Abstract
Cardiovascular disease (CVD) remains the leading cause of mortality in westernized countries, despite optimum medical therapy to lower LDL cholesterol. The pursuit of novel therapies to target this residual risk has focused on raising levels of HDL cholesterol in order to exploit its atheroprotective effects1. MicroRNAs have emerged as important post-transcriptional regulators of lipid metabolism, and are thus a new class of targets for therapeutic intervention2. MicroRNA-33a and b (miR-33a/b) are intronic microRNAs embedded in the sterol response element binding protein genes SREBF2 and SREBF13–5, respectively, that repress expression of the cholesterol transporter ABCA1, a key regulator of HDL biogenesis. Recent studies in mice suggest that antagonizing miR-33a may be an effective strategy for raising plasma HDL3–5 and protecting from atherosclerosis6, however extrapolation of these findings to humans is complicated by the fact that mice lack miR-33b which is present only in the SREBF1 gene of higher mammals. Here we show in African green monkeys that systemic delivery of an anti-miR oligonucleotide that targets both miR-33a and miR-33b increases hepatic expression of ABCA1 and induces a sustained increase in plasma HDL over 12 weeks. Notably, miR-33 antagonism in this non-human primate model also increased the expression of miR-33 target genes involved in the oxidation of fatty acids (CROT, CPT1A, HADHB, PRKAA1) and reduced genes involved in fatty acid synthesis (SREBF1, FASN, ACLY, ACACA), resulting in a marked suppression of plasma VLDL triglyceride levels, a finding not previously observed in mice. These data establish, in a model highly relevant to humans, that pharmacological inhibition of miR-33a and b is a promising therapeutic strategy to raise plasma HDL and lower VLDL triglycerides for the treatment of dyslipidemias that increase cardiovascular disease risk.
Keywords: microRNA, metabolic syndrome, HDL, triglyceride
Recent advances in the understanding of lipid metabolism have revealed that the genetic loci coding for the transcription factors SREBP1 and SREBP2 contain the microRNAs miR-33b and miR-33a, respectively, which regulate cholesterol and fatty acid homeostasis in concert with their host genes3–5,7–9. Although miR-33a and b differ by only 2 nucleotides in the mature form, they are identical in their seed sequence, and thus they are predicted to repress the same subset of genes. Notably, miR-33a has been highly conserved throughout evolution, whereas miR-33b is present only in the SREBF1 gene of large mammals. This difference between mice and humans may be particularly relevant under conditions in which the transcription of SREBF1, and thus miR-33b, is highly upregulated, such as insulin resistance9. Recently, we and others reported that silencing of miR-33a in mice using modified anti-sense oligonucleotides4,6, viral delivery of hairpin inhibitors3,5 or targeted deletion10, increased hepatic ABCA1 and circulating HDL by as much as 40%. While these studies highlighted the therapeutic promise of miR-33 inhibitors for raising plasma HDL, the absence of miR-33b in mice limits the translational relevance of these findings. Thus, to gain a comprehensive understanding of the effects of inhibiting both miR-33a and miR-33b in a model highly related to humans, we treated African green monkeys (Chlorocebus aethiops) with a 2’fluoro/methoxyethyl (2’F/MOE) phosphorothioate-backbone modified anti-miR33, that we show is equally effective at inhibiting both miR-33a and miR-33b in vitro (Supplementary Fig. 1a). Six animals per group were subcutaneously administered a clinically-relevant dose of 5 mg/kg anti-miR-33 or a mismatch control11, twice weekly for the first two weeks, and then weekly for the remainder of the study (Fig. 1a). Quantification of hepatic anti-miR levels by ion-pairing (IP) HPLC-ES/MS after 4 and 12 weeks of treatment showed equivalent delivery of anti-miR-33 and control (mismatch) oligonucleotides (Supplementary Fig. 1b). No apparent toxicity was associated with the anti-miR treatment as shown by the clinical chemistries, blood counts, coagulation markers, body weights, and serum cytokine profiles (Fig. 1b and Supplementary Fig. 1c–d), which remained within normal limits throughout the study.
