Abstract
In this study functional Magnetic Resonance Imaging (fMRI) was used to evaluate cortical motor network adaptation after a rehabilitation program for upper extremity motor function in chronic stroke patients. Patients and healthy controls were imaged when they attempted to perform shoulder–elbow and wrist–hand movements in a 1.5 T Siemens scanner. We perform fMRI analysis at both single- and group-subject levels. Activated voxel counts are calculated to quantify brain activation in regions of interest. We discuss several candidate regression models for making inference on the count data, and propose an application of a generalized negative-binomial model (GNBM) with structured dispersion in the study. The effects of inappropriate statistical models that ignore the nature of data are addressed through Monte Carlo simulations. Based on the GNBM, significant activation differences are observed in a number of cortical regions for stroke versus control and as a result of treatment; notably, these differences are not detected when the data are analyzed using a conventional linear regression model. Our findings provide an improved functional neuroimaging data analysis protocol, specifically for pixel/voxel counts.
Keywords: Functional Magnetic Resonance Imaging, Stroke, Motor function, Region of interest analysis, Number of activated voxels, Regression, Generalized negative-binomial model, Dispersion
Introduction
Functional Magnetic Resonance Imaging (fMRI) is one of few available techniques for studying human brain function non-invasively and is widely used in basic science and clinical research. In fMRI analysis, an almost unalterable step is to generate thresholded statistical maps. The analysis may stop there for simple illustration purposes. However, for most purposes further analysis of region of interest (ROI) is needed as it can be very difficult to detect the patterns of activity across conditions from an overall map in a complex factorial design (Poldrack, 2007).
In ROI analysis, two types of measurements are often used to quantify brain activation: the number of activated voxels (NAV, the number of voxels activated beyond a pre-determined statistical threshold) in an ROI, and the averaged image intensity (AII) in an ROI. Both measurements are subject to measurement error in data processing. The NAV is dependent upon the pre-determined threshold; and AII may not be a good measure to represent the level of brain activation if activation size (area or volume) is a strong interest or the distribution of intensity values in an ROI is not symmetrical. The NAV provides a spatial measurement/index to quantify the degree of activation in a cortical ROI (Luft et al., 2002), which is often used in neuroimaging studies (e.g. Benwell et al., 2007; Brodtmann et al., 2007; Carey et al., 2002; Chee et al., 2003; Liu et al., 2003; Pell et al., 2008). However, statistical methods widely used for NAV analysis in past neuroimaging studies may not be appropriate for a number of reasons.
NAVs derived from neuroimaging (e.g., fMRI or positron emission tomography) analysis represent discrete non-negative counts. It is, therefore, critical to consider the discrete nature of the measure. Without that consideration, biased estimates could occur and erroneous statistical conclusions might be reached. There have been wide applications of more sophisticated regression models for count data in the fields of biometrics (McCullagh and Nelder, 1989) and econometrics (Winkelmann, 2008). However, limited attention has been paid to the question of how to more accurately model neuroimaging count data in medical and clinical research.
The purpose of the current investigation is to apply a more sophisticated statistical model for making inferences from fMRI NAV data regarding brain function in patients who had a stroke and then were provided with stroke rehabilitation treatment.
Subjects and methods
Subjects and motor task
Data from 7 chronic (23.8±9.0 months from stroke onset) stroke patients (7 males, age=63.5±3.5 years, right-hand-dominant, left-hand affected) and 7 healthy control subjects (6 males, age=54.0±8.7 years, all right-hand-dominant) participated in the study. The experimental procedures were approved by the Institutional Review Board at the Louis Stokes Cleveland Veteran Affairs Medical Center. All subjects gave informed consent prior to the participation. During the experiment, the subjects performed two upper extremity motor tasks using the non-dominant hand (or affected hand in stroke patients) while the brain was imaged. Task 1 was a reaching movement involving simultaneous shoulder flexion and elbow extension using a wooden movement guide attached to the supporting frame of the scanner sliding board. Task 2 was a simultaneous forearm wrist–hand movement, in which subjects gently performed movements of forearm supination, wrist extension and finger extension of all digits (see details of the two tasks in Daly et al., 2008). Subjects practiced the tasks prior to the imaging experiment to make sure that they fully understood and were able to accurately perform the movements (Kimberley et al., 2008). Each task was repeated three times in each of three movement bouts for a total of nine repetitions for each of the two tasks. The motor tasks were standardized for pre- and post-treatment measurements for the stroke subjects. A strap system was used to minimize movement of the body and head during the motor performance. Two stabilization straps were secured across the chest in an X configuration, traveling from shoulder diagonally across the chest to the waist level. Both ends of each strap were firmly secured to the frame of the scanner sliding board (Daly et al., 2008).
Motor training program for stroke patients
Patients received treatment for a total of 5 h/day and 5 days/week for a total of 12 weeks. The intervention included training of coordinated movements of the shoulder, elbow, forearm, wrist and hand, based on principles of motor learning (Winstein and Schmidt, 1990), and included rest periods to ensure no difficulty in participation. Progression of exercises and task component practice were finely incrementalized, according to a hierarchy of increasing difficulty of coordination and dexterity.
fMRI data collection
Functional magnetic resonance images were collected on a SIEMENS Symphony 1.5 T system using a circularly polarized head coil and an interleaved multi-slice gradient echo EPI pulse sequence (repetition time=3 s; echo time=23 ms; flip angle=90°). The subjects lay supine in the MRI chamber and were instructed to remain as still as possible. The head was stabilized by padded restraints, and body movement was minimized by straps described previously. Both T1-weighted anatomic images and functional images were collected in the same transverse planes. Each brain volume consisted of 36 slices (3 mm slice thickness, 0.75 mm gap between slices) that covered the entire cerebrum and cerebellum (the collection of one brain volume is referred to as one scan hereafter). The in-plane resolution was 3 mm×3 mm for the fMRI images and 1 mm×1 mm for the T1-weighted images.
