV IgG treated keratinocytes (Fig. 8E–G). Note that cells expressing Dsg3.GFP exhibit well organized desmoplakin staining at cell borders, whereas desmoplakin staining is dramatically reduced in adjacent cells lacking exogenous Dsg3 expression. These data suggested that exogenous expression of Dsg3 may prevent the loss of adhesion triggered by PV IgG. Remarkably, expression of exogenous Dsg3.GFP reduced the number of fragments generated in PV IgG treated keratinocytes exposed to mechanical stress (Fig. 8H). Together, these findings indicate that exogenous expression of Dsg3 prevents the down-regulation of total cellular Dsg3 protein levels, the disruption of desmoplakin localization, and the loss of adhesion strength in response to PV IgG.
Discussion
The results of this study indicate that desmosome disassembly in response to PV IgG occurs in three sequential but temporally overlapping phases. Time lapse fluorescence microscopy indicates that the first phase of PV IgG-induced disassembly (Phase 1) is characterized by endocytosis of the non-junctional pool of Dsg3 (Fig. 1 and 3). These findings are consistent with previous studies using immunogold electron microscopy and time lapse labeling (Sato et al., 2000). The internalized Dsg3 is then delivered to early endosomes (Fig. 3), where it is most likely sorted for degradation in lysosomes (Calkins et al., 2006). This model is supported by the observation that the internalization of Dsg3 is accompanied by a decrease in the Triton soluble pool of Dsg3 as assessed biochemically (Fig. 3J). In contrast, junctional pools of Dsg3 are largely unaffected over the course of the first 1–2 hours after PV IgG binding to the keratinocyte cell surface (Fig. 1D–F). Furthermore, there is no significant loss of adhesive strength at these early time points (Calkins et al., 2006), consistent with the observation that the desmosomal pool of Dsg3 remains intact even while the non-desmosomal pool decreases. Thus, the non-desmosomal pool of Dsg3 is most susceptible to the effects of pathogenic antibody binding, and internalization of this pool from the cell surface occurs during the first phase of desmosome disassembly in response to PV IgG.
A prominent feature of keratinocytes treated for 2–6 hrs with PV IgG (Phase 2) is the rearrangement of desmosomal components into linear arrays which align with keratin filaments and extend away from cell-cell borders (Fig. 5 and Fig. 6). The linear arrays colocalize with actin and contain not only the PV IgG-Dsg3 complex, but also all other desmosomal components examined (Fig. 5 and Fig. 6). Importantly, the Dsg3 complex appears to emerge from the tips of these arrays into vesicular structures that are internalized (Fig. 4). At these intermediate time points, adherens junction components are well organized and cells remained closely apposed (Fig. 2). These data indicate that alterations in desmosomal organization precede changes in adherens junctions and alterations in cell shape. Finally, Phase 3 of disassembly (6–24 hrs) is characterized by disruption of desmosomal components and loss of adhesion strength. At these later time points, the depletion of Dsg3 from both the soluble and insoluble pools was dramatic (Fig. 1M–O, Fig.8 and Supplemental Fig. 2 and 4).
Previous studies using time lapse imaging of desmosome assembly revealed that desmosome formation also takes place in discrete phases (Bass-Zubek et al., 2008; Godsel et al., 2005). In these studies by Green and colleagues, desmoplakin and the desmosomal cadherins appear to form distinct complexes during assembly and are transported separately to sites of junction formation. This process appears analogous to that observed during disassembly. For example, Dsg3 colocalizes extensively with plakoglobin upon internalization. However, none of the other desmosomal components examined, including plakophilin-2 and desmoplakin, colocalize with the internalized pool of Dsg3 (Calkins et al., 2006). Cytoskeletal elements are also involved in desmosomal assembly and disassembly. Cytochalasin D disrupts actin filaments and impairs desmosome assembly, implicating the actin cytoskeletal network in the process of desmosome formation (Godsel et al., 2005). Similarly, we observed colocalization between the actin cytoskeleton and the linear arrays that characterize phase 2 of disassembly in response to PV IgG (Fig. 6). In addition, disruption of actin polymerization caused a dramatic increase in Dsg3 endocytosis from both the cell surface and from intercellular junctions (Fig. 7), consistent with a recent report suggesting that the actin cytoskeleton regulates desmosome disassembly in response to PV IgG (Gliem et al., 2010). These data suggest that actin plays roles in both assembly and disassembly of desmosomes, and that desmosomal protein association with the actin cytoskeleton may be most prominent and functionally relevant when desmosomal components are undergoing dynamic regulation.