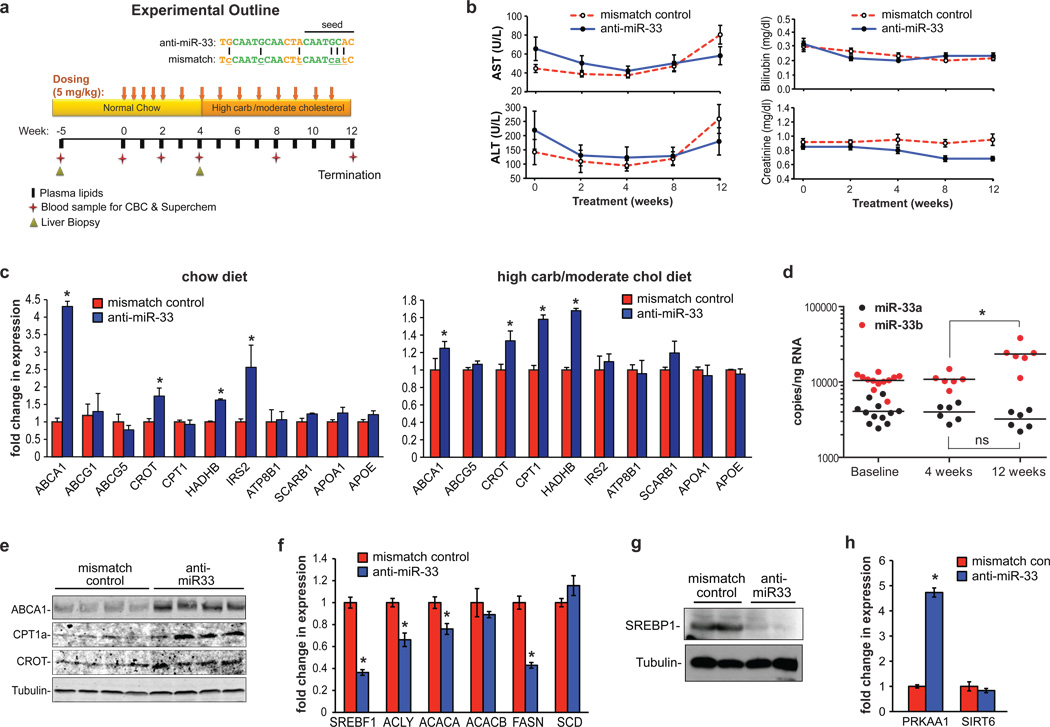
Figure 1. Silencing of miR-33a/b in non-human primates.
(a) Experimental outline of anti-miR33 or mismatch control oligonucleotide treatment in African green monkeys (n=6/group). (b) Serum transaminase (AST, ALT), bilirubin and creatinine levels. (c) Hepatic gene expression in anti-miR treated monkeys fed a chow diet (4 weeks), or a high carbohydrate, moderate cholesterol diet (12 weeks). (d) Quantitation of miR-33a and b levels in anti-miR treated monkeys. (e) Western blot for hepatic ABCA1, CPT1 and CROT following 12 weeks of anti-miR treatment. (f) Expression of hepatic SREBP1 mRNA and its downstream genes, and (g) SREBP1 protein after 12 weeks of anti-miR treatment. (h) Hepatic PRKAA1 and SIRT6 mRNA after 12 weeks of anti-miR treatment. Data are the mean ± SEM. *P ≤ 0.05.
Microarray profiling of mRNA obtained from liver biopsies after 4 weeks of treatment revealed that anti-miR-33 selectively increased the expression of miR-33 heptamer-matched genes in monkeys fed a chow diet (Supplementary Table 1). Of these, the cholesterol transporter ABCA1 was the most highly derepressed miR-33 target gene. Quantitative RT-PCR analysis confirmed the increase in ABCA1, as well as other known miR-33 target genes (target site alignment shown in Supplementary Fig 2) including two enzymes involved in fatty acid oxidation CROT and HADHB, and the insulin signaling gene IRS2 (Fig. 1c, Supplementary Fig 3). In order to assess the effects of miR-33 inhibition under different metabolic conditions, monkeys were switched after 4 weeks to a high carbohydrate, moderate cholesterol diet which increased SREBP1 mRNA 5-fold and induced a corresponding 2.2-fold increase in miR-33b, making its expression >7-fold higher than miR-33a (Fig. 1d, Supplementary Fig 3). Microarray and qRT-PCR analysis showed that the derepression of the above mentioned miR-33 target genes by anti-miR-33 was largely sustained in monkeys fed a high carbohydrate, moderate cholesterol diet (Fig. 1c, Supplementary Fig 3, Supplementary Table 2). Under these diet conditions, we observed an increase in an additional miR-33 target gene involved in fatty acid oxidation, CPT1A (Fig. 1c, Supplementary Fig 3). Although ABCG5 and ATP8B1 are predicted to contain miR-33 binding sites, no difference in their mRNA levels was observed (Fig. 1c). Furthermore, we observed no change in the expression of hepatic lipid metabolism genes lacking miR-33 binding sites, such as APOE and APOA1, as well as ABCG1, which lacks the miR-33 binding site present in the mouse gene (Fig. 1c, Supplementary Fig 3).
