Abstract
Multiple sclerosis is characterized by inflammatory demyelination and irreversible axonal injury leading to permanent neurological disabilities. Diffusion tensor imaging demonstrates an improved capability over standard magnetic resonance imaging to differentiate axon from myelin pathologies. However, the increased cellularity and vasogenic oedema associated with inflammation cannot be detected or separated from axon/myelin injury by diffusion tensor imaging, limiting its clinical applications. A novel diffusion basis spectrum imaging, capable of characterizing water diffusion properties associated with axon/myelin injury and inflammation, was developed to quantitatively reveal white matter pathologies in central nervous system disorders. Tissue phantoms made of normal fixed mouse trigeminal nerves juxtaposed with and without gel were employed to demonstrate the feasibility of diffusion basis spectrum imaging to quantify baseline cellularity in the absence and presence of vasogenic oedema. Following the phantom studies, in vivo diffusion basis spectrum imaging and diffusion tensor imaging with immunohistochemistry validation were performed on the corpus callosum of cuprizone treated mice. Results demonstrate that in vivo diffusion basis spectrum imaging can effectively separate the confounding effects of increased cellularity and/or grey matter contamination, allowing successful detection of immunohistochemistry confirmed axonal injury and/or demyelination in middle and rostral corpus callosum that were missed by diffusion tensor imaging. In addition, diffusion basis spectrum imaging-derived cellularity strongly correlated with numbers of cell nuclei determined using immunohistochemistry. Our findings suggest that diffusion basis spectrum imaging has great potential to provide non-invasive biomarkers for neuroinflammation, axonal injury and demyelination coexisting in multiple sclerosis.
Keywords: magnetic resonance imaging, diffusion tensor imaging, multiple tensor model, white matter injury, inflammation
Introduction
Inflammation is an important pathological component of complicated CNS disorders such as multiple sclerosis. While the presence of gadolinium-enhancing lesions has been regarded as a surrogate marker of inflammation in multiple sclerosis (Grossman et al., 1988), this is probably an oversimplification given a recent MRI study on ultra-small particles of iron oxide that showed that cell infiltration occurs earlier and lasts longer than lesions defined by gadolinium-enhancement in patients with relapsing–remitting multiple sclerosis (Vellinga et al., 2008). In addition to the less than ideal response to inflammation, gadolinium-enhancement also does not reflect axon or myelin pathologies. An imaging modality capable of distinguishing and quantifying co-existing inflammation, axon injury and myelin damage is required to accurately assess multiple sclerosis progression and efficacy of disease-modifying interventions.
The directional diffusivities derived from diffusion tensor imaging (DTI) describe water movements parallel to (λ||, axial diffusivity) and perpendicular to (λ⊥, radial diffusivity) axon tracts. We have previously proposed and demonstrated that decreased λ|| is associated with axonal injury and dysfunction, and increased λ⊥ is associated with myelin injury in mouse models of white matter injury (Song et al., 2002, 2003, 2005). Unfortunately, the current DTI model does not address effects of inflammation-associated vasogenic oedema or increased cellularity. Vasogenic oedema, manifested as a non-restricted isotropic diffusion, has long been recognized to result in the increased apparent diffusion coefficient, and the underestimated white matter tract diffusion anisotropy (Kuroiwa et al., 1999; Pasternak et al., 2009; Naismith et al., 2010). In contrast, effects of increased cellularity on DTI-derived indices have not been adequately investigated in multiple sclerosis or other CNS white matter disorders. With increased cell content, we expect that restricted isotropic diffusion resulting from cells would lead to a decreased apparent diffusion coefficient and underestimated diffusion anisotropy in the white matter (Anderson et al., 2000). Thus, in addition to complications from crossing fibre complications (Wheeler-Kingshott and Cercignani, 2009), DTI of CNS white matter pathology is also significantly confounded by a spectrum of isotropic diffusion tensor components resulting from inflammation (Lodygensky et al., 2010), chronic tissue loss (Kim et al., 2007) and the partial volume effect from CSF or grey matter contamination (Karampinos et al., 2008). The diffusion properties derived using DTI lose specificity and sensitivity with increasing pathological and anatomical complexity.
Herein, diffusion basis spectrum imaging (DBSI) is proposed to address DTI limitations by resolving multiple-tensor water diffusion resulting from axon injury, demyelination and inflammation. Custom-designed realistic tissue phantoms made of fixed mouse trigeminal nerves (consisting of non-crossing fibres and Schwann cells) with and without gel (mimicking oedema, CSF contamination or tissue loss) were first employed to evaluate whether DBSI is capable of separating axon fibres from baseline cellularity and oedema. Crossing nerve phantoms were constructed also using trigeminal nerves to test the feasibility of DBSI to resolve crossing fibres in the presence of vasogenic oedema. After the proof-of-concept tests using phantoms, DBSI was applied to the cuprizone-treated mouse model of CNS white matter de- and remyelination (Harsan et al., 2008; Irvine and Blakemore, 2008; Wu et al., 2008). Axonal injury, inflammation, gliosis and demyelination have been reported to coexist at 4 weeks of continuous cuprizone feeding (Matsushima and Morell, 2001; Liu et al., 2010). Our previous DTI studies on this model failed to detect corpus callosum demyelination seen by immunohistochemistry at 3–4 weeks of cuprizone feeding (Song et al., 2005; Sun et al., 2006). In this study, we compared DBSI and DTI using a rostrocaudal analysis of axonal injury, demyelination and inflammation in the corpus callosum of 4-week cuprizone-treated mice followed by immunohistochemistry.
Materials and methods
Diffusion basis spectrum imaging
We propose a novel and flexible DBSI framework, as shown in Equation (1), considering diffusion-weighted MRI data as a linear combination of multiple anisotropic (crossing myelinated and unmyelinated axons of varied directions; the first term) and a spectrum of isotropic (cells, sub-cellular structure and oedematous water; the second term) diffusion tensors (Anderson, 2005).
 |
(1) |
In Equation (1), 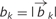 is the b-value of the k-th diffusion gradient (k = 1, 2, … , K); the direction of the k-th diffusion gradient is
is the b-value of the k-th diffusion gradient (k = 1, 2, … , K); the direction of the k-th diffusion gradient is  ; Sk is the measured diffusion-weighted signal corresponding to the k-th diffusion gradient; NAniso is the number of anisotropic tensors to be determined; Ψik is the unknown angle between the k-th diffusion gradient and the principal direction of the i-th anisotropic tensor; λ||_i and λ⊥_i are the unknown axial and radial diffusivities of the i-th anisotropic tensor under the assumption of cylindrically symmetric tensors; fi is the unknown signal intensity fraction for the i-th anisotropic tensor; and a and b are the low and high diffusivity limits for the isotropic diffusion spectrum f(D). To solve Equation (1), an algorithm for robust model selection and reliable non-linear optimization capable of handling a large number of free variables is needed (Alexander, 2005). The difficulty in model selection arises from the complexity and heterogeneity of CNS white matter pathology, where different pathology components may be present in neighbouring image voxels. In the implementation of DBSI, a two-step approach was employed to solve Equation (1).