During each experiment, the T1-weighted anatomical images were collected first followed by the functional brain images collected alternately during rest (baseline) condition (OFF) and task performance condition (ON). Each ON or OFF condition included 10 continuous scans (lasted 30 s). Each set of continuous fMRI data collection included alternating 4 OFF and 3 ON conditions (OFF-ON-OFF-ON-OFF-ON-OFF) and 3 sets of such data were acquired for each motor task. In each set, image collection started with a verbal command of “REST” and near the end of 10 OFF scans, a “MOVE” command was given after which the subjects (patients and controls) performed shoulder–elbow or wrist–hand movements; near the end of the ONscans, the “REST” command was given to instruct subjects to rest, and so on, until the image acquisition for the 4th OFF condition was completed. Subjects were trained to perform each movement in 3 s and this resulted in 10 repetitions in each ON condition. Sufficient time was provided for subjects to rest between sets and motor tasks to reduce chances of fatigue. The first 2 scans in each set of scan series were excluded from the data analysis to avoid the T1-saturation effects (Bandettini et al., 1998).
Statistical analysis of fMRI data
fMRI analysis
The fMRI data analysis was performed using the BrainVoyager software (QX Version 2.0; Brain Innovation B.V., The Netherlands; www.brainvoyager.com). Before applying statistical analysis, several preprocessing approaches were performed. First, head motion was corrected three-dimensionally relative to the images of the third OFF scan (the reference scan for head motion detection/correction and image registration). Temporal filtering and slice scan time correction were performed using the default parameters provided in the software.
For single subject analysis, general linear models (GLM) were applied to identify activated voxels. Brain activation maps were generated from the GLM in which fMRI signal was the dependent variable and the task paradigm (block design, i.e. OFF–ON–OFF–ON–OFF–ON–OFF) was the covariate. A predictor time course in the design matrix was obtained by convolving the boxcar function representing the task paradigm with the hemodynamic response function (Logothetis et al., 2001; Worsley et al., 2002). The3-D maps were automatically registered into the standard Talairach space (Talairach and Tournoux, 1988). In all cases, we applied a statistical threshold of p<0.05 corrected for multiple comparisons using the false discovery rate (FDR) procedure (Genovese et al., 2002). This adaptive procedure provides an appropriate combination of sensitivity to detecting patterns of activation, while also providing stringent control of false positives where fMRI responses were absent. Brain activation was then quantified by calculating the NAVs in each of the ROIs using a house-coded program incorporated in BrainVoyager. This was done in the Talairach space so that the Brodmann areas could be utilized automatically. The cortical fields (i.e., ROIs) being analyzed included: primary motor cortex (Brodmann area 4, [BA4]), Brodmann area 6 (corresponding to premotor and supplementary motor areas, [BA6]), cingulate gyrus (Brodmann areas 24 & 33, [BA24–33]), primary sensory cortex (Brodmann areas 1, 2, & 3, [BA1–2–3]), and Brodmann areas 5 & 7 (corresponding to association parietal cortex, [BA5–7]). Brain activation was measured from the left and right hemispheres separately.
For group analysis, brain activation maps were generated over all the subjects in control group and pre- and post-treatment measurements in the patient group. Fixed effects analysis was applied for the two tasks: shoulder–elbow and wrist–hand. Time courses from all subjects within a group were concatenated and treated as coming from a single subject. Design matrices were concatenated in the same fashion. Then the same linear model as described in single subject analysis was estimated for each voxel. To correct for multiple comparisons, a uniform probability threshold of 0.05 was used to control the FDR.
Statistical models for region of interest analysis
Activation maps from group analysis help visually identify activated locations in the brain. However, in factorial designs with multiple levels, it can often be difficult to discern the pattern of activity across conditions from an overall map. We expect to draw valid statistical conclusions in addition to visually detect the difference. There are often needs to look further into particular ROIs, such as in this stroke study. ROI analysis can help evaluate the differences of activation sizes over multiple factors (experimental conditions, ROI locations etc.), using formal statistical testing procedures.
Quantifying brain activation by the NAV in each ROI for each subject is common in neuroimaging studies. Many studies, however, performed analysis of variance (ANOVA) based on a GLM under a normal distribution assumption for the ROI analysis. In those cases, however, the NAV count data were non-negative integers and their discrete nature was ignored. Moreover, the counts often showed much greater variability (statistical dispersion) than would be expected. This phenomenon is known as overdispersion in statistics (see more discussion about the topic in Wang et al., 2007). Specifying an inappropriate probability distribution in modeling the NAV data could cause biased estimates and further lead to erroneous statistical conclusions.
Assume that we observe data (yi, xi), i=1, …, n, where the dependent variable, yi, is the NAV, and xi =(x1i, …, xpi)T is a p-dimensional vector of covariates. One is interested in the relationship between the response and the covariates. A typical GLM assumes that the response yi conditional on xi is normally distributed. Statistical inferences are to be made based on , where β=(β0, β1)=(β0, β1, …, βp) are the parameters to be estimated.
The assumption of normality of yi|xi is often invalid for the skewed non-negative integer outcomes. A possible solution to improve the GLM is to perform a transformation of the dependent variable. For count data, the log-transformation, log(yi + a), can be applied so that the transformed data appear to close to the assumptions of the statistical inference procedure that is to be applied. The a in the log-transformation is a pre-specified positive constant in order to avoid calculation of the logarithm of zero, which is undefined. However, this GLM with log transformation can perform poorly. It has been criticized by King (1989). For example, the selection of the constant a could be sensitive in a data analysis. The log transformation may still result in a skewed distribution of the response, when the data are over-dispersed (Winkelmann, 2008). Another transformation that can be used for count data is the square root transformation, which stabilizes the variance of count data. It can avoid the problem of choosing a in a logarithmic transformation since the square root of 0 is well-defined.
