Abstract
We identified complex genomic rearrangements consisting of intermixed duplications and triplications of genomic segments at both the MECP2 and PLP1 loci. These complex rearrangements were characterized by a triplicated segment embedded within a duplication in 12 unrelated subjects. Interestingly, only two novel breakpoint junctions were generated during each rearrangement formation. Remarkably, all the complex rearrangement products share the common genomic organization duplication-inverted triplication-duplication (DUP-TRP/INV-DUP) wherein the triplicated segment is inverted and located between directly oriented duplicated genomic segments. We provide evidence that the DUP-TRP/INV-DUP structures are mediated by inverted repeats that can be separated by over 300 kb; a genomic architecture that apparently leads to susceptibility to such complex rearrangements. A similar inverted repeat mediated mechanism may underlie structural variation in many other regions of the human genome. We propose a mechanism that involves both homology driven, via inverted repeats, and microhomologous/nonhomologous events.
Keywords: BIR, inversion, MMBIR, MECP2, PLP1, duplication, complex rearrangements
One of the surprising outcomes of clinical implementation of tiling-path high-resolution comparative genomic hybridization arrays (aCGH) is the frequent observation of complex genomic rearrangements, some of which include triplications. Despite the clinical relevance of genomic triplications encompassing dosage sensitive genes to both diagnosis and prognosis, the molecular mechanism(s) for triplication formation are poorly understood. Triplications have remained an enigma potentially due to both a paucity of patients reported in the literature and experimental challenges to breakpoint determination; the latter information is a prerequisite to infer mechanism. We recently reported a cohort of 30 patients with MECP2 duplications in which we identified six patients (20%) with a triplicated segment embedded within the duplication. Preliminary molecular characterization of three tandem duplications revealed microhomology in two cases (3-4 bp in length). Breakpoints from one triplication suggested potential inversion occurring concomitant with triplication formation, but the mechanism for complex duplication/triplication formation remained perplexing.
Our observations led us to hypothesize that a replication-based mechanism, such as Break-Induced Replication (BIR)1-5, Fork Stalling and Template Switching (FoSTeS)6,7 and/or Microhomology-Mediated Break Induced Replication (MMBIR)4,5 underlies formation of complex rearrangements including triplications and inversions. We also hypothesized that triplication involving dosage-sensitive genes, such as MECP2, could potentially produce a more severe clinical phenotype than duplications8. Therefore, to obtain further insight into the mechanism for triplication formation, as well as to learn how triplications impact the clinical phenotype, we studied nine patients from eight families with unique duplication and triplication rearrangements encompassing the MECP2 gene. Remarkably, our data unveiled a rearrangement product with shared structural features (Fig. 1) suggesting a common mechanism for complex duplication (DUP)/triplication (TRP) formation. Further analysis supports a role for a replication-based mechanism that relies on the presence of low copy repeats (LCRs) in an inverted orientation. The same structural pattern for rearrangement products was also observed in patients with triplications embedded in duplications at the PLP1 locus in Xq22, suggesting that the same specific mechanism might underlie triplication formation at other loci in the human genome.
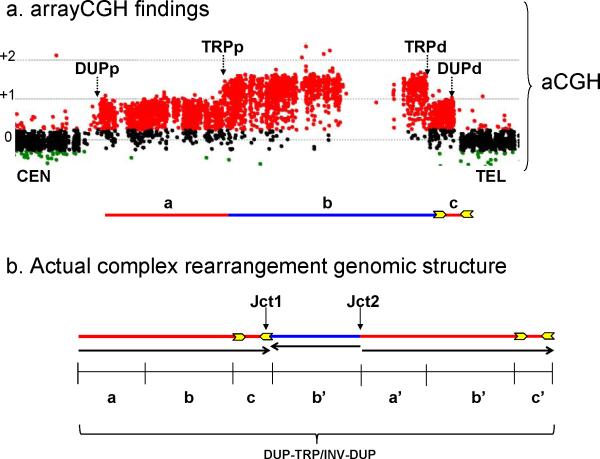
Figure 1. General genomic structure of the complex rearrangements triplications embedded in duplications.
(a) Copy number dosage alteration inferred from aCGH. A typical aCGH experiment is shown for a complex DUP-TRP-DUP rearrangement. Transitions of copy number dosage alterations are demonstrated by black vertical dotted arrows; the size of genomic segments defined by those boundaries vary in each individual complex rearrangement and are denoted a, b, c. The horizontal line below depicts the array data. Duplications are represented in red and triplications in blue; yellow arrows represent inverted repeats. (b) Representative figure of the genomic structure as determined by further analysis of copy number breakpoint junctions (jct1 and jct2) in five independent rearrangements using multiple molecular experimental approaches. The genomic segments involved are denoted a, b, c whereas the respective copy number gains are denoted as a’, b’, c’. These findings were corroborated by three additional independent rearrangements for which we obtained information for either jct1 or jct2 from two genomic loci. Change in orientation of the genomic segment (represented by black arrows) occurred at each of the junctions formed. DUPp: proximal transition/breakpoint of duplications; TRPp: proximal transition/breakpoint of triplication; DUPd: distal transition/breakpoint of duplications; TRPd: distal transition/breakpoint of triplications.
The severity of disease observed in patients with triplication positively correlates with the copy number status of MECP2 and IRAK1 in multiple patients studied, further confirming observations from case reports. Our findings elucidate a common structure DUP-TRP/INV-DUP as one potential outcome for human genome rearrangements that utilize inverted repeats as substrates for recombination. We further observed that an incremental increase of MECP2 dosage from two to three copies results in a more severe phenotype with additional novel and distinct clinical findings.
Results
Triplications embedded in duplications spanning MECP2
We previously identified six complex rearrangements (triplications embedded within duplications) in a cohort of 30 patients with MECP2 duplication by high-resolution human genome analysis using customized high-density array CGH9. An additional four subjects with a complex DUP-TRP-DUP pattern were identified by array CGH suggesting that complex rearrangements are a relatively frequently observed outcome of genomic alterations at this locus. We systematically investigated these complex rearrangements to characterize the molecular features of the rearrangement product. In total, we studied nine patients with triplications embedded in duplications; in five cases the MECP2 gene is included within the triplicated segment (Fig. 2a).