As microRNAs can mediate effects on both mRNA stability and translation, we measured hepatic ABCA1, CROT and CPT1A protein after 4 weeks of treatment. All three of these miR-33 targets were increased in the livers of monkeys treated with anti-miR-33 compared to control (Supplementary Fig. 1e). Furthermore, despite modest effects of anti-miR-33 on ABCA1 mRNA after 12 weeks, hepatic ABCA1 protein remained robustly increased, as did expression of CROT and CPT1A (Fig. 1e). Marked upregulation of ABCA1 mRNA in anti-miR-33 treated monkeys was also observed in the spleen, a macrophage rich tissue. As expected, splenic ABCG1 mRNA was not changed by anti-miR-33 treatment, as this is not a conserved target in primates (Supplementary Fig. 1f).
Notably, while we observed no difference in SREBF2 expression in anti-miR-33 and control anti-miR treated animals over the course of the study, we detected a 50% decrease in SREBF1 mRNA in the anti-miR-33 monkeys at 12 weeks (Fig. 1f and Supplementary Fig 3), which was confirmed by western blotting (Fig. 1g). We postulated that this decrease in SREBP1 may result from the derepression of negative regulators of this pathway targeted by miR-33. Consistent with this thesis, we observed a 4-fold increase in PRKAA1 (AMPK) mRNA in the livers of anti-miR-33 treated monkeys, whereas no change in SIRT6 mRNA was detected (Fig. 1h). SREBP1 plays a major role in the transcriptional regulation of fatty acid synthesis, and measurement of its downstream target genes revealed decreased mRNA levels for ATP citrate lyase (ACLY), acetyl-CoA carboxylase alpha (ACACA) and fatty acid synthase (FASN) (Fig 1f).
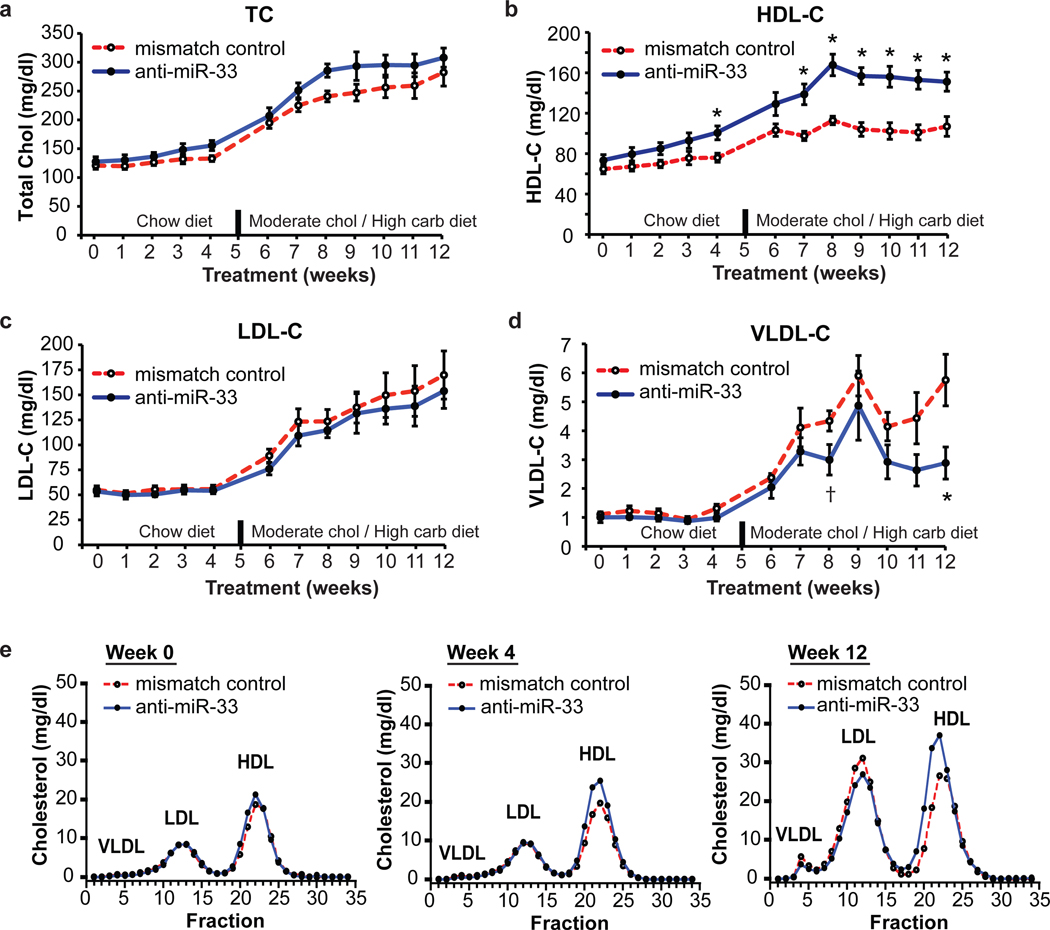
As increased hepatic expression of ABCA1 would be predicted to augment HDL biogenesis, we measured plasma lipoprotein cholesterol levels. Whereas weekly blood sampling revealed no difference in total plasma or LDL cholesterol (LDL-C) between treatment groups, there was both a significant decrease in VLDL-C and an increase in HDL-C in anti-miR-33 treated monkeys (Fig. 2a–d). A maximal HDL increase of 50% was reached after 8 weeks and was sustained throughout the remainder