Transformation is sometimes hard for investigators to interpret. In statistics, the well-known regression model for count data is Poisson regression, a special case of generalized linear models (GLIM). It assumes that the dependent variable conditional on the predictive variables follows a Poisson distribution, a discrete probability distribution that expresses the probability of a number of events occurring in specified intervals such as area, volume or time. McCullagh and Nelder (1989) discussed systematically the GLIM including logit models, Poisson models and quasi-likelihood models. Using a distribution in the Poisson family can provide a much better description of NAVs of ROIs than using a normal distribution. In a Poisson regression model, yi given xi has the probability mass function
| (1) |
where yi is a non-negative integer, and the mean parameter μi is parameterized as
| (2) |
to ensure μi > 0. Eqs. (1) and (2) jointly define the Poisson regression model. However, the Poisson model restricts the data distribution to be equal-dispersion (the conditional variance equals the conditional mean). This stringent restriction cannot handle many real applications, especially in analyzing active counts in an fMRI study. Observed counts in such a study are often over-dispersed (conditional variance exceeds the conditional mean).
The compound Poisson models provide a natural generalization of the basic Poisson model. They are often suitable to account for observed overdispersion in data and to provide a better fit. To relax the restriction of equal-dispersion, a compound Poisson model allows for unexplained randomness in μi by replacing Eq.(2) by the stochastic equation
| (3) |
where the random term νi = exp(εi) takes into account possible overdispersion. Let g(νi) denotes the probability density function of νi, then the conditional probability mass function of yi given xi becomes
| (4) |
The above expression (4) defines a compound Poisson distribution whose precise form depends on the specification of g(νi). There are a few special cases of compound Poisson models that could be suitable for over-dispersed count data.
Poisson-log-normal model
A natural selection of the distribution of νi is a log-normal distribution with mean exp(σ2/2) and variance exp(σ2)(exp(σ2)−1), where σ is the unknown parameter. When νi is log-normal distributed as above, εi = log(νi) is normally distributed with mean 0 and variance σ2. In such a case, β0 + εi in Eq. (3) can be interpreted as the random intercept, similarly as in a normal mixed model. Estimating the parameters in this Poisson-log-normal model (PLNM) can be performed using the method of maximum likelihood estimation (MLE). The log-likelihood of the model is
| (5) |
The log-likelihood in Eq. (5) is the sum of independent contributions from each observation, and each observation involves a single-dimensional integral. No closed form solution of maximizing the log likelihood is available. However, numerical integration for calculating Eq.(5) can be evaluated accurately using adaptive Gauss–Hermite quadrature techniques (Molenberghs and Verbeke, 2005). This hierarchical model is often suitable for the case of count data with mild or moderate over-dispersion.
Negative-binomial model
Another common choice for addressing the distribution characteristics of νi is a gamma distribution. Suppose that νi has a one-parameter gamma distribution Gamma(1/δ, δ) with density (for δ>0)
Using some algebraic transformation on the integral in Eq.(4), one can demonstrate that yi|xi has a negative-binomial distribution with the mean parameter μi and the dispersion parameter δ. Its probability mass function is given by
| (6) |
where . It can be shown that E(yi|xi)= μi and . The variance is a quadratic function of the mean in the negative-binomial model (NBM). The parameter δ plays a role to measure the overdispersion level. Note that the Poisson model is a special case of the NBM (as δ>0).
The log-likelihood in the NBM is simply given by . There is no integration involved in calculating the log-likelihood, so the MLE method is easier to be implemented than that in the PLNM. In practice, the NBM performs better than the PLNM when severe over-dispersion is present in a dataset.
Generalized negative-binomial model with structured dispersion
In the NBM described previously, we have a single dispersion parameter δ. In some complex real data applications, a single dispersion parameter may not be good enough to capture the complex dispersion structure in the data. We can further generalize the NBM in order to model the structure of dispersion. A generalized negative-binomial model (GNBM) with structured dispersion allows the dispersion parameter to vary observation by observation, by modeling
| (7) |
where, zi =(z1, …, zk)T, i=1, …, n is a k-dimensional vector of covariates, which could be all or a subset of the covariates xi or even completely different variables. α=(α0, α1)=(α0, α1, …, αk) is the k + 1-dimensional vector of unknown parameters (associated with dispersion) to be estimated. With this generalization, the relationship of the dependent variable yi with the covariates xi can still be modeled via the conditional expectation of yi given xi:
| (8) |
while the variation of dispersion will be taken into account by Eq. (7) simultaneously. The resulting log-likelihood of the GNBM is given by
It is straight-forward to apply the MLE method for the above log-likelihood, estimate the parameters, and make the corresponding inferences. This generalized model gives flexibility to account for heterogeneous dispersion in modeling count data.
A simulation study
We performed a simulation study to compare the models discussed previously for over-dispersed count data. Two cases of simulated data were considered: (1) Poisson-log-normal (PLN) data, where log μ i =β0 + β1ti + ui with β0 =1, β1 =2 and ui ~ N(0, 0.52) were generated and followed by the responses Yi generated by Poisson with mean μ i (i=1, …, n); and (2) negative-binomial (NB) data, where log μ i = β0 +β1ti with β0 =1, β1 =2 were generated and followed by the responses Yi generated by a NB distribution with mean μi and the dispersion parameter δ=2. In all cases, ti was generated from a uniform distribution between 0 and 1. Clearly, both cases generated over-dispersed count data, while the data from the case one were mildly over-dispersed and those from the case two were severely over-dispersed. The simulated sample size n was set to 100 and the simulation was repeated 1000 times. We considered the six types of models to fit the simulated data: NBM, Poisson model, PLNM, GLM, GLM with log transformation (a=0.5), and GLM with square root transformation.
