Abstract
Duchenne muscular dystrophy is caused by mutations in the DMD gene that disrupt the open reading frame and prevent the full translation of its protein product, dystrophin. Restoration of the open reading frame and dystrophin production can be achieved by exon skipping using antisense oligonucleotides targeted to splicing elements. This approach aims to transform the Duchenne muscular dystrophy phenotype to that of the milder disorder, Becker muscular dystrophy, typically caused by in-frame dystrophin deletions that allow the production of an internally deleted but partially functional dystrophin. There is ongoing debate regarding the functional properties of the different internally deleted dystrophins produced by exon skipping for different mutations; more insight would be valuable to improve and better predict the outcome of exon skipping clinical trials. To this end, we have characterized the clinical phenotype of 17 patients with Becker muscular dystrophy harbouring in-frame deletions relevant to on-going or planned exon skipping clinical trials for Duchenne muscular dystrophy and correlated it to the levels of dystrophin, and dystrophin-associated protein expression. The cohort of 17 patients, selected exclusively on the basis of their genotype, included 4 asymptomatic, 12 mild and 1 severe patient. All patients had dystrophin levels of >40% of control and significantly higher dystrophin (P = 0.013), β-dystroglycan (P = 0.025) and neuronal nitric oxide synthase (P = 0.034) expression was observed in asymptomatic individuals versus symptomatic patients with Becker muscular dystrophy. Furthermore, grouping the patients by deletion, patients with Becker muscular dystrophy with deletions with an end-point of exon 51 (the skipping of which could rescue the largest group of Duchenne muscular dystrophy deletions) showed significantly higher dystrophin levels (P = 0.034) than those with deletions ending with exon 53. This is the first quantitative study on both dystrophin and dystrophin-associated protein expression in patients with Becker muscular dystrophy with deletions relevant for on-going exon skipping trials in Duchenne muscular dystrophy. Taken together, our results indicate that all varieties of internally deleted dystrophin assessed in this study have the functional capability to provide a substantial clinical benefit to patients with Duchenne muscular dystrophy.
Keywords: Becker muscular dystrophy, Duchenne muscular dystrophy, nNOS, dystrophin-associated glycoprotein complex, therapy
Introduction
Duchenne muscular dystrophy is a fatal X-linked neuromuscular disorder caused by mutations in the DMD gene that disrupt the open reading frame and prevent the full translation of its protein product, dystrophin. The majority of DMD gene mutations are deletions (∼65%) although duplications (∼10%), small mutations (∼22%) and deep intronic mutations (∼2–3%) are also documented (Muntoni et al., 2003; Abbs et al., 2010). Patients develop muscle weakness in the early years of life and lose the ability to walk by their early teens; unless appropriate respiratory and cardiac treatment is initiated, affected individuals typically die before reaching their twenties.
Dystrophin is localized to the inner part of the sarcolemma of muscle fibres where it is associated with other proteins as part of the dystrophin-associated protein complex. Dystrophin plays an important role in stabilizing the muscle fibre against the mechanical forces of muscle contraction by providing a shock-absorbing connection between the cytoskeleton and the extracellular matrix and is also believed to have a role in signalling. The absence of dystrophin is thought to render muscle cells susceptible to stretch-induced damage and necrosis. Dystrophin consists of four main functional units: 1) the N-terminus contains two calponin homology domains responsible for actin-binding; 2) the central rod domain consists of 24 spectrin-like repeats with four interspersing hinge domains, which also contribute to actin binding (Ervasti, 2007; Le Rumeur et al., 2010); 3) the cysteine rich domain is required for binding to β-dystroglycan, a crucial protein of the dystroglycan complex that helps maintain the structural integrity of muscle tissue and 4) the C-terminal domain contains syntrophin and dystrobrevin binding sites (Ervasti, 2007; Le Rumeur et al., 2010). In addition to a major structural role, a specific domain of dystrophin (exons 42–45 encoding spectrin-like repeats 16 and 17 within the rod domain) is necessary for the sarcolemmal localization of neuronal nitric oxide synthase (nNOS; Lai et al., 2009), which contributes to the fine-tuning of muscle blood flow during physical activity. In the Duchenne muscular dystrophy muscle, nNOS is absent from the sarcolemma leading to paradoxical exercise-induced vasoconstriction, which contributes to the ongoing muscle damage.
A clinically milder allelic disorder, Becker muscular dystrophy, is caused by in-frame dystrophin mutations, typically involving part of the central rod domain, that preserve the reading frame permitting the translation of an internally deleted dystrophin protein. There is a large variation in the clinical severity of patients with Becker muscular dystrophy with some individuals able to experience a near normal lifestyle and lifespan while others lose the ability to walk in their late teens or early twenties (Bushby et al., 1993). Although the reasons for this variability are not fully understood, it is likely that both the functionality and levels of the internally deleted dystrophin proteins play a significant role.
Naturally occurring alternative splicing of dystrophin pre-messenger RNA can restore the reading frame in Duchenne muscular dystrophy as a result of the skipping of some exons and is thought to explain the presence of dystrophin positive ‘revertant fibres’. This phenomenon has been well characterized in the mdx mouse (Lu et al., 2000), and in Duchenne muscular dystrophy (Klein et al., 1992; Arechavala-Gomeza et al., 2010b). There has been debate regarding the functionality of both the dystrophin in revertant fibres and the internally deleted dystrophin of Becker muscular dystrophy. The milder Becker muscular dystrophy phenotype and the fact that the internally deleted proteins are correctly localized to the sarcolemma and accompanied by the dystrophin-associated protein complex suggests that they are indeed functionally competent (Matsumura et al., 1994).
There is currently no effective treatment for Duchenne muscular dystrophy; however, a promising therapeutic strategy is to transform the Duchenne muscular dystrophy phenotype into the milder Becker muscular dystrophy phenotype through the restoration of the open reading frame and the production of internally deleted, Becker muscular dystrophy-like dystrophin molecules. This can be achieved by exon skipping using oligonucleotides targeted to splicing elements (splice switching oligonucleotides) (Wood et al., 2010). We and others have previously demonstrated the local restoration of dystrophin expression in patients with Duchenne muscular dystrophy using either morpholino splice switching oligonucleotide AVI-4658 or the 2′-O-methyl phosphorothioate PRO-051, which induce the skipping of exon 51 in dystrophin messenger RNA (van Deutekom et al., 2007; Kinali et al., 2009). Skipping of exon 51 could rescue the largest group (13%) of Duchenne muscular dystrophy mutations (Aartsma-Rus et al., 2009) and the first studies aimed at assessing the biochemical efficacy and clinical safety of systemically administered splice switching oligonucleotides are now emerging (Cirak et al., 2011; Goemans et al., 2011)
Multi-exon skipping is an attractive alternative to the skipping of single exons and has the potential to rescue over 50% of all Duchenne muscular dystrophy deletions. According to bioinformatical analysis, exons 45–55 are the most optimal set of exons to skip for conversion of Duchenne muscular dystrophy to Becker muscular dystrophy (Beroud et al., 2007). Patients with Becker muscular dystrophy harbouring this deletion can have a mild phenotype, often associated with a late-onset (Ferreiro et al., 2009; Tselikas et al., 2010). There are in fact several reports describing asymptomatic Becker muscular dystrophy with a variety of deletions, including some with an end-point in exons 51 or 53 (Morrone et al., 1997; Muntoni et al., 1997; Melis et al., 1998; Torelli et al., 2004; Sanchez-Arjona et al., 2005; Saengpattrachai et al., 2006; Lesca et al., 2007; Ferreiro et al., 2009).