A key issue in pemphigus pathophysiology is the mechanism by which PV IgG binding to Dsg3 causes loss of adhesion. The data presented here, along with a number of other studies, suggest that PV IgG binding to Dsg3 is insufficient to cause loss of keratinocyte adhesion (Muller et al., 2008; Waschke, 2008). Rather, it appears that PV IgG binding disrupts the kinetics of assembly and disassembly by favoring endocytosis of Dsg3 and alterations in desmosome organization that lead to disassembly. A prediction based on this hypothesis is that increasing Dsg3 biosynthesis should counteract the increase in Dsg3 endocytosis and turnover caused by PV IgG. A shown in Figure 8, expression of exogenous Dsg3 using an adenoviral delivery system prevented the down-regulation of Dsg3, disruption of desmoplakin organization, and loss of adhesion in PV IgG treated cells. These findings suggest that induction of endocytosis and turnover of Dsg3 causes loss of adhesion, and that increased Dsg3 biosynthesis is sufficient to restore desmosome assembly and adhesion. The precise mechanism by which expression of exogenous Dsg3 prevents loss of adhesion is not fully understood, and may involve upregulation of other adhesion molecules or desmosomal plaque proteins. Interestingly, we find that keratinocytes become more resistant to the effects of PV IgG on adhesion with increasing time in high calcium medium (Saito and Kowalczyk, unpublished). This increased resistance to PV IgG is most likely due to increased Dsg3 cell surface levels and decreased Dsg3 endocytosis when cells are cultured in high calcium (Delva and Kowalczyk, 2009). These findings suggest that treatment of pemphigus patients with agents that enhance Dsg3 biosynthesis or stabilize desmosomes may be beneficial. Consistent with this notion, corticosteroids have been found to increase Dsg3 transcription (Nguyen et al., 2004). Along with the results present here, these findings suggest that corticosteroids may be beneficial in pemphigus treatment due to increase expression of adhesion molecules in addition to immune suppression.
Materials and Methods
Cell Culture
Primary human keratinocytes were isolated from neonatal foreskin and cultured in keratinocyte growth medium (Cascade Medium 154 or Gibco Defined Keratinocyte-SFM, Invitrogen, Carlsblad, CA) as described previously (Calkins et al., 2006; Delva et al., 2008). Keratinocytes, no later than passage 3, were then switched into media containing 0.55 mM or 1.2 mM calcium for 16–18 hours before experimental manipulations.
Immunofluorescence
Keratinocytes were seeded onto glass coverslips and allowed to proliferate to desired confluence. Cells were switched to 0.55 mM calcium containing media for 16–18 hours prior to treatment with NH IgG (Invitrogen, Carlsblad, CA) or PV IgG for varying amounts of time. Sera from pemphigus vulgaris patients were acquired without identifiers and IgG was affinity purified as previously described(Calkins et al., 2006; Delva et al., 2008). Data shown are representative of at least two different PV patient IgG and/or pathogenic monoclonal Dsg3 antibodies (Tsunoda et al., 2003). NH IgG or PV IgG was added to cell culture media at 100–150 µg/ml and allowed to bind to keratinocyte cell surfaces at 4°C for 30 minutes prior to incubation at 37°C. Upon completion of the incubation at 37°C, cells were washed three times with PBS+, and then fixed on ice using either −20°C methanol for 2–4 minutes or 4.0% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 minutes followed by extraction with 0.1% Triton X-100 (Roche Diagnostics, Indianapolis, IN) for 7 minutes. The localization of desmosomal components was assessed using the following antibodies: rabbit anti-desmoplakin (GeneTex, Inc., San Antonio, TX), rabbit anti- desmoplakin (NW6 a gift from Dr. Kathleen Green, Northwestern University), mouse anti-desmoplakin (Fitzgerald Industries International, Inc., Concord, MA.), mouse anti-plakoglobin (BD Transduction Laboratories, Rockville, MD), mouse anti-desmocollin-2 (7G6, a gift from Dr. James Wahl), mouse anti- plakophillin-2 (Meridian Life Sciences, Inc. Saco, ME), rabbit anti-β-catenin (Lab Vision Products, Thermo Fisher Scientific, Fremont, CA.), mouse anti-E-cadherin (BD Transduction Laboratories, Rockville, MD), and mouse anti-cytokeratin (Immunotech, Marseille, France). Secondary antibodies to appropriate species conjugated to various Alexa Fluors (Invitrogen, Carlsbad, CA.) were used for dual-label staining. Localization of actin was monitored using Alexa Fluor 555 phalloidin (Invitrogen, Carlsblad, CA). Endocytosis of Dsg3 was quantified by measuring fluorescence intensity of intracellular AK23/Dsg3 using a low pH wash to remove cell surface antibody as previously described (Delva et al., 2008). Latrunculin A (Invitrogen) was used at 250 nM.