Table 1 summarizes the average of parameter estimates (mean) and its standard deviations (SD) as well as the average of the estimates of standard errors (Mean(^ se)) from independent 1000 times of simulations for each individual model. The true parameters here are β0 =1 and β1 =2. A “well-fitted” statistical model expects that “Mean” is close to the true parameter and “Mean(≿e)” is close to “SD”. Obviously, GLM gave totally wrong estimates for the parameters. For example, the mean of β̂1 for PLN data, 18.558, was far from β1 =2. For Poisson regression, the estimates of parameters worked well but the standard errors were underestimated (far less than the standard deviation of estimated parameters). For instance, SD of β̂0, 0.354, was about four times as large as the mean of se, 0.093, using the Poisson model for NB data. The assumption in Poisson model that the variance equals the mean is not valid if overdispersion exists, and thus the estimated standard errors were too low and statistical inference from Poisson model would be mistaken. When overdispersion was small, both PLNM and NBM performed well on estimating both the parameters and their standard errors. When overdispersion was large, NBM still performed excellently but PLNM gave biased estimates (e.g. the mean of β̂0 was 0.463 using PLN model). GLM with square root transformation performs very well for data with severe overdispersion. A square root transformation is a variance stabilizing transformation for count data. The performance of GLM with log transformation was similar to that of PLNM, where it worked for data with mild overdispersion (e.g. the mean of β̂0 was 1.006), but became inaccurate for data with severe overdispersion (e.g. the mean of β̂0 was 0.469). Given the accurate results of NBM and the natural interpretation of a NB distribution for discrete count data, we used NBM/GNBM to analyze the motor function fMRI count data and the results are reported below with comparisons to results analyzed by GLM models.
Table 1.
A simulation study of count data with overdispersion. Six types of models were fit the simulated data: NBM, Poisson model, PLNM, GLM (denoted by GLM1), GLM with log transformation (denoted by GLM2), and GLM with square root transformation (denoted by GLM3). The true parameters are β0 =1, β1 =2.
| Sim data | Parameter | Estimate | Model
|
|||||
|---|---|---|---|---|---|---|---|---|
| NBM | PLNM | Poisson | GLM1 | GLM2 | GLM3 | |||
| PLN | β0 | Mean | 1.115 | 0.985 | 1.115 | 0.545 | 1.006 | 1.374 |
| SD | 0.141 | 0.175 | 0.152 | 1.012 | 0.143 | 0.171 | ||
| Mean(≿e) | 0.135 | 0.129 | 0.087 | 1.368 | 0.132 | 0.192 | ||
| β1 | Mean | 2.009 | 2.102 | 2.008 | 18.558 | 1.991 | 2.972 | |
| SD | 0.222 | 0.329 | 0.239 | 2.681 | 0.225 | 0.338 | ||
| Mean(≿e) | 0.217 | 0.214 | 0.123 | 2.364 | 0.228 | 0.333 | ||
| NB | β0 | Mean | 0.971 | 0.463 | 0.971 | 0.392 | 0.469 | 0.984 |
| SD | 0.314 | 0.542 | 0.354 | 2.127 | 0.266 | 0.326 | ||
| Mean(≿e) | 0.300 | 0.202 | 0.093 | 2.873 | 0.283 | 0.387 | ||
| β1 | Mean | 2.031 | 2.346 | 2.025 | 16.773 | 1.535 | 2.339 | |
| SD | 0.523 | 0.891 | 0.587 | 5.923 | 0.488 | 0.700 | ||
| Mean(≿e) | 0.512 | 0.336 | 0.132 | 4.967 | 0.489 | 0.669 | ||
Results
NAVs derived from analysis of single-subject activation maps were non-negative integers and not normally distributed. Initial exploratory data analysis showed that the dispersion levels from different groups (Pre vs. Post and Control vs. stroke) were very different. Thus, the GNBM with structured dispersion was applied in ROI analysis of the stroke study. The covariates incorporated in the linear mean function in Eq.(8) of the model were “Group” (control, pre- or post-treatment in stroke), “Task” (shoulder–elbow or wrist–hand movement), and “ROI” (10 different ROIs); the covariate included in the dispersion function in Eq. (7) was “Group”. Approximate F tests based on the fitted model were then conducted to evaluate the significances of covariates (Molenberghs and Verbeke, 2005). In the model fitting, we tried to add the interaction term of Group and Task. It turned out that the interaction term was not significant, hence, the term was excluded in our final reported model. In addition, for the comparison purpose, we also fit a GLM, a GLM with log transformation (a=0.5), and a GLM with square root transformation for the NAVs.
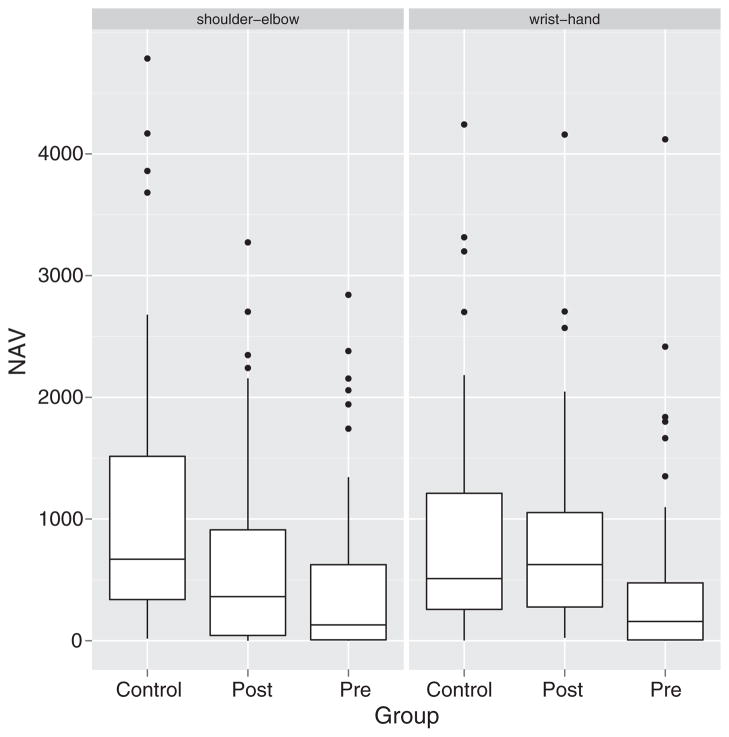
Table 2 summarizes the sample means of NAVs and Talairach coordinates of ROIs per group, task and region. Fig. 1 shows the boxplots of NAVs by different groups and tasks. The dots in the plots are outliers, which have a greater distance from the median than 1.5 times the inter-quartile range. The count data for each group and task are highly positively skewed. They are severely dispersed and the dispersion levels vary for different groups.
Table 2.