Very few thorough quantitative studies of dystrophin expression and functionality have been performed in patients with Becker muscular dystrophy (Beggs et al., 1991; Bushby et al., 1993; Nicholson et al., 1993; Comi et al., 1994; Morandi et al., 1995) and none of these studies used quantitative immunohistochemistry or had sufficient patients with deletions relevant for current exon skipping trial efforts. Several questions remain unanswered; in particular, it is not clear which exon(s) is the optimal target for removal and whether different deletions, which could all respond to the skipping of the same exon, are associated with a similar phenotype or not. To this end, we have quantitated dystrophin and several members of the dystrophin-associated protein complex in skeletal muscle biopsies from 17 patients with Becker muscular dystrophy who harbour in-frame deletions relevant to ongoing exon skipping preclinical and clinical trials for Duchenne muscular dystrophy. We investigate the association between the clinical severity of Becker muscular dystrophy with the amount of dystrophin and dystrophin-associated protein complex proteins in order to better understand and assess both the level and functional properties of the dystrophin produced by exon skipping in future clinical trials.
Materials and methods
Patients
We performed a multi-centre retrospective study of clinical data from five centres: the Dubowitz Neuromuscular Centre at the Institute of Child Health and Great Ormond Street Hospital for Children London, UK; the Institute of Neurology, Catholic University School of Medicine Rome, Italy; the University of Ferrara, Ferrara, Italy; Leiden University Medical Centre, Leiden, The Netherlands and the Institute of Genetic Medicine, Newcastle University, Newcastle, UK. A total of 17 patients were selected from a cohort with comprehensive DNA testing using the following inclusion criteria: (i) a confirmed in-frame exon deletion in the dystrophin gene by multiplex ligation-dependent probe amplification; and (ii) a deletion applicable to exons 51, 53 or 45–55 multi-exon skipping models in Duchenne muscular dystrophy. A standardized questionnaire was distributed to obtain information on gene mutation, age of onset, family history, walking ability and specific symptoms such as muscle cramps and myoglobinuria. Becker muscular dystrophy Patient 2 has been described clinically as Case A1 in a previous study (Helderman-van den Enden et al., 2010), but protein quantification was not previously studied.
Skeletal muscle biopsies were obtained with informed consent from all 17 patients with Becker muscular dystrophy along with non-myopathic controls (n = 2) and patients with Duchenne muscular dystrophy (n = 3) for comparative analysis. Muscle biopsies were taken from the quadriceps (n = 12 cases), deltoid (n = 2 cases) or tibialis anterior (n = 3 cases) muscles as indicated in Table 1. Control and Duchenne muscular dystrophy muscle biopsies (quadriceps) were obtained from the Biobank of the Medical Research Council Centre for Neuromuscular Diseases.
Table 1.
Summary of clinical features
| Patient ID |
Deletion | Centre | Biopsy | Age at study (years) | Age at onset of symptoms (years) | Symptoms at onset | Cramps | Myoglobnuria | Dilated cardiomyopathy | Family history | Ambulant | Alive | Additional comments | Severity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Model | ||||||||||||||
| 1 | 51 | 45–51 | L | Q | 12 | 12 | Toe walking, falls | n | n | n | y | y | y | Mild learning difficulties | Mild |
| 2 | 51 | 45–51 | LE | TA | 48 | 4 | Calf hypertrophy and cramps | y | n | n | y (grand-father) | y | y | Calf hypertrophy and childhood cramps, intolerance to exercise | Mild |
| 3 | 51 | 45–51 | R | Q | 23 | NA | Incidental discovery of high creatine kinase | n | n | n | y (uncle) | y | y | Diagnosis made due to high creatine kinase | Asymptomatic |
| 4 | 51 | 48–51 | L | Q | 10 | 2 | Delayed motor milestones | n | n | n | n | y | y | Learning difficulties | Mild |
| 5 | 51 | 48–51 | N | Q | 12 | 2 | Muscle cramps and myalgia | y | n | n | n | y | y | Learning difficulties | Mild |
| 6 | 51 | 48–51 | F | TA | 25 | NA | Incidental discovery of high creatine kinase | n | n | Mild | n | y | y | Epilepsy, during hospitalization high creatine kinase detected | Asymptomatic |
| 7 | 51 | 48–51 | R | Q | 16 | 8 | Fatigue, no weakness | y | n | n | y (brother, uncle) | y | y | Mild TA contractures; calf and quadriceps hypertrophy | Mild |
| 8 | 51 | 48–51 | R | D | 34 | NA | NA | y | n | n | y (cousin) | y | y | Diagnosis made due to family history and high creatine kinase | Asymptomatic |
| 9 | MS | 45–55 | R | Q | 34 | 4 | Fatigue, cramps after exercise | y | y (frequently) | n | y (grand-father, Patient 10) | y | y | Migraine; calf hypertrophy | Mild |
| 10 | MS | 45–55 | R | D | 76 | 55 | Walking difficulties | n | n | n | y (grandson, Patient 9) | n (age 73, femur fracture) | N (2003 age 76) | Stroke (1996); hypertension; diabetes; peripheral arteriopathy | Mild |
| 11 | MS | 45–55 | R | Q | 23 | 4 | Fatigue, mild difficulty in running | n | n | n | N | y | y | Neck flexors weakness | Mild |
| 12 | MS | 45–55 | R | Q | 29 | 9 | Myalgia, myoglobinuria | y | y | n | Y (two uncles) | y | y | Calf and quadriceps hypertrophy | Mild |
| 13 | 53 | 42–53 | L | Q | 14 | 4 | Learning difficulties, ADHD | n | n | n | n | y | y | Normal muscle function, only high creatine kinase | Mild |
| 14 | 53 | 45–53 | L | Q | 25 | Un | Unable to run | n | n | n | n | y | y | Significant proximal weakness, Gower's time 3 s, inability to run, positive Trendeleburg sign | Severe |
| 15 | 53 | 45–53 | L | Q | 21 | 10 | Myalgia | n | n | n | n | y | y | Calf hypertrophy | Mild |
| 16 | 53 | 45–53 | L | Q | 14 | NA | Incidental finding of high creatine kinase | n | n | n | n | y | y | Mild TA contractures | Mild |
| 17 | 53 | 52–53 | F | TA | 13 | n.a | Incidental finding of high creatine kinase | n | n | n | n | y | y | Asymptomatic, only high creatine kinase | Asymptomatic |
ADHD = attention deficit/hyperactivity disorder; D = deltoid; F = Ferrara; L = London; LE = Leiden; n = no; N = Newcastle; NA = not applicable; Q = quadriceps; R = Rome; TA = tibialis anterior; y = yes; un = unavailable. Severity classification was based on the skeletal muscle phenotype.
Immunohistochemistry
Muscle biopsies were taken using standard techniques and mounted in OCT (optimal cutting temperature) medium then frozen by immersion in isopentane cooled in liquid nitrogen. Immunohistochemistry and subsequent quantitative analysis of all samples was performed as described previously (Arechavala-Gomeza et al., 2010a) at the same centre (London) to minimize variability. Primary monoclonal antibodies used were MANDYS106 (MDA monoclonal antibody resource, Prof. Glenn Morris, 1:100), Dys 2 (Novacastra, 1:20), α-sarcoglycan (Novacastra, 1:50), β-dystroglycan (Novacastra, 1:20) and β-spectrin (Novacastra, 1:20). A rabbit polyclonal antibody was used to detect n-NOS (Santa Cruz Biotechnology, 1:50). Sections were evaluated using a Leica DMR microscope interfaced to MetaMorph (Molecular Devices) and intensity measurements performed as previously described (Arechavala-Gomeza et al., 2010a). The Mann–Whitney test was used for statistical analysis, significance was set at P = 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
Western blotting
Western blot analysis was performed at the same centre (London) to minimize variability. Proteins were homogenized from either snap frozen muscle tissue or cryostat cut sections in sodium dodecyl sulphate lysis buffer containing protease inhibitors. For snap frozen muscle samples, 40 µg of total protein was loaded; sections were lysed in a total volume of 50 µl of lysis buffer and 11 µl was loaded. Western blotting was performed with NuPAGE® Novex Tris–Acetate 3–8% gels (Invitrogen) and polyvinylidene fluoride membranes. Membranes were incubated overnight at 4°C with Dys2 (1:50, Novocastra) and sarcomeric α-actinin (as a loading control, 1:10 000, Sigma) primary antibodies. After washing, membranes were incubated with biotinylated anti-mouse IgG (Amersham GE Healthcare) for 1 h followed by horseradish peroxidase-conjugated streptavidin (Dako) for 1 h. Membranes were visualized using chemiluminescence (ECL Plus, Amersham GE Healthcare) and a Typhoon scanner (Amersham GE healthcare). For quantification, dystrophin intensity was normalized to α-actinin using ImageJ software and expressed as per cent of control.