; Sk is the measured diffusion-weighted signal corresponding to the k-th diffusion gradient; NAniso is the number of anisotropic tensors to be determined; Ψik is the unknown angle between the k-th diffusion gradient and the principal direction of the i-th anisotropic tensor; λ||_i and λ⊥_i are the unknown axial and radial diffusivities of the i-th anisotropic tensor under the assumption of cylindrically symmetric tensors; fi is the unknown signal intensity fraction for the i-th anisotropic tensor; and a and b are the low and high diffusivity limits for the isotropic diffusion spectrum f(D). To solve Equation (1), an algorithm for robust model selection and reliable non-linear optimization capable of handling a large number of free variables is needed (Alexander, 2005). The difficulty in model selection arises from the complexity and heterogeneity of CNS white matter pathology, where different pathology components may be present in neighbouring image voxels. In the implementation of DBSI, a two-step approach was employed to solve Equation (1).
In the first step, the number of anisotropic tensors (NAniso) and associated principal directions (Ψik) in a 3D space were determined in each image voxel. To achieve this, a recently reported diffusion basis decomposition approach was employed with two crucial modifications (Ramirez-Manzanares et al., 2007). The original diffusion basis decomposition approach modelled diffusion-weighted magnetic resonance signals as the linear combination of a discrete set of cylindrically symmetric anisotropic diffusion basis functions with fixed axial and radial diffusivities, uniformly distributed in a 3D space (Ramirez-Manzanares et al., 2007). The advantage of the discrete diffusion basis decomposition method is that it required only a small number of diffusion-weighted images (as few as 23) without ultra-high diffusion weighting (maximal b value 1250 s/mm2). The disadvantage is that diffusion basis with fixed diffusivities does not reflect different diffusivities known to exist between injured and non-injured axons. In addition, this original approach did not deal with the spurious fibre orientations and worsening angular resolution resulting from the isotropic partial volume effects of grey matter or CSF, or increased cellularity and vasogenic oedema associated with inflammation (Dell'acqua et al., 2010). While preserving the advantage of diffusion basis decomposition approach, DBSI fits the diffusion-weighted MRI signal in each image voxel to discrete diffusion basis sets with variable diffusivities and an isotropic diffusion tensor representing the averaged effect of the isotropic diffusion arising from contaminations of cells, grey matter, CSF and oedema, Equation (2):
| (2) |
where λ||, λ⊥ and diso are the unknown axial, radial and isotropic diffusivity of the diffusion basis tensor; ci (i = 1, 2, … , N) is the unknown signal intensity fraction for the i-th diffusion basis tensor; N = 95 is the number of diffusion basis tensors used in this study; cN+1 is the unknown signal intensity fraction of the averaged isotropic diffusion; θik is the known angle between the k-th diffusion gradient and the principal direction of the i-th diffusion basis. Determining optimal λ||, λ⊥, diso and ci (i = 1, 2, … , N + 1) values for the diffusion-weighted magnetic resonance signal Sk is a challenging non-linear optimization problem involving a multitude of variables. To simplify the solution of Equation (2), randomly selected initial values of λ||, λ⊥ and diso were used to start the optimization procedure. For a given set of λ||, λ⊥ and diso values, a non-linearly regularized least-squares analysis of data Sk according to Equation (2) was performed to obtain corresponding signal intensity fraction ci (Kadah et al., 2005) by incorporating a priori information that fibre crossings are sparse and that diffusion signal contributions should be non-negative as the penalty term to stabilize the solution (Ramirez-Manzanares et al., 2007). The fitting error for the initial set of λ||, λ⊥, diso and corresponding ci were analysed by generalized pattern search algorithm (Lewis and Torczon, 1999; Audet and Dennis, 2003) to determine the optimal selection of λ||, λ⊥ and diso recursively until the generalized pattern search algorithm converged at the global minimum. The number of anisotropic tensors (NAniso) and their principle directions (Ψik) in Equation (1) were derived based on the optimally solved ci (i = 1, 2, … , N + 1) values (Ramirez-Manzanares et al., 2007).
After solving Equation (2), NAniso and Ψik were estimated and applied to Equation (1). The second step was performed to further determine λ||_i, λ⊥_i and fi of each anisotropic tensor (i = 1, 2, … NAniso) along with the isotropic diffusion spectrum f(D) in Equation (1) using a regularized least-squares cost function. This is another non-linear optimization problem. Similar to the first step solving Equation (2), only the λ||_i and λ⊥_i values were selected as optimization variables to compute the corresponding fi and f(D) for enhanced computational efficiency. Generalized pattern search algorithm was employed to avoid converging to local minima. In brief, randomly selected initial values of λ||_i and λ⊥_i were used to start the optimization procedure. For a given set of λ||_i and λ⊥_i values, a non-linear least-square fit of the data to Equation (1) was simplified as the linear least-square fit of Equation (3) where f(D) is uniformly discretized at frequency L and associated intervals of v = (b-a)/(L-1) within the limit [a, b] = [a, a + ν, … , a + (L-2)v, b], to allow efficient computational implementation.
 |
(3) |
Equation (3) was solved using the regularized non-negative least-squares analysis. The a priori information of non-negative signal intensities (MacKay et al., 2006; Madler et al., 2008) and finite signal energy (Borgia et al., 1998, 2000) was incorporated to prevent the non-negative least-squares analysis from over-fitting the noisy data while retaining the numerical accuracy of the solution. The resulting regularized non-negative least-squares problem formulation is given as:
| (4) |
The first and second terms of Equation (4) deal with the fitting error (data fidelity) and regularization (penalty), respectively. Larger values of µ result in more regularization at the cost of increasing fitting error. At µ = 0, the fitting error achieves its minimal value at the risk of overfitting the measurement noise. In this study, µ was empirically selected as 0.01, resulting in robust fits in the presence of experimental noise. Similar regularized non-negative least-squares fitting problems have appeared previously in multiple-component nuclear magnetic resonance (NMR) relaxation data (MacKay et al., 1994). The non-negative least-squares fitting error was used by generalized pattern search to optimize the selection of λ||_i and λ⊥_i recursively until the generalized pattern search converges at the global minimum. Upon the convergence of generalized pattern search, Equation (1) is fully solved with λ||_i, λ⊥_i (i = 1, 2, … , NAniso), fj (j = 1, 2, … , L + NAniso) optimally determined. The two-step DBSI analysis was performed multiple times with different randomly chosen initial values to ensure that generalized pattern search generated consistent optimization results.
The anisotropic diffusion component represents the intra-axonal water molecules and those outside but adjacent to axon fibres (Alexander et al., 2010). We denote these anisotropic signal intensity fractions (fi) as fibre ratios. The diffusion of highly restricted water molecules inside cells or sub-cellular structures is isotropic appearing stationary (Alexander et al., 2010). This fraction of f(D) was denoted as cell ratio to assess cellularity. The rest of f(D) describes water molecules with less restriction than those associated with fibre and cell components. This tensor fraction was tentatively denoted as water ratio (Alexander et al., 2010). In the present study, DBSI employed 99 diffusion-encoding gradient directions selected from the uniformly spaced Cartesian grid used in diffusion spectrum imaging (Wedeen et al., 2005; Kuo et al., 2008). There were nine distinct b values distributed from 0 to 1000 s/mm2 for in vivo measurements of cuprizone mice and 3200 s/mm2 for phantoms.
Fixed trigeminal nerve phantoms
Trigeminal nerves from normal male C57BL/6 mice (The Jackson Laboratory) were isolated after fixation. The first group of eight nerves underwent baseline DBSI measurements. After the baseline data acquisition, five of the eight nerves were juxtaposed with gel (made of 2% agarose water gel) and scanned again. The second group of six trigeminal nerves was divided into three pairs, and each pair of nerves was aligned at crossing angles of 32°, 58° and 91°, respectively, juxtaposed with gel to model crossing fibres with oedema.