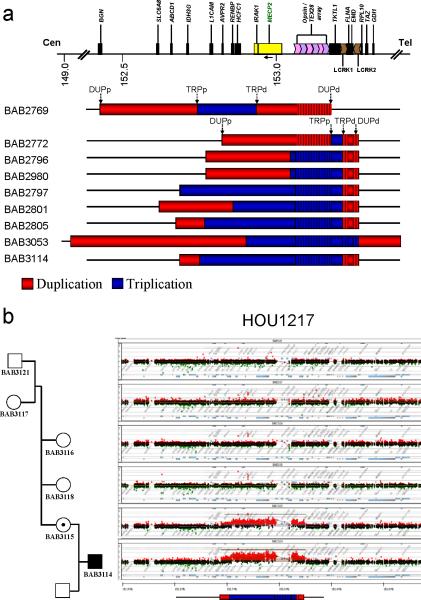
Figure 2. Individuals carrying complex triplications of Xq28.
(a) Genomic region harboring the alterations involving MECP2. Duplications are represented in red and triplications in blue. Arrows on top of the BAB2769 and BAB2772 rearrangements indicate the position of the transitions to gain according to array-based Comparative Genomic Hybridization (aCGH); in 6 out of 8 cases (BAB2796 and BAB2980 are brothers), the distal transition/breakpoint of both duplications (DUPd) and triplications (TRPd) (indicated by arrows) cluster within a pair of LCRs termed K1 and K2 contrasting with the scattered nature of the proximal breakpoints of both duplications (DUPp) and triplications (TRPp) in the same group of patients. Vertical lines embedded within the rearrangement bars represent low copy repeat regions (LCRs) for which the copy numbers were not inferred due to poor probe coverage. (b) aCGH (Agilent Technologies, Santa Clara, CA) result for family HOU1217. Carrier mother BAB3115 with a de novo complex triplication that was transmitted to her son (BAB3114).
Both triplication and duplication sizes are unique in each family and ranged from 41 kb to 537 kb and from 444 kb to 5.7 Mb, respectively. The triplicated region includes the entire MECP2 gene in five patients: BAB2797, BAB2801, BAB2805, BAB3053 and BAB3114 (Fig. 2a). Oligonucleotide-array CGH revealed that all of the complex rearrangements were inherited from a carrier mother, except for BAB3053 who harbors a translocation to Yq11.22. The breakpoint at Yq11.22 was not precisely mapped due to the paucity of unique sequences on the Y chromosome.
We independently confirmed genomic triplications in each of the nine families by both Multiplex Ligation-Dependent Probe Amplification (MLPA) and Fluorescence in situ Hybridization (FISH) (Supplementary Fig. 1 and data not shown). The mothers and grandmothers when available for study were tested by both CGH and MLPA and were shown to carry the same complex rearrangement as their sons or grandsons in all but one family (pedigree HOU1217, Fig. 2b), in which aCGH studies revealed that the complex rearrangement was a de novo event in the mother (BAB3115) (Fig. 2b). X-chromosome inactivation (XCI) studies revealed 100% advantageously skewed XCI patterns in all carrier females tested (data not shown); i.e. consistent with preferential inactivation of the X chromosome harboring the complex rearrangement. Family pedigrees are displayed in Supplementary Fig. 2.
Duplicated and triplicated segments likely originate from the same chromosome
To examine for potential interchromosomal exchanges during rearrangement product formation, we evaluated marker haplotypes from the genomic interval spanning the complex rearrangement using the Illumina HumanOmni1-Quad microarray. All patients except BAB3053 were notable for absence of heterozygosity for all SNPs tested using this platform, including SNPs localized to both duplicated and triplicated genomic intervals (Supplementary Fig. 3, Supplementary Table 1). Subject BAB3053 carries a translocation of MECP2 sequences to Yq11 and shows multiple heterozygous SNPs, suggesting that this complex rearrangement was generated by a distinct mechanism.
The absence of heterozygosity observed in all nontranslocation DUP-TRP-DUP products suggests that the substrate(s) for these complex genomic rearrangements originated from a single chromosome. This contention is supported by the results obtained in family HOU1217, in which the patient BAB3114 inherited the complex rearrangement from his mother (BAB3115) who is a de novo carrier. SNP array analysis revealed that the segment to which the rearrangement maps was inherited from his maternal grandfather (Supplementary Fig. 4 and Table 2), suggesting a premeiotic event during male gametogenesis.
Triplicated segments are inserted in inverted orientation amid the duplications
Complex rearrangements can be defined by multiple breakpoint junctions or join points that juxtapose discreet genomic segments. The genomic rearrangement complexity was revealed by aCGH; however, aCGH provides neither orientation nor genomic positional information for the complex rearrangement but rather only copy number information. Based on aCGH results that demonstrated distinct transitions at gains of genomic intervals (i.e. duplication versus normal, triplication versus duplication), we initially hypothesized the existence of at least four potential breakpoints per patient; two for transitions to and from duplications (proximal and distal) and two for transitions to and from triplications (proximal and distal) (Fig. 1a and 2a). However, the simplest hypothesis is that each of the two duplication/triplication breakpoints was joined during rearrangement formation, ultimately producing only two breakpoint junctions, designated breakpoint junction 1, jct1 and breakpoint junction 2, jct2 (Fig. 1b).
To test this latter hypothesis, we first sought to obtain breakpoint junctions using both conventional and long-range PCR and by attempting to use primer pairs in all possible orientations; i.e. inwardly-facing, outwardly-facing, forward primer pairs, and reverse primer pairs. These primers were designed at the apparent boundaries, as denoted by transitions signifying a gain of each duplicated or triplicated segment relative to the reference genome as inferred from the aCGH results. In cases of failure to obtain breakpoint junctions using this assay, alternative experimental approaches were attempted. These alternative approaches included inverse PCR (iPCR) or Southern analyses, both of which have the advantage of not relying on any preconceived notion of genome structure for the rearrangement.