Average voxel counts and Talairach coordinates of activated regions per group, task and region.
| Group | Region | Task=shoulder–elbow
|
Task=wrist– hand
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Counts | X | Y | Z | Counts | X | Y | Z | ||
| Control | BA4 Left | 171 | 166.2 | 149.6 | 79.7 | 145 | 168.2 | 147.8 | 82.6 |
| BA4 Right | 607 | 92.7 | 148.8 | 78.1 | 388 | 96.9 | 151.6 | 73.7 | |
| BA6 Left | 1792 | 150.7 | 130.3 | 79.1 | 1394 | 150.9 | 129.3 | 78.9 | |
| BA6 Right | 1907 | 101.1 | 131.7 | 79.9 | 1668 | 102.0 | 130.6 | 81.4 | |
| BA24–33 Left | 1143 | 134.0 | 129.2 | 104.0 | 936 | 136.2 | 136.9 | 98.6 | |
| BA24–33 Right | 1133 | 118.8 | 139.3 | 104.2 | 895 | 120.5 | 139.9 | 100.4 | |
| BA1–2–3 Left | 667 | 169.9 | 154.1 | 80.5 | 400 | 177.1 | 150.8 | 83.8 | |
| BA1–2–3 Right | 816 | 91.0 | 155.1 | 77.8 | 562 | 91.0 | 156.7 | 77.7 | |
| BA5–7 Left | 1076 | 142.2 | 182.9 | 76.3 | 913 | 143.1 | 187.7 | 76.8 | |
| BA5–7 Right | 1192 | 113.1 | 183.5 | 76.8 | 1217 | 109.8 | 183.7 | 76.0 | |
| Post | BA4 Left | 185 | 158.6 | 150.5 | 72.1 | 274 | 162.2 | 148.1 | 73.1 |
| BA4 Right | 340 | 90.8 | 146.4 | 79.7 | 379 | 94.0 | 149.9 | 79.3 | |
| BA6 Left | 1123 | 144.0 | 138.5 | 75.5 | 1621 | 146.6 | 132.5 | 74.8 | |
| BA6 Right | 1254 | 98.5 | 136.5 | 82.7 | 2282 | 97.3 | 127.7 | 81.4 | |
| BA24–33 Left | 1030 | 135.0 | 130.5 | 111.8 | 673 | 135.7 | 138.4 | 94.9 | |
| BA24–33 Right | 772 | 116.5 | 141.7 | 101.3 | 495 | 114.2 | 142.5 | 101.8 | |
| BA1–2–3 Left | 288 | 168.3 | 151.3 | 78.5 | 508 | 169.4 | 152.0 | 74.1 | |
| BA1–2–3 Right | 457 | 81.1 | 151.8 | 86.9 | 639 | 89.4 | 155.7 | 78.1 | |
| BA5–7 Left | 302 | 143.0 | 186.5 | 79.5 | 880 | 141.1 | 177.1 | 76.1 | |
| BA5–7 Right | 1084 | 112.6 | 180.3 | 77.6 | 920 | 105.4 | 179.4 | 73.9 | |
| Pre | BA4 Left | 80 | 168.4 | 143.6 | 85.0 | 168 | 165.7 | 147.2 | 78.1 |
| BA4 Right | 256 | 95.8 | 150.3 | 76.5 | 264 | 100.7 | 152.5 | 70.6 | |
| BA6 Left | 861 | 151.5 | 136.3 | 81.6 | 905 | 138.7 | 138.0 | 74.0 | |
| BA6 Right | 645 | 91.7 | 133.2 | 88.1 | 513 | 101.2 | 133.2 | 82.0 | |
| BA24–33 Left | 719 | 135.7 | 144.2 | 97.5 | 349 | 136.6 | 145.7 | 96.1 | |
| BA24–33 Right | 491 | 116.3 | 134.5 | 99.3 | 195 | 116.1 | 147.2 | 97.2 | |
| BA1–2–3 Left | 133 | 175.6 | 149.5 | 86.4 | 90 | 167.4 | 152.1 | 77.1 | |
| BA1–2–3 Right | 287 | 86.3 | 154.7 | 81.2 | 323 | 97.8 | 158.3 | 73.9 | |
| BA5–7 Left | 394 | 142.6 | 182.6 | 76.0 | 581 | 143.6 | 182.5 | 78.8 | |
| BA5–7 Right | 487 | 109.9 | 182.3 | 74.5 | 569 | 105.5 | 185.9 | 76.5 | |
Fig. 1.
Boxplots of NAVs by different groups and tasks. The dots in the plots are outliers, which have a greater distance from the median than 1.5 times the inter-quartile range. The count data are highly positively skewed and severely dispersed. The dispersion levels of the counts vary among the groups.
Table 3 displays the statistical results in analysis of voxel counts using the GNBM. The estimates of parameters and their standard errors are shown and the p-values of approximate F tests for each factor are also presented. The GNBM unveiled that Group and Region were significant factors, but Task was not a significant factor, according to, NAV. The results supported that, although the brain activation was located in the same ROIs for the control, pre and post patient groups, the volume of activations was significantly different across the three groups (p<0.0001). In addition, the statistical test for the effect of dispersion structure as a function of Group was significant (p<0.0001). This supported our initial exploratory analysis of heterogeneous dispersion over Group, and confirmed that the structured dispersion function component was necessary in the GNBM.
Table 3.