Diffusion-weighted spectroscopy of fixed trigeminal nerve phantoms
A diffusion-weighted spin-echo spectroscopy sequence was employed to acquire diffusion-weighted data with 99-direction diffusion encodings. A maximum diffusion weighting factor of b max = 3200 s/mm2 was employed. Other acquisition parameters were: 2 s repetition time, 49 ms echo time, 20 ms between gradient pulses (Δ), 8 ms gradient pulse duration (δ), four averages, 18 gauss/cm maximum diffusion strength. The total acquisition was 16 min and 40 s. A T2-weighted MRI with the same repetition time and echo time was acquired at the conclusion of each measurement. For the crossing fibre phantoms, a diffusion spectrum imaging data set employing 515 different diffusion encodings on a 3D grid (Kuo et al., 2008) with the maximum diffusion weighting factor of b = 6000 s/mm2 was also acquired in 1 h and 40 min.
Cuprizone treatment
Five male C57BL/6 mice (The Jackson Laboratory) were fed ad libitum a diet of 0.2% (w/w) cuprizone (Sigma-Aldrich) mixed into milled chow pellets (Harlan Teklad) beginning at 8 weeks of age. Cuprizone administration continued for 4 weeks, which has been shown to result in acute axonal injury, cell infiltration and demyelination (Hiremath et al., 1998). Five age-matched control C57BL/6 male mice were fed a normal diet.
In vivo magnetic resonance imaging
Diffusion-weighted MRI experiments were conducted on a 4.7T Varian DirectDrive spectrometer (Varian, Inc.) equipped with a 15-cm inner diameter, actively shielded gradient coil with maximum strength of 60 G/cm and 270 µs rise time (Magnex). After appropriate anaesthesia with a mixture of isoflurane/O2 (4.5% induction and 1–1.5% for maintenance during imaging), the mouse was immobilized in a custom magnetic resonance-compatible head holder, and placed on a pad of circulating warm water to maintain body temperature at 37°C. Body temperature was continuously monitored (SA instruments). An actively decoupled surface-coil receiver with a 1.7-cm outer diameter was placed above the mouse head. The mouse was then positioned inside of an actively decoupled volume transmit coil of 8-cm inner diameter.
Coronal images at the caudal, middle and rostral sites of the corpus callosum were selected on the sagittal scout (Fig. 1A). Although sagittal images would be ideal correlating with histology, coronal images were acquired due to hardware limitations. Corresponding immunohistochemistry was performed at regions of corpus callosum co-localized within the coronal image slice (Fig. 1B, yellow boxes). In each corpus callosum region, three rectangular areas were chosen for axon, myelin and cell nucleus counting (Fig. 1B, white boxes). The Stejskal–Tanner gradient pair was incorporated in a multi-echo spin-echo diffusion-weighted sequence to acquire image data. The additional two echoes were, acquired on the same k-space line as the first echo with negligible additional diffusion weighting, combined with the first echo to increase signal-to-noise ratio. The diffusion-weighting scheme was identical to that in the phantom study. Maximal diffusion weighting factor was b max = 1000 (s/mm2). Other acquisition parameters were: 1.5 s repetition time, 36 ms echo time, 20 ms Δ, 8 ms δ, 1 average, 0.75 mm slice thickness, 1.5 × 1.5 cm2 field-of-view and 128 × 128 data matrix. The total acquisition was 5 h and 20 min.
Figure 1.
Coronal images of the mouse brain were obtained from the sagittal scout (A). The width of the yellow rectangles represents the actual slice thickness of the image. The immunohistochemistry was performed on the sagittal slices for the ease of axon and myelin counting. The corresponding immunohistochemistry counting regions (white rectangles) are displayed with the location and thickness of one image voxel (yellow squares; B). In vivo fractional anisotropy maps of rostral (C), middle (D) and caudal (E) corpus callosum (from a control mouse; colour-coded full view) reveal the structure of white matter tracts. The region of interest for DBSI analysis is marked in yellow rectangles shown in the expanded view for both control and cuprizone-fed mice.
Immunohistochemistry
Trigeminal nerves were embedded in paraffin and transverse 5 -µm thick slices prepared for histology following diffusion spectroscopic acquisition. After deparaffin and rehydration, antigen was retrieved with 1 mM EDTA at 95–100°C. For the cuprizone studies, immediately following in vivo MRI acquisition, mice were perfused with 4% paraformaldehyde in phosphate-buffered saline. Brains were excised and post-fixed in 4% paraformaldehyde/phosphate-buffered saline for 24 h, then transferred to 30% sucrose/phosphate-buffered saline at 4°C for cryoprotection. Cryoprotected tissues were embedded in optimal cutting temperature medium (West Chester), and frozen using dry ice. The frozen tissues were cut parasagittally at 10 -µm thick for immunohistochemistry.
All sections were blocked in 1% bovine serum albumin and 0.5% Triton-X100 for 1 h at room temperature to prevent non-specific binding and to increase permeability of antibodies. Sections were then incubated with monoclonal anti-phosphorylated neurofilament H antibody (SMI-31; 1:3000; Sternberger Monoclonals) at 4°C overnight. After rinsing, goat anti-mouse IgG conjugated with Alexa 488 (1:1500; Invitrogen) was applied to visualize immunoreactivity. Sections were covered in Vectashield Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc.).
Histological slides were examined with a Nikon Eclipse 80i fluorescence microscope equipped with a ×60 objective, and images were captured with a black-and-white CCD camera using MetaMorph software (Universal Imaging Corporation). Axons stained with SMI-31 were counted using the software CellC (http://www.cs.tut.fi/sgn/csb/cellc/) (Selinummi et al., 2005). Intact myelin sheath labelled with anti-myelin basic protein (1:500; Invitrogen) was counted with the CellC software customized by employing an adaptive threshold algorithm to remove the effects of non-uniform foreground illumination.
Data analysis
Both tissue phantom spectroscopy and in vivo corpus callosum image data were analysed using a DBSI analysis package developed in-house with Matlab® (MathWorks). Conventional DTI analysis was also performed on the 99-grid point diffusion-weighted spectroscopy and imaging data. All data were expressed as mean ± standard deviation (SD).
For the trigeminal nerve phantoms without gel, the magnetic resonance signal intensity ratio of each diffusion tensor component was estimated and compared with axon and nucleus counts determined by immunohistochemistry. For the phantoms with gel, the signal intensity of fibre and juxtaposed gel were measured from T2-weighted images using ImageJ. The DBSI-derived fibre and gel intensity fractions, i.e. fibre and water ratios, were then compared with the signal intensity ratios measured from the T2-weighted images (defined as fibre or gel intensity divided by the total signal intensity) The diffusion parameters λ|| and λ⊥ extracted from phantoms with and without gel were assessed using Bland–Altman limit of agreement analysis (Bland and Altman, 1986).
For in vivo corpus callosum image data, the region of interest was manually selected on the colour-coded fractional anisotropy maps derived using the conventional DTI (Fig. 1). The DBSI model analysis was performed on the selected region of interest. Averaged parameters of voxels within the region of interest analysed using DTI were compared with those analysed using the DBSI model. Linear repeated measures models were used to estimate the effect of treatment (control/cuprizone), measurement location (caudal/middle/rostral) and the differential effect of treatment at each location. Treatment and location were treated as fixed effects, and linear contrasts were used to test hypotheses. Adjusted means were estimated with 95% confidence intervals. Spearman's rank correlation was used to test for presence of monotone increasing or decreasing association between the immunohistochemistry measures and the DTI or the DBSI results.