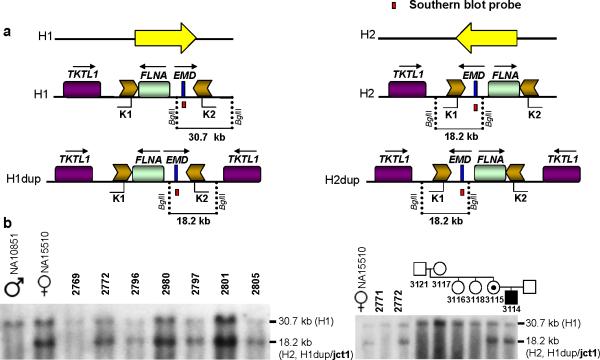
Southern blotting was used to analyze the recurrent breakpoint junctions mapping to the inverted repeat pair of low copy repeats (LCRs) K, which is involved in six our of eight independent complex rearrangements in our cohort (Fig. 2a). This assay was performed as described previously10; for males, the expected result was either a 30.7 kb band corresponding to a reference size structural variation haplotype (H1) or an 18.2 kb band corresponding to a polymorphic inversion of the region flanked by the LCR K1 and the LCR K2 that is present in 18% of the population of European-descent10 (H2) (Fig. 3a). Females could potentially carry either one allele (the 30.7 kb or 18.2 kb) in the homozygous state or both alleles as heterozygotes (NA15510, Fig. 3b). To our surprise, all male samples carrying dup/trip involving the LCR K1 and the LCR K2 (BAB2772, BAB2796/BAB2980, BAB2797, BAB2801, BAB2805 and BAB3114) yielded the same pattern consisting of two bands, 18.2 kb and 30.7 kb, corresponding to those usually observed with the H2 and H1 inversion haplotype structures, respectively. We surmised that the unexpected presence of both bands in all male patients was a result of rearrangement formation which suggests that all seven samples have a common jct1 structure. In addition, an 18.2 kb band is expected if the centromeric-flanking region (which contains the TKTL1 gene) is duplicated and inverted whilst still flanking the LCR K1 and the LCR K2 on either the reference (H1dup) or the inverted structure (H2dup) on the ancestral chromosome (Fig. 3a). Therefore, we hypothesized that the 18.2 kb band corresponds to jct1 and, by inference that the 30.7 kb band corresponds to the ancestral state (H1 structure) in these chromosomes. We confirmed this hypothesis using the haplotype data obtained from SNP arrays; all patients in our cohort carry the SNP haplotype associated with the H1 structure (Supplementary Fig. 5).
Figure 3. Southern blot analysis of the region flanked by LCRs K1 and K2 at the Xq28 chromosome.
LCRs K1 and K2 are approximately 11.3 kb in length, and are located ~38 kb apart in inverted orientation. Their 99% nucleotide sequence identity is likely maintained by frequent gene conversion34. These LCRs flank two genes FLNA and EMD. (a) Yellow arrows indicate an inversion: cent-FLNA/EMD-tel (H1) and the alternative genomic orientation cent-EMD/FLNA-tel (H2). An 18.2 kb band is expected to be produced in either inversion haplotype background (H1dup or H2dup) upon duplication and inversion; we hypothesize that the 18.2 kb band includes the breakpoint junction 1 (jct1). To test the haplotype of our cohort and to map the duplication breakpoints, we performed a Southern blot assay as shown here. Genomic DNA was digested with BglII; EMD was targeted using a PCR-based probe. The reference genome H1 produces a 30.7 kb band whereas the inversion haplotype (H2) yields an 18.2 kb band. (b) Southern blot results for patients carrying triplications embedded within duplications: BAB2769, BAB2772, BAB2796/BAB2980, BAB2797, BAB2801, BAB2805 (left) and family HOU1217 (right). NA10851: male carrying the reference haplotype (H1); NA15510 heterozygous female carrying both reference and inversion haplotypes (H1 and H2). BAB2771: patient carrying MECP2 duplication not involving LCR K1 and LCR K2 (H1).
Three important conclusions can be drawn from these experimental observations: 1) the inverted LCRs K1 and K2 likely mediated the rearrangements; 2) the new segment copied (containing the TKTL1 gene) was inserted in an inverted orientation with respect to the original copy; and 3) a second event, likely represented by jct2, must have occurred in order to “reverse” the inversion process. Supporting our experimental observations, a de novo complex rearrangement occurring in association with sporadic disease in family HOU1217 revealed a novel formation of the 18.2 kb band in addition to the 30.7 kb band already present in all family members. As anticipated, BAB2769 has a 30.7 kb band size corresponding to the reference structural H1 haplotype but no 18.2 kb band, consistent with the fact that BAB2769 is the only sample for which the complex rearrangement does not include the LCRs K1 and K2.
Jct1 for patient BAB2769 was obtained by sequencing across the junction using reverse primer pairs positioned at the proximal ends of the duplication and triplication, respectively. Remarkably, the junction consists of two identical 149 bp segments, present as two small inverted repeats (856 bp) located 317.8 kb apart from each other in the haploid reference human genome sequence (Fig. 4). These inverted repeats are 98% identical in sequence. Thus, in all seven cases in which jct1 were identified, an inverted repeat was located at the breakpoint junction.
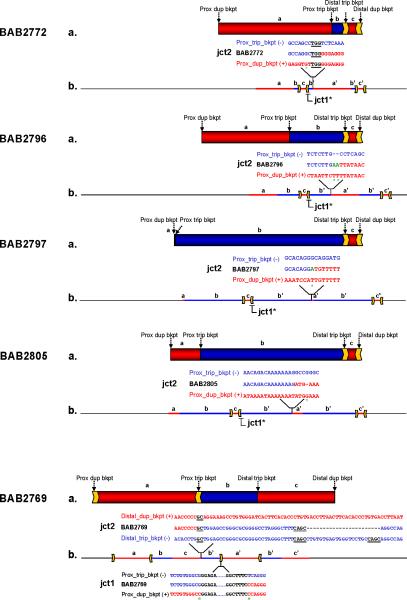
Figure 4. Rearrangement structure for patients BAB2769, BAB2772, BAB2796/BAB2980, BAB2797 and BAB2805 based on aCGH, Southern blotting and breakpoint sequencing.