The GNBM results showing significant difference for group overall and for each ROI according to NAV.
| Parameter | Estimate | Standard error | Approximate F test | |
|---|---|---|---|---|
| Intercept | 6.2404 | 0.2419 | ||
| Group | Control | 0.7894 | 0.1732 | <0.0001 |
| Post | 0.6147 | 0.2050 | ||
| Pre | 0 | – | ||
| Task | Shoulder–elbow | 0.1177 | 0.1193 | 0.3246 |
| Wrist–hand | 0 | – | ||
| Region | BA4 Left | −1.7957 | 0.2652 | <0.0001 |
| BA4 Right | −0.8858 | 0.2639 | ||
| BA6 Left | 0.3198 | 0.2620 | ||
| BA6 Right | 0.3985 | 0.2621 | ||
| BA24–33 Left | −0.1365 | 0.2621 | ||
| BA24–33 Right | −0.2676 | 0.2623 | ||
| BA1–2–3 Left | −0.9195 | 0.2625 | ||
| BA1–2–3 Right | −0.5711 | 0.2620 | ||
| BA5–7 Left | −0.2313 | 0.2621 | ||
| BA5–7 Right | 0 | – | ||
| Dispersion | Intercept | 1.2263 | 0.1083 | |
| Group (Control) | −1.5392 | 0.1547 | <0.0001 | |
| Group (Post) | −0.9292 | 0.1782 | ||
| Group (Pre) | 0 | – |
To further evaluate the pairwise difference over groups, we performed contrast tests based on the GNBM (Table 4). We found that there was a significant difference between pre and post tests in the stroke group (p<0.0001). And there was a significant difference between stroke pre-test and control group (p=0.0058). However, there was no significant difference between stroke post-test and control group (p=0.2494). The results suggested that the training program was effective since the stroke patients recovered cortical activation volume close to that in the control group. Table 4 also lists the analysis results based on a standard GLM (GLM1), a GLM with log transformation (GLM2), and a GLM with square root transformation (GLM3). For the GLM1, the difference between stroke pre-test and control group was non-significant (p=0.1202). This non-significant p-value may be questionable since the estimated parameters and standard errors are biased using a GLM for count data. Although GLM2 and GLM3 also detected the significant difference between pre and control, the parameter estimates and their standard errors of these two models might be inaccurate. Our scientific findings in the paper relied on the GNBM.
Table 4.
P-values for pairwise comparison of the different groups overall according the number of active voxels. The p-values were calculated based on the GNBM, GLM (denoted by GLM1), GLM with log transformation (denoted by GLM2), and GLM with square root transformation (denoted by GLM3).
| GNBM | GLM1 | GLM2 | GLM3 | |
|---|---|---|---|---|
| Pre vs Post | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Pre vs Control | 0.0058 | 0.1202 | 0.0096 | 0.0444 |
| Post vs Control | 0.2494 | 0.8800 | 0.4808 | 0.7585 |
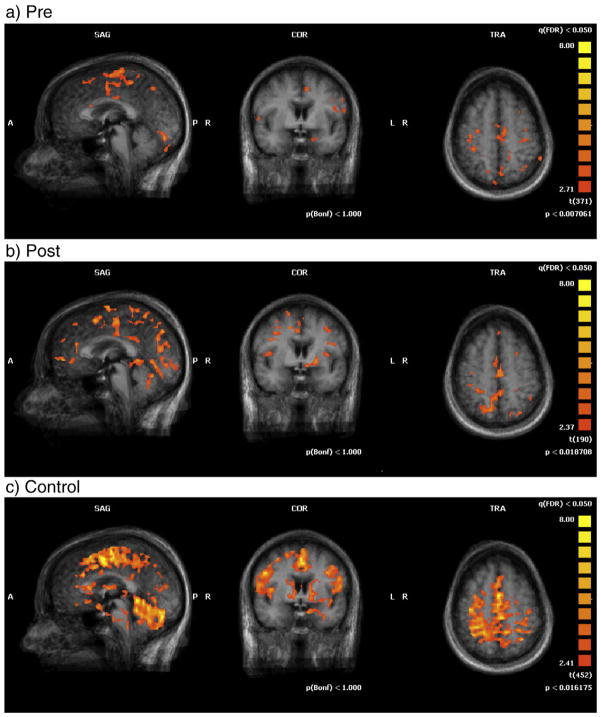
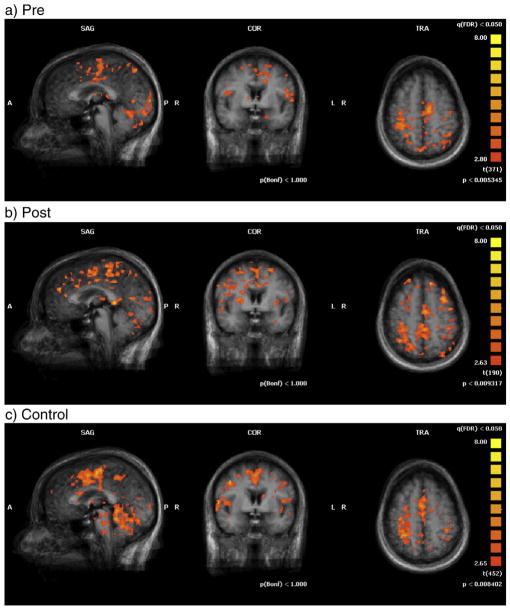
Figs. 2 and 3 show multi-subject statistical mapping illustrating the brain activation patterns in control group and pre- and post-tests in stroke patients for shoulder–elbow (Fig. 2) and hand–wrist (Fig. 3) tasks at Talairach coordinate (0, 0, 0). The activated areas (i.e. NAVs) were small for the patient group before training but the volume of activation increased after training. Statistical results of ROI analysis supported these graphical illustrations (Table 4).
Fig. 2.
Activation maps for multi-subject fMRI analysis with shoulder–elbow movement at Talairach coordinate (0, 0, 0).
Fig. 3.
Activation maps for multi-subject fMRI analysis with wrist–hand movement at Talairach coordinate (0, 0, 0).
Discussion
In this study, fMRI data were analyzed to evaluate brain activation adaptations after an upper extremity movement rehabilitation program in stroke patients. A family of compound Poisson models was discussed for analyzing the activated voxel counts in the ROI analysis. The results suggest that erroneous statistical inferences could result from specifying an inappropriate statistical model that ignores the nature of data.