Results
Diffusion properties of fixed trigeminal nerves
For the first group of eight trigeminal nerves, the DBSI model analysis identified one anisotropic fibre component representing 71.9 ± 3.6% of the non-diffusion-weighted signal intensity, a restricted isotropic cell component representing 11.6 ± 1.3% of the non-diffusion-weighted signal intensity, and an isotropic water component representing 16.6 ± 3.2% of the non-diffusion-weighted signal intensity. The SMI-31-positive axon counts and the DAPI-positive nucleus were 25 434 ± 8505 per mm2 and 4109 ± 629 per mm2, respectively. The SMI-31-to-DAPI count ratio was 6.20 ± 1.81, mirrored the DBSI-derived fibre-to-cell signal intensity ratio of 6.26 ± 0.93 (Fig. 2).
Figure 2.
The representative result of DAPI (A) and SMI-31 (B) staining from a fixed mouse trigeminal nerve. Both nucleus and axon counts were performed using CellC program (C). The fraction of MRI-derived cell and fibre component was determined by DBSI (D). The ratio of nucleus and axon counts by immunohistochemistry (IHC) (6.20 ± 1.81) was comparable to the ratio of DBSI-derived cell and fibre percentage (6.26 ± 0.93; n = 8). DBSI = diffusion basis spectrum imaging.
Trigeminal nerve diffusion parameters determined in the presence of gel
For the trigeminal nerve phantoms with gel (mimicking vasogenic oedema, CSF contamination, or tissue loss) (Fig. 3A), DTI-derived FA was smaller than that of the same tissue without gel while directional diffusivities (λ|| and λ⊥) were larger. In contrast, the DBSI-derived fractional anisotropy, λ|| and λ⊥ were comparable for phantoms with and without gel (Fig. 3A) as demonstrated by Bland–Altman limit of agreement analysis (Bland and Altman, 1986) in Fig. 3B and C. In addition, the DBSI model-derived water ratios strongly agreed with that determined by T2-weighted intensity fractions (inset of Fig. 3D).
Figure 3.
DTI failed to correctly estimate the diffusion parameters (λ||, open circles; λ⊥, open squares; fractional anisotropy, open triangles) due to the partial volume effect of the added gel (the more gel added the more significant deviation). DBSI correctly estimated diffusion parameters (λ||, solid circles; λ⊥, solid squares; fractional anisotropy, solid triangles) by separating and quantifying the gel (A). Comparable λ|| (B) and λ⊥ (C) derived from trigeminal nerves with and without gel were further confirmed by Bland–Altman analysis. Representative DBSI-derived isotropic diffusion spectra for fibre only (grey dashed line) and for fibre with gel phantom (black solid line) are compared (D). The diffusivity component near 0 µm2/ms was assigned to the cell. The higher diffusivity component in fibre only (grey dashed line) was assigned to the inter-axonal and extracellular water. The higher diffusivity component in the fibre with gel (black solid line) was assigned to inter-axonal, extracellular and free diffusion gel water. DBSI-derived gel fractions agreed with that determined by T2-weighted intensity fractions (D inset). DTI = diffusion tensor imaging; DBSI = diffusion basis spectrum imaging; FA = fractional anisotropy.
Spectral analysis of isotropic diffusion tensor compartments
Representative isotropic diffusion spectra of trigeminal nerves with (black solid line) and without (grey dashed line) gel exhibited two distinct distributions of isotropic diffusion (Fig. 3D). The total T2-weighted signal intensity of the phantom with gel doubled that of the phantom without gel. Spectral components with isotropic diffusivity close to 0 µm2/ms were assigned to cells (Fig. 3D). The second spectral component reflects isotropic diffusion from the surrounding environments. The DBSI-estimated cell ratio in the phantom with gel was ∼6%, compared to ∼13% for the phantom without gel, correctly reflecting the dilution factor of the added gel.
Resolution of crossing fibres
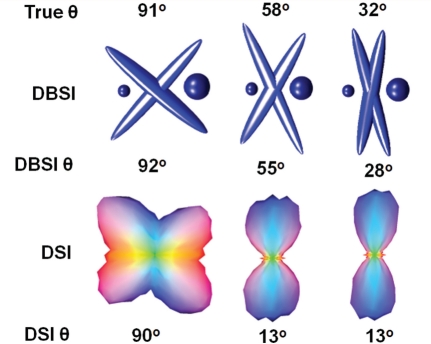
DBSI accurately estimated crossing fibre angles, comparing favourably with those derived using orientation density functions by diffusion spectrum imaging (Fig. 4) (Wedeen et al., 2005). DBSI-determined mean fibre λ|| = 1.14 ± 0.06 µm2/ms, and λ┴ = 0.12 ± 0.02 µm2/ms agreed with the single fibre without gel λ|| = 1.07 ± 0.05 µm2/ms, λ┴ = 0.14 ± 0.02 µm2/ms (P > 0.05). For 91°, 58° and 32° phantoms, DBSI-derived gel percentages were 15, 14 and 50%, in close agreement with T2-weighted MRI determined 18, 13 and 45%. Diffusion spectrum imaging failed to resolve crossing fibres in the presence of gel at angles of 58° and 32°, worse than the previously reported angular resolutions of diffusion spectrum imaging without gel contamination (Yeh et al., 2010, 2011).
Figure 4.
Resolution of crossing fibres by diffusion spectrum imaging and DBSI. Although, the goal of this study was to demonstrate the confounding effect of increased cellularity in white matter lesions, we would also like to demonstrate the capability of DBSI to resolve crossing fibres deriving crossing angles, fraction of fibres and diffusion parameters of individual fibres. Gels were added to mimic oedema/CSF contamination/tissue loss. For 91°, 58° and 32° phantoms, DBSI determined angles were 92°, 55° and 28°; diffusion spectrum imaging-derived angles were 90°, 13° and 13°. DBSI also correctly recovered the directional diffusivity of the crossing fibres removing the effect of gel (λ|| = 1.14 ± 0.06 µm2/ms, and λ┴ = 0.12 ± 0.02 µm2/ms). The fraction of the extent of gel was also estimated by DBSI.
Rostrocaudal analysis of corpus callosum in cuprizone-treated mice
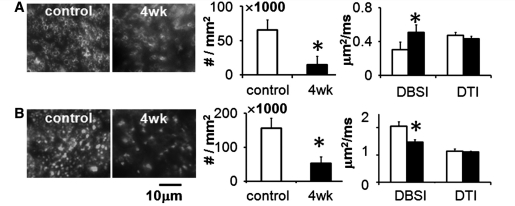
Significant axonal injury and demyelination evidenced by the significant reduction of myelin basic protein and SMI-31-positive axon counts were consistently reflected by both DTI and DBSI in the caudal corpus callosum (Table 1). Similar to caudal corpus callosum, immunohistochemistry showed significant axonal injury and demyelination in middle corpus callosum (Fig. 5). Accordingly, a 70% increase in the DBSI-derived λ⊥ was observed in the cuprizone-treated mice (P < 0.0001) compared with the control, suggesting demyelination, and 28% decrease in DBSI-derived λ|| (P < 0.0001), suggesting axonal injury. However, DTI-derived λ|| and λ⊥ did not show a difference between the cuprizone-treated and the control mice. Although rostral corpus callosum is less severely injured in this model, demyelination was detected by both immunohistochemistry and DBSI, while DTI failed to detect significant increase in λ⊥ between the control and the cuprizone-treated group (Table 1).
Table 1.