Results
To avoid any potential bias, patients were selected exclusively by their genotype and grouped according to corresponding exon skipping models for Duchenne muscular dystrophy. Individual patient's deletions and their groupings are listed in Table 1. Of the 17 patients, eight had deletions ending with exon 51 (model 51) whose skipping would rescue 13% of all Duchenne muscular dystrophy mutations (Aartsma-Rus et al., 2009). Four patients had a large deletion of exons 45–55 (model MS), multi-exon skipping of these exons in Duchenne muscular dystrophy could rescue up to 63% of Duchenne muscular dystrophy deletions (Beroud et al., 2007). Finally, five patients with Becker muscular dystrophy have deletions ending with exon 53 (model 53) whose skipping would account for 7.7% of all Duchenne muscular dystrophy mutations (Aartsma-Rus et al., 2009).
Clinical characteristics
All the patients in our study had a clinical diagnosis of Becker muscular dystrophy. According to their skeletal muscle phenotype, the 17 patients were classified as asymptomatic, mild or severe (Table 1). Asymptomatic patients with Becker muscular dystrophy were individuals with no detectable muscle weakness and in whom the only pathological feature was the isolated elevation of serum creatine kinase. Mild Becker muscular dystrophy was defined by evidence of mild proximal muscle weakness in patients in whom autonomous ambulation and running ability was retained; individuals with more severe weakness and in whom running abilities had either never been acquired or were lost by the end of adolescence were classified as severe. Of the 17 individuals studied, 12 were classified as mild, four asymptomatic and one severe. All patients were ambulant at the time of study with the exception of Patient 10 who lost independent ambulation following a femur fracture at the age of 73 years.
The average age of the 17 patients at the time of study was 25 years and ranged between 10 and 76 years; the average age of onset was 10 years and ranged between 2 and 55 years, although this information could not be precisely established in several patients (Patients 3, 6, 8, 14, 16 and 17). Seven patients were below the age of 20 years at the time of the study, six of whom were classified as mild Becker muscular dystrophy; the four asymptomatic patients were 13, 23, 25 and 34 years of age at the time of study and the severe patient (Patient 14) was 25 years old. Thus, the differences in clinical severity do not merely reflect differences in age although an influence of age cannot be fully discounted. The patient classified as severe Becker muscular dystrophy (Patient 14, deletion 45–53) had proximal weakness with a Gowers’ time of 3 s, a positive Trendelenburg sign and an inability to run.
Cramps were reported in six out of the 17 patients and recurrent myoglobinuria was described in one case. Overall, the clinical severity was heterogeneous within and between the three groups of deletions; patients in the model 51 group were either asymptomatic (three patients), or mildly affected (five patients). In the model 53 group, three patients were classified as mild; one asymptomatic and one severe; all four patients in the model MS group were classified as mild. Within our cohort, only Patient 6 (aged 25 years, 48–51 deletion) suffered from mild dilated cardiomyopathy at the time of the study, detected upon routine surveillance.
Quantification of dystrophin expression
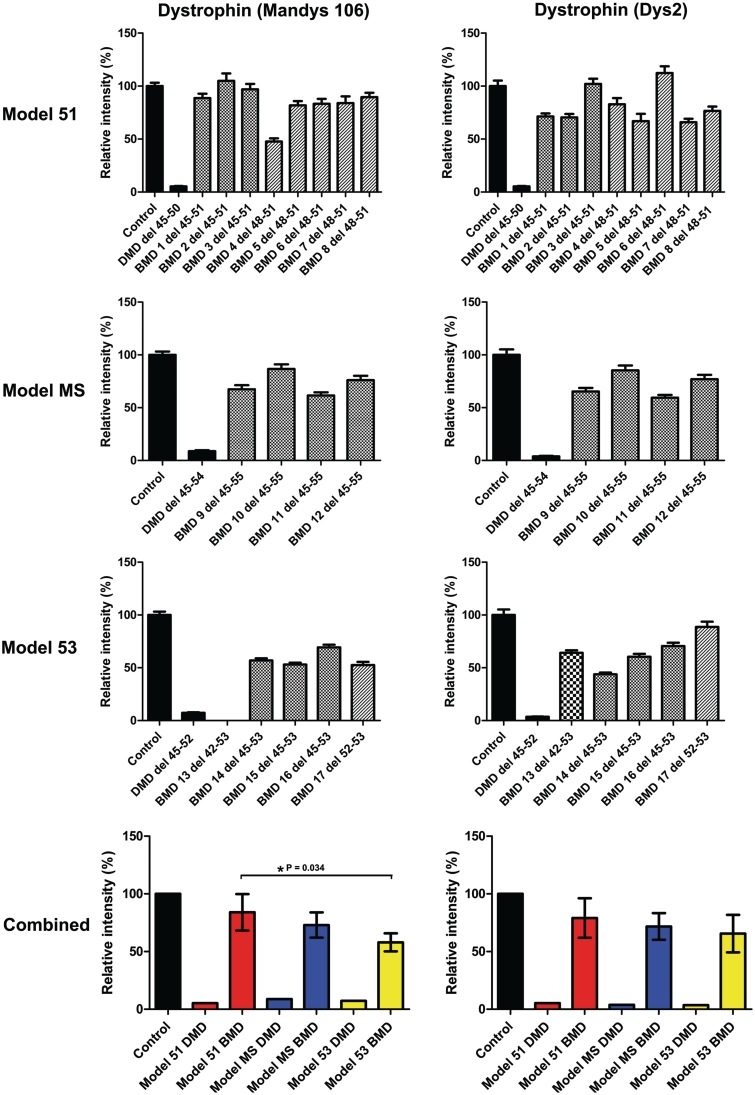
Dystrophin expression was assessed in all 17 patients with Becker muscular dystrophy by immunohistochemistry using both MANDYS106 (exon 43) and Dys2 (last 17 C-terminal amino acids) dystrophin antibodies and semi-quantitative analysis was performed as previously described (Figs 1 and 2) (Arechavala-Gomeza et al., 2010a). For each group, a Duchenne muscular dystrophy patient with an equivalent out of frame deletion was included for comparative analysis; as expected dystrophin was severely reduced in these three patients with Duchenne muscular dystrophy, with levels ranging between 4 and 9% of control. We observed reduced dystrophin staining at the sarcolemma (Fig. 2) which has been reported as a pathological feature of Becker muscular dystrophy (Beggs et al., 1991; Muntoni et al., 1993). Dystrophin levels across all 17 patients with Becker muscular dystrophy were variable ranging from ∼50% to ∼100% of control as determined by semi-quantitative analysis. In the model 51 group, which includes three patients with a 45–51 deletion and five patients with a deletion of exons 48–51, the mean dystrophin expression was ∼80% of control with both MANDYS106 (range: 48–104%) and Dys2 antibodies (range: 66–112%). The mean level of dystrophin in the model MS group was ∼70% (MANDYS106 range: 62–87%, Dys2 range: 59–85%), which decreased to ∼56% in the model 53 group (MANDYS106 range: 0–69%, Dys2 range: 44–89%). The difference in dystrophin levels between the model 51 and model 53 groups was significant when tested using the MANDYS106 antibody (P = 0.034). Intragroup variability was lowest within the model MS group, perhaps reflecting the fact that this group contains four patients harbouring the same deletion (exons 45–55). It is interesting to note that the clinically most severe patient, Becker muscular dystrophy Patient 14 in the model 53 group, had the lowest dystrophin expression with the Dys2 antibody and also one of the lowest with MANDYS106 (Dys 2: 44%, MANDYS106: 57%). On the contrary, the four asymptomatic patients belonging to the model 51 and 53 groups have comparatively higher dystrophin levels with both antibodies. This is illustrated in Fig. 5 where the difference in dystrophin expression between asymptomatic and mild patients with Becker muscular dystrophy is significant with the Dys2 antibody (P = 0.013). Statistical analysis was restricted to asymptomatic and mild patients as only one patient in our cohort was classified as severe.