Time-Lapse Microscopy
Immunofluorescence analysis and time lapse microscopy were carried out using an inverted Leica DMI-6000B microscope equipped for wide field and confocal microscopy. Confocal microscopy was carried out using two solid state lasers (491nm and 561nm) and a VT Infinity 2D array scanner (VisiTech International, Sunderland, UK). Images were captured using a Hamamatsu electron multiplier deep cooled CCD camera (C9100-12). An automated stage and image acquisition were driven by Simple PCI software (Hamamatsu Corporation, Sewickley, PA). For live cell analysis, temperature control was achieved using an environmental control chamber (Pecon Incubator ML) and heated stage insert (Pecon Heating Insert P). For time lapse experiments, keratinocytes were seeded into chamber slides (Lab-Tek/Nunc, Rochester, NY) and then switched to medium containing 0.55mM calcium for 16–18 hours prior to the launch of the experiment. Keratinocytes were placed on ice and labeled with Alexa Flour 555-tagged AK23 or AK15 monoclonal antibodies directed against Dsg3 (Tsunoda et al., 2003) for 20–30 minutes. Unbound AK23 or AK15 antibody was removed using a PBS+ wash and fresh media containing PV IgG was added and then to the cells were warmed to 37° C. Time lapse images and movies represent projection images of the entire cell z-plane of the cell. Retrospective analysis was achieved by removing the cells from the microscope chamber, immediately fixing in 4.0 % paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 minutes followed by 0.1% Triton X-100 (Roche Diagnostics, Indianapolis, IN) extraction and staining with appropriate primary antibody. Upon completion of staining, fixed keratinocytes were placed back on the microscope in the exact location where time lapse imaging occurred.
Sequential Detergent Extraction and Western Blot Analysis
For western blot analysis, keratinocytes were grown in 35mm or 4 well tissue culture plates and extracted sequentially in Triton buffer (1% Triton X-100, 10mM Tris, pH 7.5, 140 mM NaCl, 5mM EDTA, 2 mM EGTA, with protease inhibitor cocktail tablets from Roche Diagnostics, Indianapolis, IN) followed by extraction with Urea-SDS buffer (1% SDS, 8 M Urea, 10mM Tris-HCL, pH 7.5, 5 mM EDTA, 2 mM EGTA) as described previously (Calkins et al., 2006). Rabbit anti-Glyceraldehyde 3-phosphate dehydrogenaseA (GAPDH, Santa Cruz), mouse anti-cytokeratin and anti-plakoglobin (mentioned above) were used as loading controls for the Triton soluble and insoluble fractions. Horseradish peroxidase-conjugated secondary antibodies (Bio-Rad, Hercules, CA) were detected using enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Dispase Cell Dissociation Assay
Keratinocytes were grown to 100 % confluence in 35 mm tissue culture plates. Cells were switched into 1.2 mM calcium containing media 16–18 hours prior to the addition of NH or PV IgG at approximately 100 µg/ml. After 24 hours, keratinocytes were washed with PBS+ and incubated with 1 unit/ml dispase (Roche Diagnostics, Indianapolis, IN) for 30 minutes to an hour. Following release from the culture plate, the cell sheets were transferred to 15-ml conical tubes containing 11 ml final volume of PBS+ and subjected to mechanical stress by 50 inversion cycles on a rocker panel. Fragments were counted using a dissecting microscope. Adenovirus expressing Dsg3.GFP or empty vector adenovirus (GFP alone) was added to cells 24 hours prior to the addition of PV IgG or NH IgG.
Adenovirus Expression
A full length GFP tagged Dsg3 adenoviral expression constructed was generated as follows. The cDNA encoding mouse Dsg3 was subcloned into pEGFP-N1 (Clontech, Mountain View, CA). GFP tagged Dsg3 (Dsg3.GFP) was further subcloned into pENTR11 vector (Invitrogen, Carlsblad, CA). A pAd-CMV-Dsg3.GFP expression vector was then generated with pENTR11- Dsg3.GFP using the Gateway system. PacI digested pAd-CMV-Dsg3.GFP was transfected into 293 cells and recombinant Dsg3.GFP adenovirus was generated and purified as described previously (Delva et al., 2008).