DTI, DBSI immunohistochemistry results
| Caudal corpus callosum |
Middle corpus callosum |
Rostral corpus callosum |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | 4 week | Test | Control | 4 week | Test | Control | 4 week | Test | |
| MBP count | 63 657 ± 11 543 | 6515 ± 7546 | *** | 65 609 ± 14 804 | 14 830 ± 12 407 | *** | 58 627 ± 9971 | 34 012 ± 14 257 | *** |
| DTI λ┴ | 0.32 ± 0.03 | 0.45 ± 0.04 | *** | 0.47 ± 0.04 | 0.43 ± 0.03 | * | 0.39 ± 0.06 | 0.43 ± 0.04 | * |
| DBSI λ┴ | 0.31 ± 0.05 | 0.53 ± 0.1 | *** | 0.31 ± 0.05 | 0.51 ± 0.09 | *** | 0.30 ± 0.02 | 0.42 ± 0.04 | ** |
| SMI31 count | 146 607 ± 21 786 | 55 098 ± 22 805 | *** | 155 832 ± 29 934 | 53 685 ± 18 712 | *** | 135 333 ± 44 379 | 116 386 ± 22 379 | * |
| DTI λ|| | 1.60 ± 0.08 | 0.90 ± 0.12 | *** | 1.14 ± 0.09 | 1.11 ± 0.03 | * | 1.27 ± 0.09 | 1.15 ± 0.11 | * |
| DBSI λ|| | 1.69 ± 0.05 | 1.13 ± 0.19 | *** | 2.05 ± 0.17 | 1.47 ± 0.09 | *** | 1.67 ± 0.23 | 1.50 ± 0.18 | * |
| DAPI count | 3473 ± 582 | 16 543 ± 3523 | *** | 3333 ± 622 | 13 852 ± 3868 | *** | 3122 ± 542 | 6986 ± 3011 | ** |
| DBSI cell (%) | 3 ± 3 | 19 ± 5 | *** | 51 ± 9 | 16 ± 3 | ** | 20 ± 16 | 18 ± 14 | * |
Counts in numbers per square millimetre; directional diffusivity in µm2/ms.
*P > 0.05; **P < 10−2; ***P < 10−4.
MBP = myelin basic protein.
Figure 5.
The middle corpus callosum after 4 weeks of cuprizone treatment was examined using in vivo diffusion MRI (analysed using both DTI and DBSI model) followed by post-mortem immunohistochemistry using (A) the antibody against myelin basic protein, and (B) the antibody against phosphorylated neurofilament (SMI-31), to quantify the extent of demyelination (A), and axonal injury (B). All data were derived from the same region of interest from the middle corpus callosum as marked in Fig. 1B and D. Due to the partial volume effect originating from the surrounding grey matter, DTI failed to depict the extent of demyelination or axonal injury in this region, overestimating λ⊥ while underestimating λ|| of the control corpus callosum. The myelinated axon counts significantly decreased by 77% (65 609 ± 14 804 versus 14 830 ± 12 407 per mm2) in the cuprizone-treated mice compared with that of the control. The λ⊥ increased by 70% measured using DBSI (0.31 ± 0.05 versus 0.51 ± 0.09 µm2/ms) in the cuprizone-treated mice compared with that of the control. However, DTI determined λ⊥ values were not different between the cuprizone-treated (0.43 ± 0.03 µm2/ms) and the control mice (0.47 ± 0.04 µm2/ms). The SMI-31-positive axon counts significantly decreased in the cuprizone-treated corpus callosum (155 832 ± 29 934 versus 53 685 ± 18 712 per mm2) compared with that of the control. The DBSI derived λ|| significantly decreased by 28% (2.05 ± 0.17 versus 1.47 ± 0.09 µm2/ms) while no difference was observed in that derived using DTI (1.14 ± 0.09 versus 1.11 ± 0.03 µm2/ms) in the cuprizone-treated mice compared with that of the control.
In the control corpus callosum, the DAPI nucleus count from the caudal, middle and rostral corpus callosum regions suggested no rostrocaudal inhomogeneity (Table 1). In the 4-week cuprizone-treated mice, rostrocaudal differences in nucleus counts were seen with decreasing inflammation from caudal to rostral corpus callosum (Table 1). Nucleus counts decreased by ∼19% from caudal to middle corpus callosum (P = 0.032), and by ∼45% (P < 0.0001) from middle to rostral corpus callosum. This was reflected by DBSI, where estimated cell ratio decreased by ∼16% from caudal to middle corpus callosum, and by ∼35% from middle to rostral corpus callosum.
Correlating diffusion basis spectrum imaging and diffusion tensor imaging with immunohistochemistry
To compare the DBSI and DTI measurements with the immunohistochemistry findings, data from the caudal, middle and rostral corpus callosum regions were pooled and the correlation among all parameters was examined. Both DBSI (r = 0.76, P < 0.0001) and DTI (r = 0.62, P = 0.0003) derived axial diffusivities correlated with the SMI-31-positive axon counts. The DBSI-derived radial diffusivities correlated far better with the myelinated axon counts (r = −0.76, P < 0.0001) than DTI-derived radial diffusivity correlated with the myelinated axon counts (r = −0.38, P = 0.036).
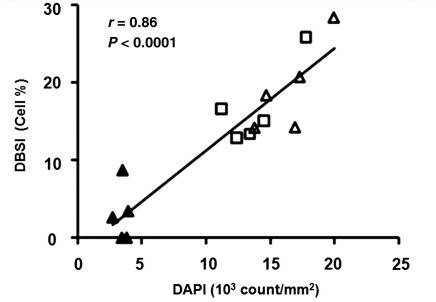
In the control middle and rostral corpus callosum, and 4-week cuprizone-treated rostral corpus callosum (Fig. 1A and B), DBSI estimated cell ratio reflected not only cells in corpus callosum quantified by DAPI counts, but also nuclei from neuronal cells of grey matter within the same imaging voxel. In contrast, the grey matter involvement in control caudal corpus callosum, 4-week cuprizone-treated caudal and middle corpus callosum was negligible. Thus, only data from control caudal, cuprizone-treated caudal and middle corpus callosum were employed to correlate DBSI-estimated cell ratio with DAPI counts of corpus callosum (Fig. 6; r = 0.86, P < 0.0001).
Figure 6.
Cell densities quantified based on DAPI-positive nucleus counts linearly correlating with the restricted diffusion (assigned to cells) ratio derived by DBSI. Data from both the control group (caudal corpus callosum: filled triangle) and the 4-week cuprizone-treated group (caudal corpus callosum: open triangle; middle corpus callosum: open square) were analysed by linear regression. The significant correlation (r = 0.86, P < 0.0001) supported that DBSI-derived cell ratio can potentially be used as a novel non-invasive index of inflammation or gliosis. DBSI = diffusion basis spectrum imaging.
Discussion
The effect of typical CNS white matter pathology on conventional DTI analysis (without considering fibre crossing) can be conceptually visualized by considering the signal contributions from tissue and/or pathology components within an image voxel or region of interest (Fig. 7). Conventional DTI performs reasonably well at identifying axonal injury and demyelination when magnetic resonance signals from coherent axon bundles dominate in an image voxel (Fig. 7A). This has largely been the case in previously reported CNS injury rodent models (Budde et al., 2007, 2009; Xie et al., 2010). However, during inflammation with increased numbers of cells coexisting with axon and myelin injury (Fig. 7B), DTI may underestimate the extent of demyelination (less increase in λ⊥) while exaggerating the extent of axonal injury (greater decrease in λ||). This is due to the increased cellularity, with its highly restricted diffusion (small circular ellipsoids) (Song et al., 2005; Sun et al., 2006; Xie et al., 2010). In chronic disease, axonal loss is common (Fig. 7C), and the increased isotropic diffusion component (the large circular ellipsoid) biases DTI measurements, exaggerating both λ|| and λ⊥ (Kim et al., 2007). The diffusion MRI measurement can be further complicated by coexistent inflammation, axonal loss, axonal injury and demyelination (Fig. 7D), where DTI can no longer discern the different underlying pathologies. Due to the inherent low sensitivity of MRI, these partial volume effects will always adversely affect the accuracy of DTI measurements.