Figure 1.
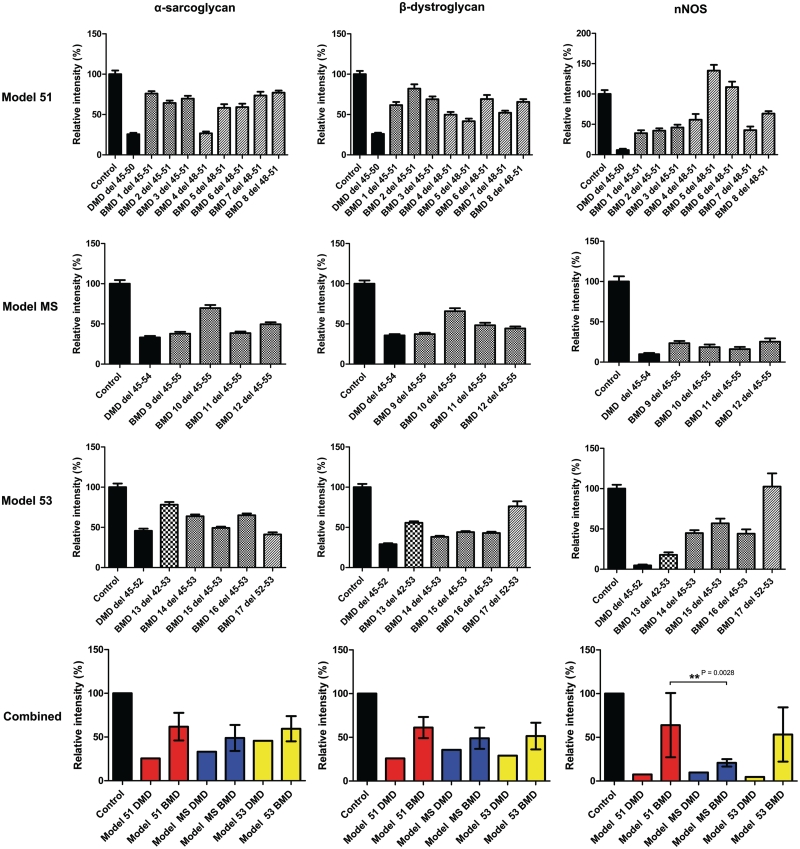
Comparative immunohistochemical analysis of dystrophin expression in 17 patients with Becker muscular dystrophy with in-frame deletions. Control, Duchenne muscular dystrophy and Becker muscular dystrophy transverse muscle sections were immunolabelled for β-spectrin and with MANDYS106 (exon 43) and Dys2 (C-terminal, last 17 amino acids) antibodies against dystrophin. Expression was quantified relative to control muscle in 40 muscle fibres and normalized to β-spectrin expression. Values represent means ± SEM except for the bottom graphs where values represent the mean expression level for each group ± SD of the difference between sample means. In the bottom two graphs patients with Becker muscular dystrophy were grouped according to corresponding exon skipping models for Duchenne muscular dystrophy: exon 51 skipping (Model 51, red bars), multi-exon skipping (model MS, blue bars) and exon 53 skipping (Model 53, yellow bars).
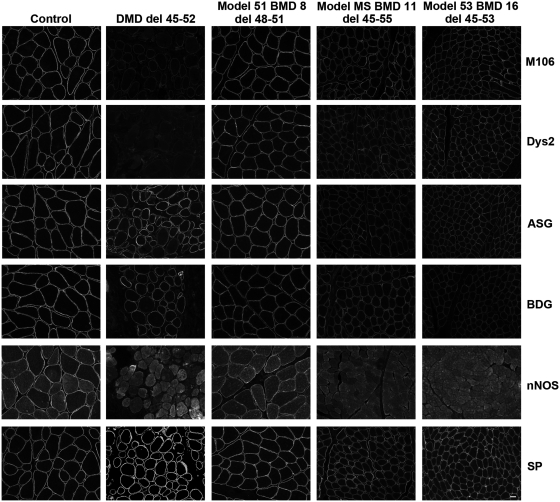
Figure 2.
Representative immunohistochemistry images of dystrophin and dystrophin-associated proteins in patients with Becker muscular dystrophy (BMD) with in-frame deletions. Unfixed, frozen transverse muscle sections (7 µm) from control, Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (Patients 8, 11 and 16 are shown) patients were immunolabelled with MANDYS106 (M106, exon 43) and Dys2 (last 17 amino acids of the C-terminus) antibodies against dystrophin and with antibodies against α-sarcoglycan (ASG), β-dystroglycan (BDG), neuronal nitric oxide synthase (nNOS) and β-spectrin (SP). Scale bar = 20 µm.
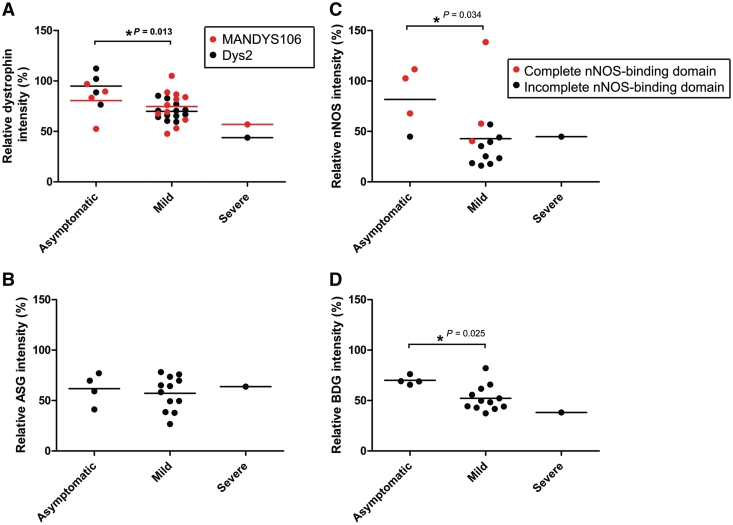
Figure 5.
Correlation of dystrophin and dystrophin-associated protein expression with clinical severity. Clinical severity correlated to dystrophin expression with both MANDYS106 (red) and Dys2 (black) antibodies (A), α-sarcoglycan (ASG) (B), nNOS (C) and β-dystroglycan (BDG) (D) expression. Lines represent mean expression of each group. P-values are shown for statistically significant differences and were determined using a two-tailed Mann–Whitney test.
The dystrophin quantities detected by MANDYS106 and Dys2 antibodies do not always perfectly correlate. For example, the deletion of Becker muscular dystrophy Patient 13 includes the MANDYS106 epitope in exon 43 and therefore zero dystrophin was detected with this antibody, confirming the high specificity of the MANDYS106 antibody (this data point was excluded from statistical analysis as the zero value does not fairly represent the dystrophin content in this patient). Only one patient, Becker muscular dystrophy Patient 4 (deletion 48–51), showed an unexpectedly large difference between the two antibodies. This individual's dystrophin level was 48% of control with MANDYS106 and 83% with Dys2.