(a) Genomic region harboring duplications and triplications spanning chromosome Xq28 according to aCGH: duplications are represented in red, triplications in blue. Arrows on top of the rearrangements indicate the position of the breakpoints; inverted repeats involved in the rearrangement are represented as yellow arrows. Letters a, b and c represent the segments with copy-number gain. (b) Individual genomic structure of the region involved in the rearrangement as inferred by analysis of breakpoint junction 1 (jct1) and breakpoint junction 2 (jct2) for each patient. Jct1* represents those junctions analyzed by Southern blotting (please refer to Fig. 3); all others were sequenced. Genomic positions of each of the junctions are shown. Breakpoint junction sequences are color-coded to highlight their segment of origin in the reference genome (duplications in red and triplications in blue). The triplicated segment (b’) is inserted amid a normal (a, b, c) and a duplicated copy (a’, c’) in inverted orientation as supported by jct1 and jct2 analysis. This structure was further confirmed by FISH for patient BAB2805 (Supplementary Fig. 6). Microhomologies observed at the junctions are represented by underlined black letters; insertions or mismatches at the junctions are represented in green; deletions are represented by dashes; mismatches between the reference sequences and patient sequences are marked with a green asterisk underneath.
Jct2 in five out of eight rearrangements (BAB2769, BAB2772, BAB2796/BAB2980, BAB2797 and BAB2805) was obtained by PCR (regular, long-range or inverted PCR). For patients BAB2772, BAB2796/BAB2980, BAB2797 and BAB2805, the breakpoints were obtained using reverse primers at the proximal ends of the duplication and the triplication, respectively. Jct2 in patient BAB2769 was obtained using forward primer pairs designed at the distal ends of the duplication and triplication, respectively. Routine PCR was attempted first followed by sequencing of the PCR products. One junction was obtained by iPCR (BAB2805); three samples (BAB2801, BAB3053 and BAB3114) were refractory to all attempts to amplify a unique breakpoint junction.
Analysis of the breakpoint sequences of jct2 revealed that the triplicated segment is inverted relative to the duplicated segment in all patients (BAB2769, BAB2772, BAB2796/BAB2980, BAB2797 and BAB2805). Microhomologies of 2 to 4 nucleotides were observed in two out of five cases (BAB2772 and BAB2769, Fig. 4); in two cases, one nucleotide A or two nucleotides AA were inserted at the junction (BAB2796/BAB2980 and BAB2797); in one case (BAB2805), the junction was perfectly joined. In all five cases, one of the breakpoints occurred within or adjacent to a repetitive sequence element such as a SINE or a LINE (Table 1). A few nucleotide dissimilarities flanking the junctions were observed in two cases (BAB2772: transversion C=>G; BAB2805: deletion of one G, Fig. 4); we interpret these dissimilarities to be likely population polymorphisms that are not yet deposited in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). Alternatively, there is evidence that the polymerase(s) involved in break-induced-replication (BIR) are ‘error prone’, with poor processivity2 at initiation followed by lower replication fidelity compared to normal DNA replication11. The jct2 for subject BAB2769 reveals a break that we interpret as two template-switching events. The first event is represented by a GC microhomology that connects the distal duplication breakpoint to the distal triplication breakpoint; the second event is represented by a microhomology of CAGC accompanying a deletion of 23 bp on the distal triplication side (Fig. 4).
Table 1.
Presence of inverted repeats at the breakpoint junctions of genomic triplications observed in the present study and in the literature
| Chromosomal alteration | Patient ID | Size* | Method used to detect complexity | Complexity pattern | Jct1 | Jct2 | Reference |
|---|---|---|---|---|---|---|---|
| dup PLP1 | BAB1290 | 800 kb | aCGH | dup-trpa-dup | IRs at dist brkpt | NA | 6 |
| dup PLP1 | BAB1612 | 125 kb | aCGH | dup-trp/inva-dup | IRs at dist brkpt | Microhomology ACCT, Prox trip: L2 | 35This study |
| dup PLP1 | BAB2389 | 4.0 Mb | aCGH | dup-trpa-dup | IRs at dist brkpt | NA | 6 |
| dup MECP2 | BAB2769 | 651 kb | aCGH Brkpt seq Southern | dup-trp/inv-dup | IRs at prox brkpt | Microhomologies GC, CAGC, 23 nt del Dist dup: MER73 | This study |
| dup MECP2 | BAB2772 | 444 kb | aCGH Brkpt seq Southern | dup-trp/inv-dup | IRs at dist brkpt | Microhomology TGG Prox trip: AluSx Prox dup: low complexity | This study |
| dup MECP2 | BAB2796 BAB2980 |
497 kb | aCGH Brkpt seq Southern | dup-trp/inv-dup | IRs at dist brkpt | Insertion AA Prox trip: AluSx | This study |
| dup MECP2 | BAB2797 | 586 kb | aCGH Brkpt seq Southern | dup-trp/inv-dup | IRs at dist brkpt | Insertion A Prox dup: LIM5 | This study |
| dup MECP2 | BAB2801 | 653 kb | aCGH Southern | dup-trp/inv-dup | IRs at dist brkpt | NA | This study |
| dup MECP2 | BAB2805 | 597 kb | aCGH/Brkpt seq Southern | dup-trp/inv-dup | IRs at dist brkpt | No microhomology Prox trip: LIM4 Prox dup: AluSx/LIMB4 | This study |
| dup MECP2 | BAB3114 | 586 kb | aCGH Southern | dup-trp/inv-dup | IRs at dist brkpt | NA | This study |
| dup MECP2 | BAB2681 | 558 kb | aCGH | dup-trpa-dup | IRs at dist brkpt | NA | 9 |
| dup MECP2 | BAB2727 | 430 kb | aCGH Brkpt seq | dup-trp/inv-dup | IRs at dist brkpt | Microhomologies GG CA, ACA insertion Prox trip: AluSx | 9 |
| dup MECP2 | Family 57 | 492 kb | aCGH | dup-trpa-dup | IRs at dist brkpt | NA | 17 |
| Trip 2q11.2-q21 | - | 36 Mb to 41 Mb | Karyotype and FISH | trp/inv | IRs at 2q21 | - | 25 |
| Trip SNCA | - | 1.6 Mb to 2.0 Mb | Real-time PCR FISH | trpa | IRs at prox brkpt | - | 36 |
| Trip 15q11-q13 | - | 10 Mb to 14 Mb | Karyotype and FISH | trp/inv | IRs at 15q13 | - | 24 |
| Trip 7q11.23 | - | ~1.25 Mb | aCGH | trpa | IRs at dist brkpt | - | 37 |
N.A., not available
Prox trip, proximal triplication junction; Dist trip, distal triplication junction; Prox dup, proximal duplication junction; Dist dup, distal duplication junction; IRs, inverted repeats.