Figure 7.
Different white matter pathologies associated multiple tensor representation and DTI simplification. Grey ellipsoid represents the diffusion tensor profile for normal myelinated white matter with λ|| > λ⊥. Black drawings represent the diffusion profiles for multiple tensor representation or DTI simplification: (A) co-existing axon and myelin injury of coherent pure myelinated axons; (B) axon and myelin injury with cell infiltration; (C) axon and myelin injury with axonal loss; and (D) axon and myelin injury, cell infiltration or proliferation and axonal loss.
Cuprizone intoxication is a widely used mouse model for evaluating the mechanism of CNS white matter de- and remyelination (Torkildsen et al., 2008). Its pathology includes significant inflammation and gliosis (Hiremath et al., 1998; McMahon et al., 2002). Thus, it represents a more complicated pathology than de- or remyelination alone. To date, no MRI method capable of non-invasively detecting the extent of cell content in CNS tissues has been reported. The present study revealed that cell ratios estimated by in vivo DBSI correlated well with DAPI positive nucleus counts in the cuprizone-treated mouse (Fig. 6). Thus, this new method may be able to quantify increased cellularity in white matter relating to the inflammation commonly seen in CNS diseases such as multiple sclerosis, for which the cuprizone-treated mouse serves as a model.
Both DBSI and DTI consistently detected cuprizone-induced axonal injury and demyelination in caudal corpus callosum where injury was most severe (Fig. 1A; Table 1) (Wu et al., 2008; Xie et al., 2010). The situation in the middle corpus callosum was more complicated due to the significant grey matter contamination (Fig. 1A) arising from the narrower corpus callosum at this location (Fig. 1D) in the control group. In this case, the partial volume effect contributed from grey matter resulted in DTI underestimating λ|| by 45% and overestimating λ⊥ by 50% compared with the more accurate DBSI measurements in the control middle corpus callosum. After 4-week cuprizone treatment, grey matter contamination of the middle corpus callosum was significantly reduced due to tissue swelling from the increased cellular content (Xie et al., 2010). DTI estimated λ⊥ and λ|| of the fibre component were 16 and 25% lower, respectively, compared to those measured by DBSI (Fig. 5) due to increased cellularity. Failed detection of demyelination and axonal injury by DTI at the middle corpus callosum was mainly due to the 50% overestimated control λ⊥ and the 45% underestimated control λ|| by DTI, and partially due to the underestimation of λ||, λ⊥ in the treated group. Similar to middle corpus callosum, in rostral corpus callosum the partial volume effect from grey matter in controls again played an important role masking the demyelination missed by DTI (Table 1).
Non-parametric diffusion methods such as diffusion spectrum imaging (Wedeen et al., 2005), Q-ball imaging (Tuch, 2004), persistent angular structure MRI (Jansons and Alexander, 2003) and numerous others (Johansen-Berg and Behrens, 2009) have been proposed in the last decade to resolve non-Gaussian diffusion resulting from multiple crossing fibres for accurate fibre tracking. The isotropic diffusion components resulting from cells and unrestricted tissue water observed in many types of pathology as well as in normal tissues have been infrequently addressed and were recently recognized as a factor affecting the accuracy to resolve crossing fibres and conduct fibre tracking (Dell'acqua et al., 2010; Yeh et al., 2011). The similar effect from isotropic diffusion components was demonstrated in our comparison of diffusion spectrum imaging versus DBSI using the crossing fibre phantom where the inclusion of gel (mimicking vasogenic oedema or tissue loss) significantly reduced the ability of diffusion spectrum imaging to resolving the crossing angle (Fig. 4) in addition to its innate inability to estimate diffusion parameters or the content of the fibres.
In addition to non-parametric methods, parametric diffusion methods provide an alternative to resolve and quantify crossing fibres. Spherical deconvolution (Tournier et al., 2004) approximates measured diffusion-weighted signals as the linear combination of a set of predetermined ‘ideal single fibre’ distributed in a 3D space. Spherical deconvolution-derived orientation distribution function reveals the underlying structure of crossing fibres. Similarly, the dendrite density was recently estimated by modelling axons/dendrites as a distribution of anisotropic components with the addition of a single isotropic component to represent the overall background isotropic diffusion (Jespersen et al., 2007). Several other parametric models with predetermined tissue components have also been used to describe crossing fibres and the CSF partial volume effect (Pasternak et al., 2009; Caan et al., 2010). CHARMED was proposed to characterize the tissue structure based on diffusion processes as hindered and restricted water diffusion in the brain (Assaf et al., 2004). AxCaliber was developed based on the CHARMED model (Assaf et al., 2008) to estimate the distribution of the axonal diameters. Glial cells have previously been modelled as a highly restricted isotropic component in a three-component NMR analytical model (Stanisz et al., 1997) without including crossing-fibres. Most recently, a four-tensor model was proposed to include the restricted isotropic diffusion resulting from cell and the extracellular water components (Alexander et al., 2010). The authors quantified the orientationally invariant indices of axonal diameter and density without solving the cell component or fibre diffusivities on coherent white matter.
DBSI adopts various features from the aforementioned parametric methods modelling diffusion magnetic resonance signal as a linear combination of anisotropic and isotropic diffusion components. DBSI is philosophically similar to CHARMED (Assaf et al., 2004), modelling diffusion magnetic resonance signal based on diffusion processes without assumption of tissue structures. However, DBSI and CHARMED are very different in that CHARMED assumes that intracellular water, in glial and other cells, exchanges freely with hindered diffusion compartment under commonly used parameters, thus prohibiting CHARMED to distinguish restricted diffusion inside cells from the hindered compartment. In contrast, DBSI assumes no exchange between components, as do most parametric models, enabling the assessment of restricted isotropic diffusion inside cells. Inclusion of isotropic diffusion in addition to anisotropic diffusion of fibres in DBSI is similar to Jespersen et al. (2007), Caan et al. (2010) and Pasternak et al. (2009). The incorporation by DBSI of the concept of restricted isotropic diffusion was primarily motivated by the work of Stanisz et al. (1997), similar to Alexander et al. (2010). The uniqueness of DBSI compared with the aforementioned existing methods includes (i) DBSI resolves crossing fibres and quantifies directional diffusivities and fractions of resolved fibres; and (ii) DBSI incorporates a spectrum of isotropic diffusion components allowing quantification of diffusivities, and fractions of components of the isotropic diffusion spectrum, allowing the estimation of cellularity and oedema contents.
Diffusion-weighted MRI signals reflect an ensemble-averaged diffusion effect arising from various microscopic structures (Novikov and Kiselev, 2010). Accurately deciphering the underlying microscopic structures based on the diffusion-weighted MRI signal is challenging if not impossible (Mulkern et al., 2009). The goal of DBSI is to determine the fractional distribution and diffusivity of water molecules associated with axonal fibres (including both intra- and extra-axonal water appearing anisotropic), and those inside cells and extracellular spaces (appearing isotropic) based on the overall diffusion patterns without attempting to definitively define the underlying microscopic structures. Our results on both phantoms and in vivo cuprizone-treated mouse corpus callosum suggest that DBSI is capable of ‘imaging’ CNS inflammation through quantifying the extent of vasogenic oedema and increased cellularity, thus hypothesized to be an effective marker of inflammation. Most importantly, DBSI improves the quantification of MRI metrics, the axial and radial diffusivity, that distinguish and reflect axonal versus myelin injury. We believe the current testing and validation of DBSI portends a significant and substantial advance in the tools available to clinicians for the management of multiple sclerosis.