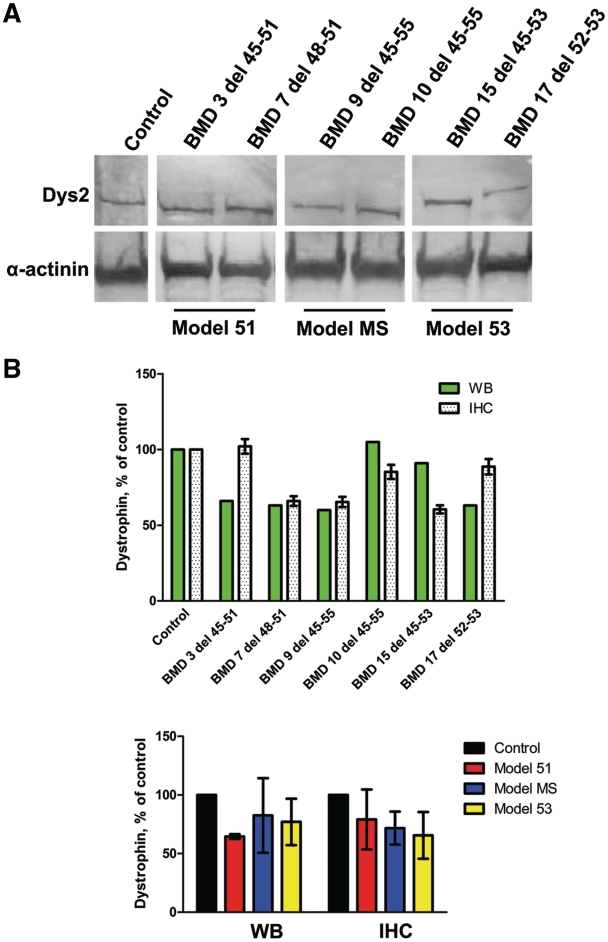
To confirm the immunohistochemical findings, we next analysed dystrophin expression by western blotting with the Dys2 antibody in two patients with Becker muscular dystrophy from each group (Fig. 3A). Dystrophin bands were detected for all patients at the expected molecular mass, with the patients from the MS model having the lowest molecular mass owing to their larger deletions. Semi-quantitative analysis revealed that model 51 patients with Becker muscular dystrophy had a mean dystrophin expression of 65% (SD ± 2.1) of control, model MS and model 53 had 83% (SD ± 31.8) and 77% (SD ± 19.8), respectively (Fig. 3B). Although a robust comparison between dystrophin quantification methods was not the aim of this study, in most instances, we observed comparable expression levels between the two techniques (Fig. 3B). However, for model 51, the dystrophin expression level is substantially lower for Becker muscular dystrophy Patient 3 when calculated by Western blotting, possibly reflecting variability in muscle preservation in different blocks of the muscle biopsy studied with the different techniques. Nonetheless, by two independent methods, we have demonstrated that our cohorts of asymptomatic or mild patients with Becker muscular dystrophy have dystrophin levels of at least 40% of control.
Figure 3.
Western blot analysis of dystrophin expression in patients with Becker muscular dystrophy (BMD) with in-frame deletions. (A) Western blot of total protein extracts from skeletal muscle biopsy of control and patients with Becker muscular dystrophy. Blots were probed with the C-terminal Dys2 antibody for dystrophin and a sarcomeric α-actinin antibody as a loading control. (B) Western blot semi-quantification using ImageJ software. Data are normalized to α-actinin and both individual and grouped patient data are presented as per cent of control. Immunohistochemistry analysis of the same patients is presented alongside for comparison. Values represent means; for the graph of individual patients the error bars signify the SEM and for the grouped sample graph the error bars represent the SD of the difference between sample means. IHC = immunohistochemistry; WB = western blot.
Expression of proteins of the dystrophin-associated protein complex
To further assess the functional properties of the Becker muscular dystrophy dystrophin proteins and to correlate this to clinical severity, we quantified the expression of α-sarcoglycan, β-dystroglycan and nNOS in the 17 patients with Becker muscular dystrophy by immunohistochemistry (Figs 2 and 4). α-Sarcoglycan and β-dystroglycan expression was reduced in all patients with Becker muscular dystrophy in comparison to controls, and in several cases the level was comparable with that observed in patients with Duchenne muscular dystrophy. α-Sarcoglycan expression varied from 27–78% of control, while β-dystroglycan expression varied from 37–82% of control. Expression of both α-sarcoglycan and β-dystroglycan was lower in patients belonging to the model MS and model 53 groups; interestingly dystrophin levels determined by immunohistochemistry were also lower in these groups compared to the model 51 group. Higher levels of β-dystroglycan expression were associated with asymptomatic skeletal muscle phenotypes (asymptomatic versus mild, P = 0.025), while no association was found with α-sarcoglycan expression levels (Fig. 5). Clinically severe Patient 14 had 64% α-sarcoglycan levels but only 38% β-dystroglycan, while mild Becker muscular dystrophy Patient 10 had the highest α-sarcoglycan and β-dystroglycan levels (70% and 66%, respectively) within the model MS group and also the highest dystrophin levels of the group (MANDYS106: 86% and Dys2: 85% of control). Thus, in this cohort of patients, β-dystroglycan expression was significantly associated with mild clinical severity while α-sarcoglycan expression was not.
Figure 4.
Comparative immunohistochemical analysis of dystrophin-associated protein expression in 17 patients with Becker muscular dystrophy (BMD) with in-frame deletions. Control, Duchenne muscular dystrophy and Becker muscular dystrophy transverse muscle sections were immunolabelled with antibodies against α-sarcoglycan, β-dystroglycan, neuronal nitric oxide synthase (nNOS) and β-spectrin. Expression was quantified relative to control muscle in 40 muscle fibres and normalized to β-spectrin expression. Values represent means ± SEM except for graphs on the bottom where values represent the mean expression level for each group ± SD of the difference between sample means. In the bottom two graphs patients with Becker muscular dystrophy are grouped according to corresponding exon skipping models for Duchenne muscular dystrophy: exon 51 skipping (model 51, red bars), multi-exon skipping (model MS, blue bars) and exon 53 skipping (model 53, yellow bars).
Of the dystrophin-associated protein complex proteins studied, nNOS showed the most variable expression across all patients (between 16% and 139% of controls). The dystrophin nNOS-binding domain is encoded by exons 42–45 and so the presence of nNOS at the sarcolemma of Becker muscular dystrophy muscle is expected to be significantly influenced by the patient's deletion. Indeed, we demonstrate that patients with Becker muscular dystrophy with disrupted dystrophin nNOS-binding domains (11 out of 17 patients) have lower sarcolemmal nNOS expression despite relatively high levels (>50% of control) of dystrophin (Figs 4 and 5). This is particularly noticeable with the model MS group in which all patients have incomplete nNOS-binding domains and the nNOS levels for all four patients were <25% of controls. Within the model 53 group, asymptomatic Patient 17 with an intact nNOS-binding domain has the highest nNOS expression of the whole cohort (102% of control); similarly Becker muscular dystrophy Patients 5, 6 and 8 have the highest nNOS expression within the model 51 group, Patients 6 and 8 being classified as asymptomatic. The effect of dystrophin nNOS-binding domain integrity on nNOS expression is further illustrated by the large SD between the sample means of the model 51 group which contains five patients with a complete nNOS-binding domain and three with incomplete binding domains. The clinically most severe patient (Becker muscular dystrophy Patient 14) has a low nNOS expression, 45% of control; this clinical association is illustrated in Fig. 5, where the difference in nNOS expression between asymptomatic and mild patients is significant (P = 0.034). As indicated by the coloured circles in Fig. 5, three out of the four asymptomatic patients have complete nNOS-binding domains and mild patients with Becker muscular dystrophy with complete nNOS-binding domains have a relatively higher nNOS expression level than those with incomplete nNOS-binding domains. Mild Becker muscular dystrophy Patients 4 and 7 (deletion 48–51) with complete binding domains have lower than expected nNOS levels suggesting that an intact dystrophin nNOS-binding domain might not be the only variable to determine sarcolemmal nNOS expression, as recently suggested by others (Miyagoe-Suzuki and Takeda, 2001; Finanger Hedderick et al., 2011). Of the six patients who suffered frequent cramps, three had a complete nNOS-binding domain; thus at least in our cohort, the occurrence of cramps does not necessarily depend on the integrity of the nNOS-binding domain.