As defined in this present study, jct1 and jct2 are join points of duplications and triplications produced in the same molecular event. In this table, they were arbitrarily assigned to each of the two duplication/triplication join points of complex rearrangements published by other groups.
This is the size of the whole region involved in the complex rearrangement.
** aCGH was performed in the mother only.
The orientation of the triplicated segment is unknown.
bThere is no evidence for duplication.
In summary, analysis of two breakpoint junctions (jct1 and jct2) from each of five unrelated patients with triplications embedded within duplications at the Xq28 chromosome reveals a common structure in that the triplication was inserted in an inverted orientation within the duplication (i.e. DUP-TRP/INV-DUP). FISH experiments in patient BAB2805 reveal a pattern consistent with this DUP-TRP/INV-DUP genomic structure (Supplementary Fig. 6). Furthermore, in all cases one of the junctions of the rearrangement involved an inverted repeat pair, with the inverted genomic segments either closely approximated (38 kb) or separated by a sizable distance (>300 kb). These shared genomic architectural features are observed at breakpoints of all complex duplication/triplication alterations at the MECP2 locus analyzed herein.
Inverted repeats mediate triplication at the PLP1/Xq22 region
We have provided evidence to demonstrate that inverted repeats between 856 bp and 11.3 kb in length with at least 98% sequence identity and separated by ~38 kb to ~318 kb can mediate complex triplications (DUP-TRP/INV-DUP) at the MECP2 locus; and that the genomic rearrangement likely involved only one chromosome homologue. We applied these “rules” to reanalyze the breakpoint junctions of previously published DUP-TRP-DUP cases involving the PLP1 locus at Xq226,12 (see Table 1 and Supplementary Fig. 7). Remarkably, all three cases present the same pattern observed in DUP-TRP-DUP cases at Xq28: clustering of distal duplication and triplication breakpoints at a pair of inverted repeats (jct1) with high identity between each paralagous segment (~98.9% nucleotide identity, in this case separated by ~64 kb) plus scattered proximal breakpoints (jct2). In addition, sequencing of the proximal triplication breakpoint junction (jct2) in patient BAB1612 demonstrated inversion in regard to the reference genome and connection to the proximal duplicated segment consistent with a DUP-TRP/INV-DUP structure.
Phenotypic consequences of DUP-TRP/INV-DUP
The complex DUP-TRP/INV-DUP products vary in size for both triplicated and duplicated intervals (Fig. 2a). In the case of complex Xq28 genomic rearrangements, the MECP2 gene was either duplicated or triplicated. This distinction provided a unique opportunity to assess the phenotypic consequences of incremental increases in MECP2 gene dosage. The MECP2 gene was entirely mapped within the triplicated genomic interval in five patients with the complex DUP-TRP/INV-DUP rearrangement. Similar to observations in a previous case report8 and observations in patients without precise breakpoint junction mapping13, the phenotype associated with MECP2 triplication was more severe than that observed for MECP2 duplication.
The most salient clinical findings are summarized in Supplementary Table 3 (for complete clinical descriptions, please see Supplementary Note). Note that early respiratory insufficiency with an oxygen or ventilation requirement, early dysphagia and requirement for a feeding tube, hearing loss, and minor cardiac defects are much more commonly observed with MECP2 triplication (100%) compared with MECP2 duplication (0% to 25%), a robust observation even when compared to the collective published data on boys with MECP2 duplication13. Moreover, polyhydramnios and intestinal pseudoobstruction were observed clinically only in subjects with triplication. Interestingly, patients BAB2805 and BAB3114 were reported to have Xq28/MECP2 duplications by the diagnostic laboratories that performed their clinical chromosome microarray analysis. We correctly anticipated that the MECP2 gene was triplicated in patients BAB2805 and BAB3114 based on the observed clinical phenotype. Routine clinical diagnostic testing correctly identified Xq28/MECP2 triplication in the remaining three children with MECP2 triplications.
Discussion
We demonstrate from repeated independent cases of complex rearrangement at the MECP2 locus that a particular genomic rearrangement structure DUP-TRP/INV-DUP is associated with a specific and common pattern of underlying genomic architecture, namely the presence of inverted repeats separated by distances of up to hundreds of kb, including one pair too small (i.e. < 1kb) to be called segmental duplication under the current definition14. We show that these complex rearrangement events appear to involve a single X-chromosome homologue, likely in the male germline, generating carrier daughters and affected grandsons. The involvement of a single homologue is consistent with studies of copy number gain in other X-chromosome loci including duplications involving the DMD locus15 and PLP116. Furthermore, we provide evidence that DUP-TRP/INV-DUP occurring at the PLP1 locus is also associated with underlying inverted repeat genomic architecture.
Whereas many genomic disorders have been shown to result from CNV due to either duplication or deletion at a given locus, our data clearly show that triplication of MECP2 conveys a more severe, distinct and clinically recognizable syndrome.
Triplications embedded in duplications may share the same general genomic structure
We observed triplications embedded in duplications in at least 20% of the rearrangements involving MECP2 copy number gain9. This observation is supported by two recent reports in which triplications were observed in 2 out of 9 patients17 and in 2 out of 4 patients18. Our data show that triplications embedded in duplications at Xq28 share a common structure and reveal a potential common formation mechanism. This mechanism: i) requires two breakpoint junctions: one (jct1) invariably maps within inverted repeats with at least 98% sequence identity that can be separated by up to hundreds of kb, the second (jct2) is scattered and does not occur at sites of sequence homology; ii) the triplicated segment is inserted in an inverted orientation between duplicated sequences in direct orientation (one of the copies in direct orientation corresponds to the original copy); iii) the second breakpoint junction presents no extensive homology although some microhomologies may be found at the junction (e.g. BAB2769); iv) all extra segments (duplications and triplications) apparently originated from only one chromosome. The same pattern was also observed in patients carrying triplications embedded in a duplication reported at the PLP1 locus (Table 1 and Supplementary Fig. 6), suggesting that the mechanism producing triplication at Xq28 is also responsible for triplication formation elsewhere in the genome.