Supplementary Material
Acknowledgements
The authors acknowledge insightful comments and advice from members of the Kowalczyk laboratory as well as Drs. Kathleen Green and Victor Faundez. The authors also gratefully acknowledge Dr. Laura Delong for assistance with statistical analysis and Mr. Christopher Caughman for technical assistance. This work was supported by funding from the NIH/NIAMS (R01 AR048266). MDK was supported by T32AR007587, DKT was supported by T32GM008367, and ED was supported by F31CA110278.
Abbreviations
- PV IgG
pemphigus vulgaris IgG
- NH IgG
normal human IgG
Footnotes
Conflict of Interest: The authors state no conflict of interest.
References
- Amagai M. Pemphigus vulgaris and its active disease mouse model. Curr Dir Autoimmun. 2008;10:167–181. doi: 10.1159/000131453. [DOI] [PubMed] [Google Scholar]
- Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5:258–267. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass-Zubek AE, Hobbs RP, Amargo EV, Garcia NJ, Hsieh SN, Chen X, et al. Plakophilin 2: a critical scaffold for PKC alpha that regulates intercellular junction assembly. J Cell Biol. 2008;181:605–613. doi: 10.1083/jcb.200712133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Christiano AM. Broken hearts, woolly hair, and tattered skin: when desmosomal adhesion goes awry. Curr Opin Cell Biol. 2007;19:515–520. doi: 10.1016/j.ceb.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz P, Hu P, Liu Z, Diaz LA, Enghild JJ, Chua MP, et al. Desmosome signaling. Inhibition of p38MAPK prevents pemphigus vulgaris IgG-induced cytoskeleton reorganization. J Biol Chem. 2005;280:23778–23784. doi: 10.1074/jbc.M501365200. [DOI] [PubMed] [Google Scholar]
- Berkowitz P, Hu P, Warren S, Liu Z, Diaz LA, Rubenstein DS. p38MAPK inhibition prevents disease in pemphigus vulgaris mice. Proc Natl Acad Sci U S A. 2006;103:12855–12860. doi: 10.1073/pnas.0602973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins CC, Setzer SV, Jennings JM, Summers S, Tsunoda K, Amagai M, et al. Desmoglein endocytosis and desmosome disassembly are coordinated responses to pemphigus autoantibodies. J Biol Chem. 2006;281:7623–7634. doi: 10.1074/jbc.M512447200. [DOI] [PubMed] [Google Scholar]
- de Bruin A, Caldelari R, Williamson L, Suter MM, Hunziker T, Wyder M, et al. Plakoglobin-dependent disruption of the desmosomal plaque in pemphigus vulgaris. Exp Dermatol. 2007;16:468–475. doi: 10.1111/j.1600-0625.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- Delva E, Jennings JM, Calkins CC, Kottke MD, Faundez V, Kowalczyk AP. Pemphigus vulgaris IgG-induced desmoglein-3 endocytosis and desmosomal disassembly are mediated by a clathrin- and dynamin-independent mechanism. J Biol Chem. 2008;283:18303–18313. doi: 10.1074/jbc.M710046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E, Kowalczyk AP. Regulation of cadherin trafficking. Traffic. 2009;10:259–267. doi: 10.1111/j.1600-0854.2008.00862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai BV, Harmon RM, Green KJ. Desmosomes at a glance. J Cell Sci. 2009;122:4401–4407. doi: 10.1242/jcs.037457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Gliem M, Heupel W-M, Spindler V, Harms GS, Waschke J. Actin reorganization contributes to loss of cell adhesion in pemphigus vulgaris. Am J Physiol Cell Physiol. 2010;299:C606–C613. doi: 10.1152/ajpcell.00075.2010. [DOI] [PubMed] [Google Scholar]
- Godsel LM, Hsieh SN, Amargo EV, Bass AE, Pascoe-McGillicuddy LT, Huen AC, et al. Desmoplakin assembly dynamics in four dimensions: multiple phases differentially regulated by intermediate filaments and actin. J Cell Biol. 2005;171:1045–1059. doi: 10.1083/jcb.200510038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochim Biophys Acta. 2007;1773:69–77. doi: 10.1016/j.bbamcr.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Heupel WM, Zillikens D, Drenckhahn D, Waschke J. Pemphigus vulgaris IgG directly inhibit desmoglein 3-mediated transinteraction. J Immunol. 2008;181:1825–1834. doi: 10.4049/jimmunol.181.3.1825. [DOI] [PubMed] [Google Scholar]