Discussion
We have quantitatively assessed the levels of dystrophin and dystrophin-associated protein expression in 17 patients with Becker muscular dystrophy grouped by deletion according to currently studied exon skipping models for Duchenne muscular dystrophy. Using two independent methods, we demonstrate that all the patients with Becker muscular dystrophy in our cohort have reduced dystrophin expression levels of at least 40% of controls. A previous study of families with X-linked dilated cardiomyopathy determined that dystrophin levels of 30% of control can be sufficient to avoid muscle weakness in these families, although these patients expressed lower levels of normal dystrophin (Neri et al., 2007). Thus, our data indicate that the production of internally deleted dystrophin proteins by exon skipping should provide a substantial benefit to patients with Duchenne muscular dystrophy.
Overall, patients in the model 51 group had higher dystrophin and dystrophin-associated protein complex levels than patients in the other two groups with significant differences in dystrophin and nNOS expression. This lends further support to the ongoing exon 51 skipping clinical trials for Duchenne muscular dystrophy (Kinali et al., 2009; Cirak et al., 2011; Goemans et al., 2011). Intragroup variability was lowest with the model MS group, which is the only group to be comprised entirely of patients harbouring the same deletion (exons 45–55). However, moderate variability was observed in dystrophin and dystrophin-associated protein complex levels between patients of the same deletion.
The two different deletions within the model 51 group have comparable dystrophin and dystrophin-associated protein complex expression levels as well as a mild clinical phenotype. There is a greater variability in the clinical phenotypes between the patients of the model 53 group; our results also indicate a possible benefit for the retention of the nNOS-binding domain in internally deleted dystrophin.
We observed variability in dystrophin protein levels between patients with the same deletion; this could stem from the differences of individual intronic breakpoints in patients, which may affect alternative splicing and/or translation efficiency. Other explanations for intra and intergroup variability could be the differential stability of the internally deleted dystrophins (Krieger et al., 2010; Henderson et al., 2011) and/or the endogenous splicing of other exons. For example, exon 44 is known to spontaneously skip in patients with deletions of the surrounding exons (van Vliet et al., 2008). Although we have not measured this in our population, its occurrence could affect the levels of the dystrophin protein observed. Other genetic modifiers of dystrophin translation efficiency could also be involved in these discrepancies; for example a mutation in the promoter region of the gene encoding the extracellular matrix protein, osteopontin, identified osteopontin genotype as a genetic modifier of Duchenne muscular dystrophy severity (Pegoraro et al., 2011). Additionally the microRNA, miR-31, was recently shown to repress dystrophin expression through binding to the 3′ untranslated region of dystrophin RNA (Cacchiarelli et al., 2011).
The model 51 group contained three out of the four asymptomatic patients, while the model 53 group consisted of severe, mild and asymptomatic patients; thus there is no clear-cut genotype–phenotype correlation in this latter group of patients with Becker muscular dystrophy. We do, however, demonstrate for the first time that in Becker muscular dystrophy, higher expression levels of dystrophin, β-dystroglycan and nNOS are significantly associated with a milder skeletal muscle phenotype.
Our results suggest that the milder phenotypes in our cohort of patients with Becker muscular dystrophy are associated with higher expression levels of dystrophin and those dystrophin-associated protein complex members that bind directly to dystrophin. We found that β-dystroglycan and nNOS expression correlates with clinical severity, while α-sarcoglycan expression does not. This could be due to the direct binding of β-dystroglycan and nNOS to dystrophin; while α-sarcoglycan binds dystrophin indirectly (Ervasti, 2007; Ozawa, 2010). Our findings are further supported by mouse model data that demonstrate that the skipping of exons that preserve the nNOS-binding domain results in a more favourable clinical outcome (Lai et al., 2009); this information should be considered when developing exon skipping strategies.
While it is generally established that the 45–55 deletion is associated with a favourable prognosis, this deletion is also associated with X-linked dilated cardiomyopathy (Beroud et al., 2007; Nakamura et al., 2008; Ferreiro et al., 2009; Miyazaki et al., 2009). In our study, the four patients with the 45–55 deletion were classified as mild and none had developed dilated cardiomyopathy, including one patient who died at age 76.
In fact, a large percentage of patients with Becker muscular dystrophy develop dilated cardiomyopathy (Melacini et al., 1993, 1996; Bushby et al., 2003) and mutations that disrupt the phasing of the helical spectrin repeats encoded by exons 45–49 are thought to lead to an earlier-onset of dilated cardiomyopathy as a result of altered dystrophin structure (Kaspar et al., 2009). The majority of the internally deleted dystrophin proteins studied within our cohort additionally remove the proline-rich hinge 3 region encoded by exons 50 and 51; the hinge 3 region is thought to be helical but does not necessarily form a stable tertiary structure (Bhasin et al., 2005). One study designed nano-constructs of dystrophin based on in-frame Becker muscular dystrophy and therapeutic deletions including exons 45–51 (Krieger et al., 2010); our cohort included three patients with this deletion in the model 51 group. The secondary structure prediction for the abnormal ‘linker’ created by the joining of exons 44 to 52 revealed that this region folds into a stable repeat domain that maintains its helicity (Krieger et al., 2010). These findings and our results provide further support and guidance for exon skipping within the rod domain. However, a recent study has demonstrated that the removal of hinge 2 through to spectrin-like repeat 19 causes a significant loss in protein stability (Henderson et al., 2011); thus caution is needed when removing large segments of the rod domain such as with multi-exon skipping models. Multi-exon skipping is also technically very challenging; only very minimal skipping has been detected at low frequency in preliminary cell culture work with a cocktail of antisense oligonucleotides for skipping of exons 45–55 (van Vliet et al., 2008).
It has been hypothesized that patients with Becker muscular dystrophy with rod domain deletions suffer muscle cramps following exercise due to the mislocalization of nNOS from the sarcolemma with subsequent disruption of blood flow during exercise (Thomas et al., 1998; Sander et al., 2000; Kobayashi et al., 2008). Our quantitative analysis of 17 patients with Becker muscular dystrophy does not completely support this hypothesis. Firstly, several patients with deleted nNOS-binding domains and/or low sarcolemmal expression levels do not suffer cramps and perhaps more convincingly, Becker muscular dystrophy Patients 5, 7 and 8 report cramps even though their deletions do not span the nNOS-binding domain. In fact, Patients 5 and 8 have near normal nNOS expression levels. The aetiology of cramps in Becker muscular dystrophy is complex and not necessarily or exclusively related to blood flow, but to physical exercise. Thus, mild individuals with preserved strength may have frequent cramps because they endure more physical activity, on the other hand growth and development may also play a role. A limitation of our conclusion is the relatively young age of a number of the patients, and we cannot rule out the possibility that some patients might develop cramps later on in life. Moreover, the recruitment of nNOS to the sarcolemma might also be affected by structural abnormalities located outside the nNOS-binding domain. Indeed, a complex correlation between sarcolemmal nNOS localization and muscle activity, oxidative stress and calcium signalling has been recently demonstrated (Pietri-Rouxel et al., 2010; Finanger Hedderick et al., 2011), suggesting the presence of multiple independent regulators of nNOS localization in muscle.