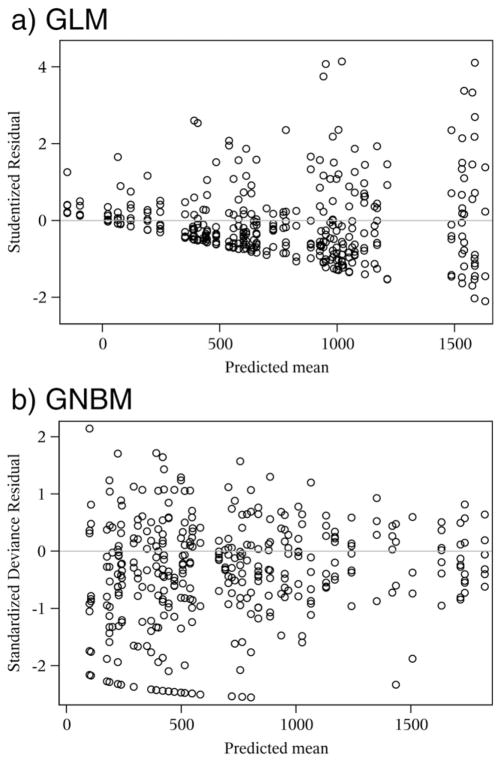
Residual plots are very useful and are popular diagnostic tools for statistical regression models. Fig. 4 displays the diagnostic residual plots from GLM and GNBM in the stroke study. Typically, a residual plot shows the residuals on the vertical axis and the predicted mean value on the horizontal axis. If the points in a residual plot are randomly dispersed around the horizontal axis, a regression model is appropriate for the data; otherwise, one may question the validity of the model. Conventionally, the studentized residual is used for normal linear models, while the standardized deviance residual is used for compound Poisson models.
Fig. 4.
Diagnostic plots for the statistical models in ROI analysis of the fMRI stroke study: (a) the residual plot based on the GLM, and (b) the residual plot based on the GNBM.
From the upper plot in Fig. 4, we can see that the residuals from GLM were not randomly dispersed. The plot showed the heterogeneity, where the residual variance increased as the predicted mean increased. Note that the predicted mean from GLM cannot ensure the voxel counts are always positive. Some negative predicted values were identified from the GLM. In contrast, the residual plot of GNBM displayed a random pattern around the horizontal axis, which supports that the model was appropriate (the lower plot in Fig. 4). The predicted means did not have the issue of negative values. These results from examination of the residual plots further supported our concern regarding use of the standard linear regression model for fMRI count data.
For the purpose of analyzing active voxel counts in a neuroimaging study, our experiences suggest that NBM/GNBM is a more appropriate statistical model compared with Poisson model and PLNM, because active voxel counts often show a characteristic feature of severe overdispersion. One should proceed with caution in using a GLM directly, or a GLM with a transformation for fMRI count data. The assumption of GLM is invalid for count data; and linear regression models with nonlinear transformed response could be difficult for biomedical scientists to explain. All of the compound Poisson models discussed can be programmed and calculated using commercially-validated statistical software. For instance, SAS proc nlimixed can be used to specify the models.
Clinically, our results supported the notion that motor training alters brain activation towards the normal pattern in stroke patients and this change is essential for motor function recovery. The training program resulted in a more normal volume of activation, which was correlated with improvement of upper limb motor function (Daly et al., 2006). However, based on the analysis using conventional GLM, this effect could not be detected. Our results underscore the importance of selecting appropriate statistical models for analyzing count data in neuroimaging research.
Although quantifying brain activation by the NAV is common in neurophysiological studies, criticism has been raised about its stability as the measure of signal magnitude in fMRI. Cohen and DuBois (1999) found that in their study the voxel counting was unstable. This issue could be viewed as the response measurement error problem (Carroll et al., 2006; Wang and Wang, 2011). The response measurement error increased the variability of the fitted function, and thus decreased the power of the model. A potential remedy of the measurement error problem is to get repeated measurements for each subject if possible. Modeling the repeated NAVs together can increase the statistical power and reduce the effect of measurement error. In addition, Cohen and DuBois(1999) evaluated the brain activation as the slope of the regression line between the modeled time course and the actual data, and found the measured slope was largely independent of the contrast-to-noise ratio. One may use the slope as the activation measurement and fit a GLIM with an appropriate distribution. Other approaches, such as clustering methods, may work as well in ROI analysis of fMRI data.
Acknowledgments
We are grateful to the reviewers for their valuable comments. This research was supported by the NIH grant UL1 RR024989 and the NIH grant R01 NS035130.
Appendix A
List of abbreviations
| AII: averaged image intensity ANOVA: analysis of variance BA1-2-3: Brodmann areas 1, 2, & 3 BA24-33: Brodmann areas 24 & 33 BA4: Brodmann area 4 BA5-7: Brodmann areas 5 & 7 BA6: Brodmann area 6 FDR: false discovery rate GLM: general linear model GLIM: generalized linear model GNBM: generalized negative-binomial model MLE: maximum likelihood estimation NAV: number of activated voxels NB: negative-binomial NBM: negative-binomial model PLNM: Poisson-log-normal model ROI: region of interest SD: standard deviation |
SAS code
The following is the SAS 9.2 programming code we used to fit the GNBM.