Towards a mechanism of formation of triplications embedded in duplications
We propose that DUP-TRP/INV-DUP complex rearrangements are formed by a combination of homology-directed BIR with microhomology-mediated BIR or non-homologous end joining (NHEJ) as described in Fig. 5. BIR is a mechanism that uses homologous recombination to restart a collapsed (broken) replication fork1-5. During this process, a 3’-tail at the broken DNA end invades the sister molecule from which it broke. The 3’-end primes DNA synthesis and forms a replication fork. BIR, in the cases discussed here, occurred non-allelically (ectopically), using the homology of the inverted LCR. This has the effect of synthesizing a length of chromatid back in the opposite direction from that in which the fork had been traveling, forming a large inverted duplication. This is unlikely to result in a healthy viable cell unless a second compensating inversion event occurs. If the reversed replication fork again collapses, or if there is a double-strand break in the chromatid carrying the inverted duplication, then there exists a new DNA end. This end could again be repaired by BIR. However, in the patients we studied, rejoining did not use homologous sequence. Instead, ends joined in inverted orientation to the unchanged chromatid by non-homologous end joining19 (if there was a second break), or by a replicative mechanism such as MMBIR4 or break-induced serial replication slippage (BISRS)20. These mechanisms are suggested to substitute for BIR when, for any reason, homologous recombination repair is unavailable. Such mechanisms yield the non-homologous or microhomologous joints that we see, including the complexities that are characteristic of events attributed to MMBIR. The complex microhomologous events observed in BAB2769 (Fig. 4) are characteristic of events attributed to the MMBIR mechanism4,6,9,21,22.
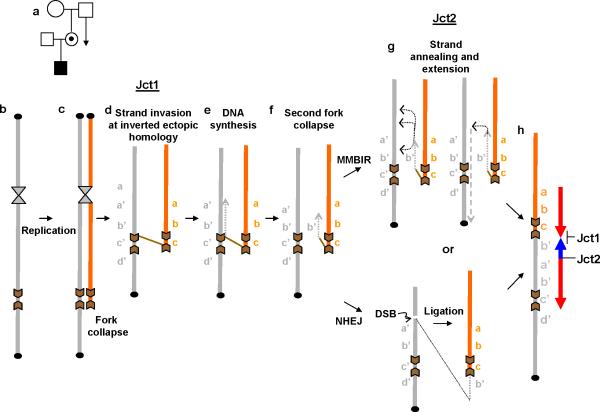
Figure 5. Proposed model for generation of the common DUP-TRP/INV-DUP rearrangement product.
(a) The rearrangement may have occurred during spermatogenesis in the ancestral male on his X chromosome, likely during S or G2 phase. (b) During replication of the sister chromatid, the replication fork may collapse and induce break-induced replication (BIR) (c) BIR uses homologous recombination to re-establish a new fork using ectopic homology provided by inverted repeats forming jct1. (d) This event initiates replication that forms a length of chromatid back in the opposite direction from that in which the fork had been traveling before the collapse, forming a large inverted duplication (e). If the reversed replication again collapses, or if there is a double-strand break in the chromatid carrying the inverted duplication, then there is a new DNA end (f). In our patient cohort, this new end rejoining (jct2 formation) occurred by either non-homologous end joining (NHEJ) requiring a double-strand break (DSB) on the original strand followed by ligation of that segment to the end of the rearranged newly replicated strand, or by a replication mechanism such as microhomology-mediated break-induced replication (MMBIR) (g) that requires a new strand annealing and extension to the end of the replicon or the chromosome. (h) Representative structure obtained after two steps of homologous and nonhomologous mechanisms. Duplicated and triplicated segments are represented in red and blue colors, respectively.
If this second inverted join links the duplication-carrying chromatid to the intact sister chromatid in direct orientation so that it compensates for the first inversion, and if the joint occurs beyond the length already duplicated, then a triplication embedded in duplication will result (Fig. 5). Six out of seven of the events reported here are interpreted as having initiated from replication forks oriented away from the centromere, and the seventh event commenced from a fork oriented towards the centromere. However, the sequence of events in the two configurations is the same. This mixture of orientations indicates that a dicentric intermediate was not necessary in this process. Thus, the model we propose is a two-step mechanism: BIR followed by a non-homologous or microhomologous mechanism, probably occuring during phase S or G2 in a single pre-meiotic cell in a male gonad.
The relevance of this model for formation of triplications in other parts of the genome as well as for formation of novel inversions is still to be unveiled. Duplications embedded in triplications inserted in inverted orientation have been reported; such as triplication encompassing chromosome 9q3423 and large interstitial triplications associated with inversions detected by FISH including 15q11-q1324, 2q11.2-q2125 and 13q12q2226 (Table 1). The observation of co-occurrence of triplications and inversions in other genomic disorders suggest that this mechanism may underlie the triplication formation at other sites of the human genome.
Inverted repeat genome architecture
Evidence that inverted repeats and palindromes can interfere with the replication process and lead to chromosomal rearrangements has been accumulating. Lebofsky and Bensimon27 studied the replication of the palindromic-laden human rDNA gene array using DNA molecular combing in HeLa cells and observed fork arrest associated with the presence of palindromic structures. Inverted Alu repeats close enough to form hairpins can cause a replication blockage in E. coli, yeast and mammalian cells in a homology-dependent manner28. The inverted repeats involved in the rearrangements observed in our cohort are not palindromes, as the spacer distance is too long; therefore, thus far there is no evidence that in these cases secondary structures such as hairpin and cruciform are causing fork stalling or fork collapse. However, our data add evidence that inverted repeats, even at a distance, can lead to rearrangements, and can contribute to local instability in the human genome.