In summary, we demonstrate that all patients with the genotypes we have studied in this report had Becker muscular dystrophy and not Duchenne muscular dystrophy, in keeping with the reading frame hypothesis. While this not surprising, concerns that a significant number of patients with these genotypes could have Duchenne muscular dystrophy or severe Becker muscular dystrophy have been raised, although the source of the information relied on old studies when multiplex ligation-dependent probe amplification was not available and the end-points of the deletions were not systematically assessed (Yokota et al., 2009). In fact, a more recent analysis of the same Leiden database used by Yokota et al. (2009) has demonstrated that all patients with in-frame 45–51 or 50–51 deletions have a mild Becker muscular dystrophy phenotype (Helderman-van den Enden et al., 2010). Our data are further supported by two other reports; firstly Beroud et al. (2007) reported on the mild or asymptomatic phenotypes of 15 patients with a 45–55 in-frame deletion and secondly a study that focused on dilated cardiomyopathy in Becker muscular dystrophy reported the mild Becker muscular dystrophy phenotype of 24 patients with in-frame deletions that fit into one of the three models used in our study (Kaspar et al., 2009).
We show that patients with Becker muscular dystrophy who express the same internally deleted dystrophin as could be induced by exon skipping therapies are mostly associated with mild phenotypes and express dystrophin at a high enough level (at least 40% of control) to provide a functional benefit to patients with Duchenne muscular dystrophy. We report that the asymptomatic Becker muscular dystrophy phenotype is associated with significantly higher dystrophin, β-dystroglycan and nNOS expression levels than the mild phenotype and highlight exon 51 as an optimal target exon for removal. This information is encouraging, as the dystrophin levels obtained in at least some of the patients recruited into the recently completed systemic clinical trials had levels of 15% (Goemans et al., 2011) and 18% (Cirak et al., 2011) of normal levels; the latter representing ∼45% efficacy during a short 12 week study, suggesting that a significant clinical benefit from these dystrophins is a realistic possibility.
Funding
Medical Research Council (MRC) (grant to F.M.); European Union BIO-NMD FP7 grant; MRC Centre for Neuromuscular diseases at UCL and Newcastle including the MRC Neuromuscular Centre Biobank; Wellcome Trust University Award (to J.M.) and Great Ormond Street Hospital Children’s Charity (to F.M.).
Acknowledgements
The authors wish to thank the participating subjects and their families, the charities Muscular Dystrophy Campaign, Action Duchenne and the Duchenne Family Support Group for participating in the UK MDEX consortium (www.mdex.org.uk) which performed this study. We also gratefully acknowledge the support of the Duchenne Parent Project, Italy (Duchenne muscular dystrophy/Becker muscular dystrophy National Registry) and the TREAT-NMD neuromuscular network. We thank Ms Christa de Winter, Dr Valeria Ricotti and Mr Darren Chambers for their assistance in immunohistochemical analysis and the professors Gert Jan B Van Ommen and Johan T. Den Dunnen for constructive discussions. We also wish to thank the MRC Neuromuscular Centre Biobank and Prof. Glenn Morris, Oswestry, UK and the MDA Monoclonal Antibody Resource for supplying the MANDSY106 antibody.
Glossary
Abbreviations
- nNOS
neuronal nitric oxide synthase
References
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–9. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- Abbs S, Tuffery-Giraud S, Bakker E, Ferlini A, Sejersen T, Mueller CR. Best practice guidelines on molecular diagnostics in Duchenne/Becker muscular dystrophies. Neuromuscul Disord. 2010;20:422–7. doi: 10.1016/j.nmd.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Kinali M, Feng L, Brown SC, Sewry C, Morgan JE, et al. Immunohistological intensity measurements as a tool to assess sarcolemma-associated protein expression. Neuropathol Appl Neurobiol. 2010a;36:265–74. doi: 10.1111/j.1365-2990.2009.01056.x. [DOI] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Kinali M, Feng L, Guglieri M, Edge G, Main M, et al. Revertant fibres and dystrophin traces in Duchenne muscular dystrophy: implication for clinical trials. Neuromuscul Disord. 2010b;20:295–301. doi: 10.1016/j.nmd.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F, et al. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- Beroud C, Tuffery-Giraud S, Matsuo M, Hamroun D, Humbertclaude V, Monnier N, et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat. 2007;28:196–202. doi: 10.1002/humu.20428. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Law R, Liao G, Safer D, Ellmer J, Discher BM, et al. Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J Mol Biol. 2005;352:795–806. doi: 10.1016/j.jmb.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Bushby K, Muntoni F, Bourke JP. 107th ENMC international workshop: the management of cardiac involvement in muscular dystrophy and myotonic dystrophy. 7th–9th June 2002, Naarden, the Netherlands. Neuromuscul Disord. 2003;13:166–72. doi: 10.1016/s0960-8966(02)00213-4. [DOI] [PubMed] [Google Scholar]
- Bushby KM, Gardner-Medwin D, Nicholson LV, Johnson MA, Haggerty ID, Cleghorn NJ, et al. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. II. Correlation of phenotype with genetic and protein abnormalities. J Neurol. 1993;240:105–12. doi: 10.1007/BF00858726. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D, Incitti T, Martone J, Cesana M, Cazzella V, Santini T, et al. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136–41. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi GP, Prelle A, Bresolin N, Moggio M, Bardoni A, Gallanti A, et al. Clinical variability in Becker muscular dystrophy. Genetic, biochemical and immunohistochemical correlates. Brain. 1994;117(Pt 1):1–14. doi: 10.1093/brain/117.1.1-a. [DOI] [PubMed] [Google Scholar]
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–17. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Ferreiro V, Giliberto F, Muniz GM, Francipane L, Marzese DM, Mampel A, et al. Asymptomatic Becker muscular dystrophy in a family with a multiexon deletion. Muscle Nerve. 2009;39:239–43. doi: 10.1002/mus.21193. [DOI] [PubMed] [Google Scholar]
- Finanger Hedderick EL, Simmers JL, Soleimani A, Andres-Mateos E, Marx R, Files DC, et al. Loss of sarcolemmal nNOS is common in acquired and inherited neuromuscular disorders. Neurology. 2011;76:960–7. doi: 10.1212/WNL.0b013e31821043c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N Engl J Med. 2011;364:1513–22. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- Helderman-van den Enden AT, Straathof CS, Aartsma-Rus A, den Dunnen JT, Verbist BM, Bakker E, et al. Becker muscular dystrophy patients with deletions around exon 51; a promising outlook for exon skipping therapy in Duchenne patients. Neuromuscul Disord. 2010;20:251–4. doi: 10.1016/j.nmd.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Henderson DM, Belanto JJ, Li B, Heun-Johnson H, Ervasti JM. Internal deletion compromises the stability of dystrophin. Hum Mol Genet. 2011;20:2955–63. doi: 10.1093/hmg/ddr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar RW, Allen HD, Ray WC, Alvarez CE, Kissel JT, Pestronk A, et al. Analysis of dystrophin deletion mutations predicts age of cardiomyopathy onset in becker muscular dystrophy. Circ Cardiovasc Genet. 2009;2:544–51. doi: 10.1161/CIRCGENETICS.109.867242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–28. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Coovert DD, Bulman DE, Ray PN, Mendell JR, Burghes AH. Somatic reversion/suppression in Duchenne muscular dystrophy (DMD): evidence supporting a frame-restoring mechanism in rare dystrophin-positive fibers. Am J Hum Genet. 1992;50:950–9. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi YM, Rader EP, Crawford RW, Iyengar NK, Thedens DR, Faulkner JA, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456:511–5. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger CC, Bhasin N, Tewari M, Brown AE, Safer D, Sweeney HL, et al. Exon-skipped dystrophins for treatment of Duchenne muscular dystrophy: mass spectrometry mapping of most exons and cooperative domain designs based on single molecule mechanics. Cytoskeleton (Hoboken) 2010;67:796–807. doi: 10.1002/cm.20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, Long C, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119:624–35. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rumeur E, Winder SJ, Hubert JF. Dystrophin: more than just the sum of its parts. Biochim Biophys Acta. 2010;1804:1713–22. doi: 10.1016/j.bbapap.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Lesca G, Testard H, Streichenberger N, Pelissier JF, Lestra C, Burel E, et al. [Family study allows more optimistic prognosis and genetic counselling in a child with a deletion of exons 50–51 of the dystrophin gene] Arch Pediatr. 2007;14:262–5. doi: 10.1016/j.arcped.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Lu QL, Morris GE, Wilton SD, Ly T, Artem'yeva OV, Strong P, et al. Massive idiosyncratic exon skipping corrects the nonsense mutation in dystrophic mouse muscle and produces functional revertant fibers by clonal expansion. J Cell Biol. 2000;148:985–96. doi: 10.1083/jcb.148.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Tome FM, Collin H, Leturcq F, Jeanpierre M, Kaplan JC, et al. Expression of dystrophin-associated proteins in dystrophin-positive muscle fibers (revertants) in Duchenne muscular dystrophy. Neuromuscul Disord. 1994;4:115–20. doi: 10.1016/0960-8966(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Melacini P, Fanin M, Danieli GA, Fasoli G, Villanova C, Angelini C, et al. Cardiac involvement in Becker muscular dystrophy. J Am Coll Cardiol. 1993;22:1927–34. doi: 10.1016/0735-1097(93)90781-u. [DOI] [PubMed] [Google Scholar]
- Melacini P, Fanin M, Danieli GA, Villanova C, Martinello F, Miorin M, et al. Myocardial involvement is very frequent among patients affected with subclinical Becker's muscular dystrophy. Circulation. 1996;94:3168–75. doi: 10.1161/01.cir.94.12.3168. [DOI] [PubMed] [Google Scholar]
- Melis MA, Cau M, Muntoni F, Mateddu A, Galanello R, Boccone L, et al. Elevation of serum creatine kinase as the only manifestation of an intragenic deletion of the dystrophin gene in three unrelated families. Eur J Paediatr Neurol. 1998;2:255–61. doi: 10.1016/s1090-3798(98)80039-1. [DOI] [PubMed] [Google Scholar]
- Miyagoe-Suzuki Y, Takeda SI. Association of neuronal nitric oxide synthase (nNOS) with alpha1-syntrophin at the sarcolemma. Microsc Res Tech. 2001;55:164–70. doi: 10.1002/jemt.1167. [DOI] [PubMed] [Google Scholar]
- Miyazaki D, Yoshida K, Fukushima K, Nakamura A, Suzuki K, Sato T, et al. Characterization of deletion breakpoints in patients with dystrophinopathy carrying a deletion of exons 45-55 of the Duchenne muscular dystrophy (DMD) gene. J Hum Genet. 2009;54:127–30. doi: 10.1038/jhg.2008.8. [DOI] [PubMed] [Google Scholar]
- Morandi L, Mora M, Confalonieri V, Barresi R, Di Blasi C, Brugnoni R, et al. Dystrophin characterization in BMD patients: correlation of abnormal protein with clinical phenotype. J Neurol Sci. 1995;132:146–55. doi: 10.1016/0022-510x(95)00147-t. [DOI] [PubMed] [Google Scholar]
- Morrone A, Zammarchi E, Scacheri PC, Donati MA, Hoop RC, Servidei S, et al. Asymptomatic dystrophinopathy. Am J Med Genet. 1997;69:261–7. doi: 10.1002/(sici)1096-8628(19970331)69:3<261::aid-ajmg9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Di Lenarda A, Porcu M, Sinagra G, Mateddu A, Marrosu G, et al. Dystrophin gene abnormalities in two patients with idiopathic dilated cardiomyopathy. Heart. 1997;78:608–12. doi: 10.1136/hrt.78.6.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Mateddu A, Cianchetti C, Marrosu MG, Clerk A, Cau M, et al. Dystrophin analysis using a panel of anti-dystrophin antibodies in Duchenne and Becker muscular dystrophy. J Neurol Neurosurg Psychiatry. 1993;56:26–31. doi: 10.1136/jnnp.56.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–40. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Yoshida K, Fukushima K, Ueda H, Urasawa N, Koyama J, et al. Follow-up of three patients with a large in-frame deletion of exons 45–55 in the Duchenne muscular dystrophy (DMD) gene. J Clin Neurosci. 2008;15:757–63. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Neri M, Torelli S, Brown S, Ugo I, Sabatelli P, Merlini L, et al. Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human. Neuromuscul Disord. 2007;17:913–8. doi: 10.1016/j.nmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Nicholson LV, Johnson MA, Bushby KM, Gardner-Medwin D, Curtis A, Ginjaar IB, et al. Integrated study of 100 patients with Xp21 linked muscular dystrophy using clinical, genetic, immunochemical, and histopathological data. Part 2. Correlations within individual patients. J Med Genet. 1993;30:737–44. doi: 10.1136/jmg.30.9.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa E. Our trails and trials in the subsarcolemmal cytoskeleton network and muscular dystrophy researches in the dystrophin era. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:798–821. doi: 10.2183/pjab.86.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76:219–26. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri-Rouxel F, Gentil C, Vassilopoulos S, Baas D, Mouisel E, Ferry A, et al. DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 2010;29:643–54. doi: 10.1038/emboj.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengpattrachai M, Ray PN, Hawkins CE, Berzen A, Banwell BL. Grandpa and I have dystrophinopathy?: approach to asymptomatic hyperCKemia. Pediatr Neurol. 2006;35:145–9. doi: 10.1016/j.pediatrneurol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Sanchez-Arjona MB, Rodriguez-Uranga JJ, Giles-Lima M, Fernandez-Garcia R, Chinchon-Lara I, Antinolo G, et al. Spanish family with myalgia and cramps syndrome. J Neurol Neurosurg Psychiatry. 2005;76:286–9. doi: 10.1136/jnnp.2004.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Chavoshan B, Harris SA, Iannaccone ST, Stull JT, Thomas GD, et al. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2000;97:13818–23. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GD, Sander M, Lau KS, Huang PL, Stull JT, Victor RG. Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc Natl Acad Sci USA. 1998;95:15090–5. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli S, Brown SC, Jimenez-Mallebrera C, Feng L, Muntoni F, Sewry CA. Absence of neuronal nitric oxide synthase (nNOS) as a pathological marker for the diagnosis of Becker muscular dystrophy with rod domain deletions. Neuropathol Appl Neurobiol. 2004;30:540–5. doi: 10.1111/j.1365-2990.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- Tselikas L, Rodrigues E, Jammal M, Tiev K, Chayet C, Josselin-Mahr L, et al. [Late onset Becker muscular dystrophy. A case report and literature review.] Rev Med Interne. 2010;32:181–6. doi: 10.1016/j.revmed.2010.10.353. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–86. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- van Vliet L, de Winter CL, van Deutekom JC, van Ommen GJ, Aartsma-Rus A. Assessment of the feasibility of exon 45-55 multiexon skipping for Duchenne muscular dystrophy. BMC Med Genet. 2008;9:105. doi: 10.1186/1471-2350-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MJ, Gait MJ, Yin H. RNA-targeted splice-correction therapy for neuromuscular disease. Brain. 2010;133(Pt 4):957–72. doi: 10.1093/brain/awq002. [DOI] [PubMed] [Google Scholar]
- Yokota T, Takeda S, Lu QL, Partridge TA, Nakamura A, Hoffman EP. A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Arch Neurol. 2009;66:32–8. doi: 10.1001/archneurol.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]