Predicting the highly variable course of multiple sclerosis and optimizing treatment are extremely difficult (Confavreux and Vukusic, 2006; Leray et al., 2010; Scalfari et al., 2010) because clinical symptoms do not accurately reflect the progression of underlying pathologies. Axonal loss is the pathologic correlate of irreversible neurological impairment, which can occur early in multiple sclerosis (Trapp et al., 1999; Bjartmar and Trapp, 2003; DeLuca et al., 2004). Although an accurate assessment of the underlying axonal pathologies may or may not correctly reflect clinical symptoms during the early phase of the disease, it would likely predict the long-term patient disability. Current multiple sclerosis treatments follow a standard dosing regimen, with limited opportunity to adjust for individual patient responses. By quantitatively distinguishing and tracking inflammation, and axon and myelin injury, DBSI will provide the opportunity for efficacy assessment of disease-modifying interventions and allow treatment planning to reflect individual patient response. For example, patients may be imaged using DBSI to assess the extent of inflammation and axonal pathologies at the onset of multiple sclerosis to correctly dose anti-inflammatory drugs and stratify axonal preservation treatment.
Funding
This study was supported in part by National Institute of Health R01-NS047592 and R01-NS054194 (to S.-K.S.), and P01-NS059560 (to A.H.C.).
Acknowledgements
The authors would like to thank Chia-Wen Chiang and Denise Dorsey for the help in histological analysis, Dr Wen-Yih Tseng for his help on DSI analysis, and Joseph J.H. Ackerman, Joel Garbow and Junyi Ying for the insightful discussions during the writing of the manuscript.
Glossary
Abbreviations
- DBSI
diffusion basis spectrum imaging
- DTI
diffusion tensor imaging
References
- Alexander DC. Multiple-fiber reconstruction algorithms for diffusion MRI. Ann N Y Acad Sci. 2005;1064:113–33. doi: 10.1196/annals.1340.018. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJ, et al. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage. 2010;52:1374–89. doi: 10.1016/j.neuroimage.2010.05.043. [DOI] [PubMed] [Google Scholar]
- Anderson AW. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magn Reson Med. 2005;54:1194–206. doi: 10.1002/mrm.20667. [DOI] [PubMed] [Google Scholar]
- Anderson AW, Xie J, Pizzonia J, Bronen RA, Spencer DD, Gore JC. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn Reson Imaging. 2000;18:689–95. doi: 10.1016/s0730-725x(00)00147-8. [DOI] [PubMed] [Google Scholar]
- Audet C, Dennis JE. Analysis of generalized pattern searches. Siam J Optimization. 2003;13:889–903. [Google Scholar]
- Bjartmar C, Trapp BD. Axonal degeneration and progressive neurologic disability in multiple sclerosis. Neurotox Res. 2003;5:157–64. doi: 10.1007/BF03033380. [DOI] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- Borgia GC, Brown RJ, Fantazzini P. Uniform-penalty inversion of multiexponential decay data. J Magn Reson. 1998;132:65–77. doi: 10.1006/jmre.1998.1387. [DOI] [PubMed] [Google Scholar]
- Borgia GC, Brown RJ, Fantazzini P. Uniform-penalty inversion of multiexponential decay data. II. Data spacing, T(2) data, systemic data errors, and diagnostics. J Magn Reson. 2000;147:273–85. doi: 10.1006/jmre.2000.2197. [DOI] [PubMed] [Google Scholar]
- Budde MD, Kim JH, Liang HF, Schmidt RE, Russell JH, Cross AH, et al. Toward accurate diagnosis of white matter pathology using diffusion tensor imaging. Magn Reson Med. 2007;57:688–95. doi: 10.1002/mrm.21200. [DOI] [PubMed] [Google Scholar]
- Budde MD, Xie M, Cross AH, Song SK. Axial diffusivity is the primary correlate of axonal injury in the experimental autoimmune encephalomyelitis spinal cord: a quantitative pixelwise analysis. J Neurosci. 2009;29:2805–13. doi: 10.1523/JNEUROSCI.4605-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caan MW, Khedoe HG, Poot DH, den Dekker AJ, Olabarriaga SD, Grimbergen KA, et al. Estimation of diffusion properties in crossing fiber bundles. IEEE Trans Med Imaging. 2010;29:1504–15. doi: 10.1109/TMI.2010.2049577. [DOI] [PubMed] [Google Scholar]
- Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129(Pt 3):606–16. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- Dell'acqua F, Scifo P, Rizzo G, Catani M, Simmons A, Scotti G, et al. A modified damped Richardson-Lucy algorithm to reduce isotropic background effects in spherical deconvolution. Neuroimage. 2010;49:1446–58. doi: 10.1016/j.neuroimage.2009.09.033. [DOI] [PubMed] [Google Scholar]
- DeLuca GC, Ebers GC, Esiri MM. Axonal loss in multiple sclerosis: a pathological survey of the corticospinal and sensory tracts. Brain. 2004;127(Pt 5):1009–18. doi: 10.1093/brain/awh118. [DOI] [PubMed] [Google Scholar]
- Grossman RI, Braffman BH, Brorson JR, Goldberg HI, Silberberg DH, Gonzalez-Scarano F. Multiple sclerosis: serial study of gadolinium-enhanced MR imaging. Radiology. 1988;169:117–22. doi: 10.1148/radiology.169.1.3420246. [DOI] [PubMed] [Google Scholar]
- Harsan LA, Steibel J, Zaremba A, Agin A, Sapin R, Poulet P, et al. Recovery from chronic demyelination by thyroid hormone therapy: myelinogenesis induction and assessment by diffusion tensor magnetic resonance imaging. J Neurosci. 2008;28:14189–201. doi: 10.1523/JNEUROSCI.4453-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath MM, Saito Y, Knapp GW, Ting JP, Suzuki K, Matsushima GK. Microglial/macrophage accumulation during cuprizone-induced demyelination in C57BL/6 mice. J Neuroimmunol. 1998;92:38–49. doi: 10.1016/s0165-5728(98)00168-4. [DOI] [PubMed] [Google Scholar]
- Irvine KA, Blakemore WF. Remyelination protects axons from demyelination-associated axon degeneration. Brain. 2008;131(Pt 6):1464–77. doi: 10.1093/brain/awn080. [DOI] [PubMed] [Google Scholar]
- Kadah YM, Ma X, LaConte S, Yassine I, Hu X. Robust multi-component modeling of diffusion tensor magnetic resonance imaging data. In: Amini AA, editor. Proceedings of SPIE. SPIE, Bellingham, WA: 2005. pp. 148–59. [Google Scholar]
- Karampinos DC, Van AT, Olivero WC, Georgiadis JG, Sutton BP. High resolution reduced-FOV diffusion tensor imaging of the human pons with multi-shot variable density spiral at 3T. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:5761–4. doi: 10.1109/IEMBS.2008.4650523. [DOI] [PubMed] [Google Scholar]
- Kim JH, Loy DN, Liang HF, Trinkaus K, Schmidt RE, Song SK. Noninvasive diffusion tensor imaging of evolving white matter pathology in a mouse model of acute spinal cord injury. Magn Reson Med. 2007;58:253–60. doi: 10.1002/mrm.21316. [DOI] [PubMed] [Google Scholar]