Recently, Paek et al.29 observed fusion of nearby inverted repeats in budding yeast, and Mizuno et al.30 observed similar events in fission yeast. Paek et al.29 demonstrated that formation of dicentric and acentric fragments in budding yeast lead to further chromosome instability; Mizuno et al.30 also showed that formation of dicentric and acentric chromosomes followed replication fork arrest within palindromes. There was no evidence of involvement of either double-strand breaks or homologous recombination proteins in this process that is stimulated upon disruption of DNA replication29. The mechanism that Paek et al.29 proposed, termed “faulty template switching”, relies on homology between the inverted repeats to re-start a stalled fork that underwent a fork reversal; if the nascent strand pairs with the inverted nearby copy, then this will lead to an inverted repeat fusion and will result in the formation of an unstable dicentric chromosome prone to undergo further rearrangements29. Interestingly, paralleling our data, the inverted repeats involved could be several kilobases apart and share nucleotide identity as short as 20 bp. Whether fork reversal of inverted repeats, perhaps brought in proximity either by a replication factory31 or long-distance transcriptional regulatory complexes, or if collisions between ‘head-on’ and/or co-directional replication transcriptional conflicts32,33 can stimulate fork collapse potentially associated with inverted repeat directed DUP-TRP/INV-DUP formation remains unknown.
In conclusion, we document that the presence of inverted LCRs in the MECP2 vicinity are mediating genomic disorder-associated complex rearrangements that have a particular genomic structure DUP-TRP/INV-DUP. Furthermore, such a structure is also observed to occur at the PLP1 locus in association with inverted repeats. These genomic instability considerations are likely to apply wherever inverted repeats occur within a range of hundreds of kilobase pairs of DNA in the human genome. Moreover, structural variation in personal genomes may result in individual specific structural haplotypes that are more susceptible to the events reported herein. Of note, inversion during rearrangement formation can generate remarkable complexity with only two breakpoint junctions. Furthermore, multiple genic changes (e.g. gene interruptions, fusions, dosage changes, etc) can evolve with a single mutational event suggesting that complex genomic rearrangements such as DUP-TRP/INV-DUP may have an important role in evolution.
Supplementary Material
Acknowledgements
We thank all of the families who participated in this study. We gratefully acknowledge Dr. Pawel Stankiewicz for helpful discussions and Dr. Steven Scherer for providing us with the fosmids and BAC clones used in this study. This work was supported in part by NINDS grant R01 NS058529 to J.R.L., NIGMS grant R01 GM064022 to P.J.H., and NINDS grant 5K08NS062711-03 to M.B.R. Lymphoblast cell lines were developed by the BCM IDDRC cell culture core which is funded by award number P30HD024064 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS, NIGMS, NICHD or the NIH. Huda Zoghbi is an Investigator with the Howard Hughes Medical Institute.
Methods
Subjects
Families with genomic rearrangements of Xq28 including the MECP2 gene were identified by physician referral or self-referral. Informed consent for participation and sample collection was obtained using protocols H-18122, H-20268 and H-26667 approved by the Institutional Review Board for Baylor College of Medicine and affiliated hospitals.
Duplication size and genome content
To determine the size, genomic extent and gene content for each rearrangement, we designed a tiling-path oligonucleotide microarray spanning 4.6 Mb surrounding the MECP2 region on Xq28. The custom 4x44k Agilent Technologies microarray was designed using the Agilent earray website (http://earray.chem.agilent.com/earray/). We selected 22,000 probes covering ChrX: 150,000,000-154,600,000 (NCBI build 36), including the MECP2 gene, which represents an average distribution of 1 probe per 250 bp. Probe labeling and hybridization were performed as described9.
Long-Range PCR amplification
To investigate the possibility of inversions at breakpoint junctions, PCR amplification was attempted using only reverse primer pairs or forward primer pairs (relative to the reference genome) at the apparent boundaries of each duplicated or triplicated segment(s) as refined by aCGH analysis (primer sequences can be found in Supplementary Table 4). Long-range PCR was performed using TaKaRa LA Taq (Clontech, Mountain View, CA).
Inverse PCR
Genomic DNA (10 ug) was digested overnight with restriction enzyme MboI, (New England Biolabs, Ipswich, MA) and purified with a PCR Purification kit (Qiagen, Valencia, CA). The digested DNA was ligated in a volume of 1 ml with 10 U of T4 DNA ligase (Invitrogen Corporation, Carlsbad, CA) for 48 h. The ligated DNA was purified again with the PCR Purification kit and employed as a template for PCR amplification.
Multiplex Ligation-Dependent Probe Amplification (MLPA)
Dosage analysis of the IDH3G, L1CAM, IRAK1, MECP2, TKTL1, FLNA, GDI1, and FVIII genes were performed by MLPA analysis using commercially available reagents (SALSA P015-D2 probe mix) or custom-designed probes according to instructions from MRC-Holland (Amsterdam, The Netherlands). Ligation products were amplified and resolved on an ABI 3730 XL Genetic Analyzer and the data were analyzed using Genemapper software (Applied Biosystems). Patient data were normalized to gender-matched controls for copy-number differences using the Gene Marker Software (Softgenetics, State College, PA).
Fluorescence in situ Hybridization (FISH)
We harvested cultured lymphoblastoid (except in the case of BAB3053 we harvested cultured skin fibroblasts) patient cells after Colcemid (Sigma) treatment by standard methods, and dropped them onto glass slides. FISH probes (clones) were selected based on patient-specific information related to their presumed duplication or triplication status as determined by custom array CGH described herein. For interphase FISH, we selected BAC clones RP11-119A22 (test) and RP11-157E12 (control) as FISH probes to confirm triplication of the MECP2 gene in patients BAB2797, BAB2801, BAB2805 and BAB3053. Patient BAB2769 was analyzed using BAC clones RP11-846A22 (test) and RP11-157E12 (control). We labeled DNA by nick translation directly with Green-dUTP or Red-dUTP (Abbott Laboratories), and visualized the nuclei and metaphase chromosomes by fluorescence microscopy.