%let IntialParameters b_0=0 b_1=0 b_2=0 b_3=0 b_4=0 b_5=0 b_6=0 b_7=0 b_8=0 b_9=0 b_10=0 b_11=0 b_12=0 a_0=0 a_1=0 a_2=0; %let LinearFunction b_0+b_1*(Group='Control')+b_2*(Group= 'Post') + b_3*(Task='shoulder-elbow') + b_4*(Region='BA4 Left')+b_5*(Region='BA4 Right') + b_6*(Region='BA6 Left')+b_7*(Region='BA6 Right') + b_8*(Region='BA24-33 Left')+b_9*(Region='BA24-33 Right') + b_10*(Region='BA1-2-3 Left')+b_11*(Region='BA1-2-3 Right') + b_12*(Region='BA5-7 Left'); %let DispersionFunction a_0 +a_1*(Group = 'Control') + a_2* (Group='Post'); title "Generalized Negative Binomial model"; proc nlmixed data=fMRI_counts; parms &IntialParameters; eta_lambda=&LinearFunction; mean=exp(eta_lambda); eta_k=&DispersionFunction; k=exp(eta_k); loglike=(lgamma(Counts+(1/k)) − lgamma(Counts+1) − lgamma(1/k) + Counts*log(k*mean) − (Counts +(1/k))× log(1 +k*mean)); model Counts~general(loglike); contrast 'Mode' b_1, b_2; contrast 'Mode2_1' b_1; contrast 'Mode2_2' b_2; contrast 'Mode2_3' b_1-b_2; contrast 'Task' b_3; contrast 'Region' b_4, b_5, b_6, b_7, b_8, b_9, b_10, b_11, b_12; contrast 'dispersion' a_1, a_2; contrast 'dispersion2' a_1-a_2; predict eta_lambda out=eta_lambda; run;
References
- Bandettini PA, Jesmanowicz A, Van Kylen J, Birn RM, Hyde JS. Functional MRI of brain activation induced by scanner acoustic noise. Magn Reson Med. 1998;39:410–416. doi: 10.1002/mrm.1910390311. [DOI] [PubMed] [Google Scholar]
- Benwell NM, Mastaglia FL, Thickbroom GW. Changes in the functional MR signal in motor and non-motor areas during intermittent fatiguing hand exercise. Exp Brain Res. 2007;182 (1):93–97. doi: 10.1007/s00221-007-0973-5. [DOI] [PubMed] [Google Scholar]
- Brodtmann A, Puce A, Darby D, Donnan G. fMRI demonstrates diaschisis in the extrastriate visual cortex. Stroke. 2007;38 (8):2360–2363. doi: 10.1161/STROKEAHA.106.480574. [DOI] [PubMed] [Google Scholar]
- Carey J, Kimberley T, Lewis S, Auerbach E, Dorsey L, Rundquist P, Ugurbil K. Analysis of fMRI and finger tracking training in subjects with chronic stroke. Brain. 2002;125:773–788. doi: 10.1093/brain/awf091. [DOI] [PubMed] [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu C. Measurement Error in Nonlinear Models: a Modern Perspective, Second ed. Chapman Hall; New York: 2006. [Google Scholar]
- Chee M, Lee H, Soon C, Westphal C, Venkatraman V. Reproducibility of the word frequency effect: comparison of signal change and voxel counting. NeuroImage. 2003;18 (2):468–482. doi: 10.1016/s1053-8119(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Cohen MS, DuBois RM. Stability, repeatability, and the expression of signal magnitude in functional magnetic resonance imaging. J Magn Reson Imaging. 1999 Jul;10(1):33–40. doi: 10.1002/(sici)1522-2586(199907)10:1<33::aid-jmri5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Daly JJ, Fang Y, Perepezko EM, Siemionow V, Yue GH. Prolonged cognitive planning time, elevated cognitive effort, and relationship to coordination and motor control following stroke. IEEE Trans Neural Syst Rehabil Eng. 2006;14 (2):168–171. doi: 10.1109/TNSRE.2006.875554. [DOI] [PubMed] [Google Scholar]
- Daly JJ, Hrovat K, Pundik S, Sunshine J, Yue G. fMRI methods for proximal upper limb joint motor testing and identification of undesired mirror movement after stroke. J Neurosci Methods. 2008;175 (1):133–142. doi: 10.1016/j.jneumeth.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Genovese C, Lazar N, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002;15 (4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Kimberley TJ, Birkholz DD, Hancock RA, VonBank SM, Werth TN. Reliability of fMRI during a continuous motor task: assessment of analysis techniques. J Neuroimaging. 2008;18 (1):18–27. doi: 10.1111/j.1552-6569.2007.00163.x. [DOI] [PubMed] [Google Scholar]
- King G. Variance specification in event count models: from restrictive assumptions to a generalized estimator. Am J Pol Sci. 1989;33:762–784. [Google Scholar]
- Liu JZ, Shan ZY, Zhang LD, Sahgal V, Brown RW, Yue GH. Human brain activation during sustained and intermittent submaximal fatigue muscle contractions: an fMRI study. J Neurophysiol. 2003;90 (1):300–312. doi: 10.1152/jn.00821.2002. [DOI] [PubMed] [Google Scholar]
- Logothetis N, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412 (6843):150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Luft A, Smith G, Forrester L, Whitall J, Macko R, Hauser T, Goldberg A, Hanley D. Comparing brain activation associated with isolated upper and lower limb movement across corresponding joints. Hum Brain Mapp. 2002;17 (2):131–140. doi: 10.1002/hbm.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2. Chapman and Hall; London: 1989. [Google Scholar]
- Molenberghs G, Verbeke G. Models for Discrete Longitudinal Data. Springer; New York: 2005. [Google Scholar]
- Pell GS, Briellmann RS, Chan CHP, Pardoe H, Abbott DF, Jackson GD. Selection of the control group for VBM analysis: infiuence of covariates, matching and sample size. NeuroImage. 2008;41 (4):1324–1335. doi: 10.1016/j.neuroimage.2008.02.050. [DOI] [PubMed] [Google Scholar]
- Poldrack RA. Region of interest analysis for fMRI. Soc Cogn Affect Neurosci. 2007;2 (1):67–70. doi: 10.1093/scan/nsm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System — an Approach to Cerebral Imaging. Thieme Medical Publishers; New York: 1988. [Google Scholar]
- Wang XF, Sun J, Gustafson KJ, Yue GH. Modeling heterogeneity and dependence for analysis of neuronal data. Stat Med. 2007;26 (21):3927–3945. doi: 10.1002/sim.2943. [DOI] [PubMed] [Google Scholar]
- Wang XF, Wang B. Deconvolution estimation in measurement error models: the R package decon. J Stat Softw. 2011;39 (10):1–24. [PMC free article] [PubMed] [Google Scholar]
- Winkelmann R. Econometric Analysis of Count Data. 5. Springer; New York: 2008. [Google Scholar]
- Winstein CJ, Schmidt RA. Reduced frequency of knowledge of results enhances motor skill learning. J Exp Psychol Learn Mem Cogn. 1990;16 (4):677–691. [Google Scholar]
- Worsley KJ, Liao CH, Aston J, Petre V, Duncan GH, Morales F, Evans AC. A general statistical analysis for fMRI data. NeuroImage. 2002 Jan;15(1):1–15. doi: 10.1006/nimg.2001.0933. [DOI] [PubMed] [Google Scholar]