- Kuo LW, Chen JH, Wedeen VJ, Tseng WY. Optimization of diffusion spectrum imaging and q-ball imaging on clinical MRI system. Neuroimage. 2008;41:7–18. doi: 10.1016/j.neuroimage.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Nagaoka T, Ueki M, Yamada I, Miyasaka N, Akimoto H, et al. Correlations between the apparent diffusion coefficient, water content, and ultrastructure after induction of vasogenic brain edema in cats. J Neurosurg. 1999;90:499–503. doi: 10.3171/jns.1999.90.3.0499. [DOI] [PubMed] [Google Scholar]
- Leray E, Yaouanq J, Le Page E, Coustans M, Laplaud D, Oger J, et al. Evidence for a two-stage disability progression in multiple sclerosis. Brain. 2010;133(Pt 7):1900–13. doi: 10.1093/brain/awq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RM, Torczon V. Pattern search algorithms for bound constrained minimization. Siam J Optimization. 1999;9:1082–99. [Google Scholar]
- Liu L, Belkadi A, Darnall L, Hu T, Drescher C, Cotleur AC, et al. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: relevance to multiple sclerosis. Nat Neurosci. 2010;13:319–26. doi: 10.1038/nn.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygensky GA, West T, Stump M, Holtzman DM, Inder TE, Neil JJ. In vivo MRI analysis of an inflammatory injury in the developing brain. Brain Behav Immun. 2010;24:759–67. doi: 10.1016/j.bbi.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay A, Laule C, Vavasour I, Bjarnason T, Kolind S, Madler B. Insights into brain microstructure from the T2 distribution. Magn Reson Imaging. 2006;24:515–25. doi: 10.1016/j.mri.2005.12.037. [DOI] [PubMed] [Google Scholar]
- MacKay A, Whittall K, Adler J, Li D, Paty D, Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magn Reson Med. 1994;31:673–7. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- Madler B, Drabycz SA, Kolind SH, Whittall KP, MacKay AL. Is diffusion anisotropy an accurate monitor of myelination? Correlation of multicomponent T2 relaxation and diffusion tensor anisotropy in human brain. Magn Reson Imaging. 2008;26:874–88. doi: 10.1016/j.mri.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Matsushima GK, Morell P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001;11:107–16. doi: 10.1111/j.1750-3639.2001.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon EJ, Suzuki K, Matsushima GK. Peripheral macrophage recruitment in cuprizone-induced CNS demyelination despite an intact blood-brain barrier. J Neuroimmunol. 2002;130:32–45. doi: 10.1016/s0165-5728(02)00205-9. [DOI] [PubMed] [Google Scholar]
- Mulkern RV, Haker SJ, Maier SE. On high b diffusion imaging in the human brain: ruminations and experimental insights. Magn Reson Imaging. 2009;27:1151–62. doi: 10.1016/j.mri.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith RT, Xu J, Tutlam NT, Scully PT, Trinkaus K, Snyder AZ, et al. Increased diffusivity in acute multiple sclerosis lesions predicts risk of black hole. Neurology. 2010;74:1694–701. doi: 10.1212/WNL.0b013e3181e042c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov DS, Kiselev VG. Effective medium theory of a diffusion-weighted signal. NMR Biomed. 2010;23:682–97. doi: 10.1002/nbm.1584. [DOI] [PubMed] [Google Scholar]
- Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y. Free water elimination and mapping from diffusion MRI. Magn Reson Med. 2009;62:717–30. doi: 10.1002/mrm.22055. [DOI] [PubMed] [Google Scholar]
- Ramirez-Manzanares A, Rivera M, Vemuri BC, Carney P, Mareci T. Diffusion basis functions decomposition for estimating white matter intravoxel fiber geometry. IEEE Trans Med Imaging. 2007;26:1091–102. doi: 10.1109/TMI.2007.900461. [DOI] [PubMed] [Google Scholar]
- Scalfari A, Neuhaus A, Degenhardt A, Rice GP, Muraro PA, Daumer M, et al. The natural history of multiple sclerosis: a geographically based study 10: relapses and long-term disability. Brain. 2010;133(Pt 7):1914–29. doi: 10.1093/brain/awq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinummi J, Seppala J, Yli-Harja O, Puhakka JA. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques. 2005;39:859–63. doi: 10.2144/000112018. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ju WK, Lin SJ, Cross AH, Neufeld AH. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage. 2003;20:1714–22. doi: 10.1016/j.neuroimage.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17:1429–36. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- Song SK, Yoshino J, Le TQ, Lin SJ, Sun SW, Cross AH, et al. Demyelination increases radial diffusivity in corpus callosum of mouse brain. Neuroimage. 2005;26:132–40. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Stanisz GJ, Szafer A, Wright GA, Henkelman RM. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med. 1997;37:103–11. doi: 10.1002/mrm.1910370115. [DOI] [PubMed] [Google Scholar]
- Sun SW, Liang HF, Trinkaus K, Cross AH, Armstrong RC, Song SK. Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn Reson Med. 2006;55:302–8. doi: 10.1002/mrm.20774. [DOI] [PubMed] [Google Scholar]
- Torkildsen O, Brunborg LA, Myhr KM, Bo L. The cuprizone model for demyelination. Acta Neurol Scand Suppl. 2008;188:72–6. doi: 10.1111/j.1600-0404.2008.01036.x. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Bo L, Mork S, Chang A. Pathogenesis of tissue injury in MS lesions. J Neuroimmunol. 1999;98:49–56. doi: 10.1016/s0165-5728(99)00081-8. [DOI] [PubMed] [Google Scholar]
- Vellinga MM, Oude Engberink RD, Seewann A, Pouwels PJ, Wattjes MP, van der Pol SM, et al. Pluriformity of inflammation in multiple sclerosis shown by ultra-small iron oxide particle enhancement. Brain. 2008;131(Pt 3):800–7. doi: 10.1093/brain/awn009. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005;54:1377–86. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- Wheeler-Kingshott CA, Cercignani M. About ‘axial’ and ‘radial’ diffusivities. Magn Reson Med. 2009;61:1255–60. doi: 10.1002/mrm.21965. [DOI] [PubMed] [Google Scholar]
- Wu QZ, Yang Q, Cate HS, Kemper D, Binder M, Wang HX, et al. MRI identification of the rostral-caudal pattern of pathology within the corpus callosum in the cuprizone mouse model. J Magn Reson Imaging. 2008;27:446–53. doi: 10.1002/jmri.21111. [DOI] [PubMed] [Google Scholar]
- Xie M, Tobin JE, Budde MD, Chen CI, Trinkaus K, Cross AH, et al. Rostrocaudal analysis of corpus callosum demyelination and axon damage across disease stages refines diffusion tensor imaging correlations with pathological features. J Neuropathol Exp Neurol. 2010;69:704–16. doi: 10.1097/NEN.0b013e3181e3de90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Wedeen VJ, Tseng WY. Generalized q-sampling imaging. IEEE Trans Med Imaging. 2010;29:1626–35. doi: 10.1109/TMI.2010.2045126. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Wedeen VJ, Tseng WY. Estimation of fiber orientation and spin density distribution by diffusion deconvolution. Neuroimage. 2011;55:1054–62. doi: 10.1016/j.neuroimage.2010.11.087. [DOI] [PubMed] [Google Scholar]