Probe design and Southern blot hybridization
We designed probes targeting the EMD gene mapped amid the LCRs K1 and K2 using primers described in Supplementary Table 4. We amplified the DNA probes by PCR from BAC clone CTD-2238E23 (product size: 683 bp). We digested DNA with BglII restriction enzyme for 1 day at 37°C (New England Biolabs), followed by separation on a 0.7% agarose gel in 0.5X Tris–Borate–EDTA buffer. Hybridization was performed as described38.
X-chromosome inactivation studies
X-inactivation studies were performed based on the protocol described by Allen et al. with modifications39 as described previously40.
Genotyping
DNA samples were quantified using Quant-iT PicoGreen dsDNA Reagent (Invitrogen) in a Tecan GENios microplate reader (Tecan Group, Mannendorf, Switzerland). Genotyping was performed on Illumina HumanOmni1-Quad microarrays (Illumina, Inc., San Diego, CA, U.S.A.) following the manufacturer's instructions. All microarrays had call rates > 0.99. Basic quality control and analysis of the genotyping data were performed on GenomeStudio software, version 2009.1 (Illumina, Inc., San Diego, CA, U.S.A.). Haplotype analysis was performed on Haploview 41, version 4.2. CNV calls were performed using cnvPartition v2.4.4 with default parameters. The cnvPartition algorithm was not able to distinguish between duplications and triplications (except in case BAB3053) as it ‘scored’ the entire rearranged region as duplicated (data not shown). The reason for this is not clear; likely it may be explained by a technical limitation of the algorithm which takes into account both relative probe intensity, log R ratio (LRR) and B allele frequency (BAF) deviation to estimate copy number.
Footnotes
Author Contributions
C.M.B.C. conducted high-density CGH arrays, FISH, breakpoint sequencing, Southern-blotting experiments and data analysis. M.B.R. coordinated human studies, recruited patients and analyzed clinical data. D.P. and P.L. assisted with high-density CGH arrays and breakpoint sequencing. L.M.F. and J.W.B. conducted SNP genotyping. E.K.P., S.H.D., L.S., L.F. S.L., R.S. recruited and clinically characterized patients C.G.J. assisted with data analysis. P.F. conducted the X-inactivation studies. A.M. and M.W. carried out cell culture. D.D.G. conducted MLPA. S.W.C. was involved in cytogenetic and clinical array CGH studies. J.R.L. and H.Y.Z. were involved in research design and data analyses. P.J.H. was involved in data analyses. C.M.B.C., M.B.R., P.J.H and J.R.L. prepared the manuscript.
Supplementary Information
Supplementary Information includes detailed clinical descriptions of the patients, four tables, and seven figures.
References
- 1.Morrow DM, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–82. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–5. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 3.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–35. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 4.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Gaudio D, et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet. Med. 2006;8:784–92. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho CM, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 2009;18:2188–203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small K, Iber J, Warren ST. Emerin deletion reveals a common X- chromosome inversion mediated by inverted repeats. Nat. Genet. 1997;16:96–9. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- 11.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JA, et al. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59:398–403. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- 13.Ramocki MB, Tavyev YJ, Peters SU. The MECP2 duplication syndrome. Am. J. Med. Genet. A. 2010;152A:1079–88. doi: 10.1002/ajmg.a.33184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 15.Hu XY, Ray PN, Murphy EG, Thompson MW, Worton RG. Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin, and phenotypegenotype correlation. Am. J. Hum. Genet. 1990;46:682–95. [PMC free article] [PubMed] [Google Scholar]
- 16.Mimault C, et al. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher Disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am. J. Hum. Genet. 1999;65:360–9. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whibley AC, et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am. J. Hum. Genet. 2010;87:173–88. doi: 10.1016/j.ajhg.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartsch O, et al. Four unrelated patients with Lubs X-linked mental retardation syndrome and different Xq28 duplications. Am. J. Med. Genet. A. 2010;152A:305–12. doi: 10.1002/ajmg.a.33198. [DOI] [PubMed] [Google Scholar]
- 19.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 20.Sheen CR, et al. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum. Mutat. 2007;28:1198–206. doi: 10.1002/humu.20591. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsenko SA, et al. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum. Mol. Genet. 2009;18:1924–36. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vialard F, et al. Mechanism of intrachromosomal triplications 15q11-q13: a new clinical report. Am. J. Med. Genet. A. 2003;118A:229–34. doi: 10.1002/ajmg.a.10164. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, et al. Intrachromosomal triplication of 2q11.2-q21 in a severely malformed infant: case report and review of triplications and their possible mechanism. Am. J. Med. Genet. 1999;82:312–7. doi: 10.1002/(sici)1096-8628(19990212)82:4<312::aid-ajmg7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Reddy KS, Logan JJ. Intrachromosomal triplications: molecular cytogenetic and clinical studies. Clin. Genet. 2000;58:134–41. doi: 10.1034/j.1399-0004.2000.580208.x. [DOI] [PubMed] [Google Scholar]
- 27.Lebofsky R, Bensimon A. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol. 2005;25:6789–97. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A. 2008;105:9936–41. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paek AL, et al. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–75. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23:2876–86. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirkin EV, Mirkin SM. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol. 2005;25:888–95. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–7. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caceres M, Sullivan RT, Thomas JW. A recurrent inversion on the eutherian X chromosome. Proc Natl Acad Sci U S A. 2007;104:18571–6. doi: 10.1073/pnas.0706604104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JA, Cheung SW, Ward PA, Inoue K, Lupski JR. Prenatal diagnosis of PLP1 copy number by array comparative genomic hybridization. Prenat Diagn. 2005;25:1188–91. doi: 10.1002/pd.1308. [DOI] [PubMed] [Google Scholar]
- 36.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 37.Beunders G, et al. A triplication of the Williams-Beuren syndrome region in a patient with mental retardation, a severe expressive language delay, behavioural problems and dysmorphisms. J. Med. Genet. 2010;47:271–5. doi: 10.1136/jmg.2009.070490. [DOI] [PubMed] [Google Scholar]
- 38.Lee JA, et al. Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum. Mol. Genet. 2006;15:2250–65. doi: 10.1093/hmg/ddl150. [DOI] [PubMed] [Google Scholar]
- 39.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 40.Ramocki MB, et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MECP2 duplication syndrome. Ann Neurol. 2009;66:771–82. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.