- Holthofer B, Windoffer R, Troyanovsky S, Leube RE. Structure and function of desmosomes. Int Rev Cytol. 2007;264:65–163. doi: 10.1016/S0074-7696(07)64003-0. [DOI] [PubMed] [Google Scholar]
- Kitajima Y. Mechanisms of desmosome assembly and disassembly. Clin Exp Dermatol. 2002;27:684–690. doi: 10.1046/j.1365-2230.2002.01116.x. [DOI] [PubMed] [Google Scholar]
- Kottke MD, Delva E, Kowalczyk AP. The desmosome: cell science lessons from human diseases. J Cell Sci. 2006;119:797–806. doi: 10.1242/jcs.02888. [DOI] [PubMed] [Google Scholar]
- Lai-Cheong JE, Arita K, McGrath JA. Genetic diseases of junctions. J Invest Dermatol. 2007;127:2713–2725. doi: 10.1038/sj.jid.5700727. [DOI] [PubMed] [Google Scholar]
- Mao X, Choi EJ, Payne AS. Disruption of desmosome assembly by monovalent human pemphigus vulgaris monoclonal antibodies. J Invest Dermatol. 2009;129:908–918. doi: 10.1038/jid.2008.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EJ, Williamson L, Kolly C, Suter MM. Outside-in signaling through integrins and cadherins: a central mechanism to control epidermal growth and differentiation? J Invest Dermatol. 2008;128:501–516. doi: 10.1038/sj.jid.5701248. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Arredondo J, Chernyavsky AI, Kitajima Y, Pittelkow M, Grando SA. Pemphigus vulgaris IgG and methylprednisolone exhibit reciprocal effects on keratinocytes. J Biol Chem. 2004;279:2135–2146. doi: 10.1074/jbc.M309000200. [DOI] [PubMed] [Google Scholar]
- Payne AS, Hanakawa Y, Amagai M, Stanley JR. Desmosomes and disease: pemphigus and bullous impetigo. Curr Opin Cell Biol. 2004;16:536–543. doi: 10.1016/j.ceb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Sato M, Aoyama Y, Kitajima Y. Assembly pathway of desmoglein 3 to desmosomes and its perturbation by pemphigus vulgaris-IgG in cultured keratinocytes, as revealed by time-lapsed labeling immunoelectron microscopy. Lab Invest. 2000;80:1583–1592. doi: 10.1038/labinvest.3780168. [DOI] [PubMed] [Google Scholar]
- Sharma P, Mao X, Payne AS. Beyond steric hindrance: the role of adhesion signaling pathways in the pathogenesis of pemphigus. J Dermatol Sci. 2007;48:1–14. doi: 10.1016/j.jdermsci.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Shimizu A, Ishiko A, Ota T, Tsunoda K, Amagai M, Nishikawa T. IgG binds to desmoglein 3 in desmosomes and causes a desmosomal split without keratin retraction in a pemphigus mouse model. J Invest Dermatol. 2004;122:1145–1153. doi: 10.1111/j.0022-202X.2004.22426.x. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Stanley JR, Amagai M. Pemphigus, bullous impetigo, and the staphylococcal scalded-skin syndrome. N Engl J Med. 2006;355:1800–1810. doi: 10.1056/NEJMra061111. [DOI] [PubMed] [Google Scholar]
- Tsunoda K, Ota T, Aoki M, Yamada T, Nagai T, Nakagawa T, et al. Induction of pemphigus phenotype by a mouse monoclonal antibody against the amino-terminal adhesive interface of desmoglein 3. J Immunol. 2003;170:2170–2178. doi: 10.4049/jimmunol.170.4.2170. [DOI] [PubMed] [Google Scholar]
- Waschke J. The desmosome and pemphigus. Histochem Cell Biol. 2008;130:21–54. doi: 10.1007/s00418-008-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–235. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, Posthaus H, et al. Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. Embo J. 2006;25:3298–3309. doi: 10.1038/sj.emboj.7601224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Aoyama Y, Shu E, Tsunoda K, Amagai M, Kitajima Y. Anti-desmoglein 3 (Dsg3) monoclonal antibodies deplete desmosomes of Dsg3 and differ in their Dsg3-depleting activities related to pathogenicity. J Biol Chem. 2007;282:17866–17876. doi: 10.1074/jbc.M607963200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.