of the study (Fig. 2b). Correspondingly, lipoprotein separation by fast-pressure liquid chromatography (FPLC) showed increased cholesterol in the HDL fraction and a left-shifted HDL peak in anti-miR-33 treated monkeys, suggestive of larger sized HDL particles (Fig. 2e). To further characterize the HDL, we examined plasma concentrations of apolipoprotein AI, AII, and E in both total plasma and HDL fractionated by FPLC using ELISA and western blotting, respectively. By these two measures, we observed significant increases in the primary apolipoproteins carried on HDL, apoAI and AII, associated with large and very large HDL particles (Fig. 3a–b, Supplementary Fig 4). As the static measurement of HDL has inherent limitations in extrapolating to its functionality1, we examined the atheroprotective properties of anti-miR-33 generated HDL, namely its ability to promote macrophage cholesterol efflux and to protect endothelial cells from cytokine induced inflammation. Equivalent volumes of serum or polyethylene glycol (PEG) isolated HDL from anti-miR-33 treated monkeys induced greater macrophage cholesterol efflux than that from control monkeys (Fig. 3c), which correlates with the HDL concentration in these animals. When normalized for cholesterol content, the PEG-HDL from anti-miR-33 and control treated monkeys showed similar acceptor capacity for cholesterol efflux from macrophages (data not shown). Furthermore, anti-miR-33 HDL maintained its anti-inflammatory effects on endothelial cells (Supplementary Fig 4).
Figure 2. Plasma cholesterol levels in control and anti-miR33 treated monkeys.
Levels of plasma (a) total cholesterol, (b) HDL-cholesterol, (c) LDL-cholesterol and (d) VLDL-cholesterol in anti-miR treated monkeys. *P ≤ 0.05, † P ≤ 0.1. (e) Cholesterol content of FPLC fractionated lipoproteins.
Figure 3. Characterization of HDL.
(a) Plasma apoAI and apoAII in anti-miR treated monkeys. *P ≤ 0.05. (b) HDL fractions (VL=very large, L=large, M=medium and S=small) analyzed by Western blot for apoE, apoAI and apoAII. (c) Macrophage cholesterol efflux to serum (2.5%) or PEG-isolated HDL from anti-miR treated monkeys. *P ≤ 0.05.
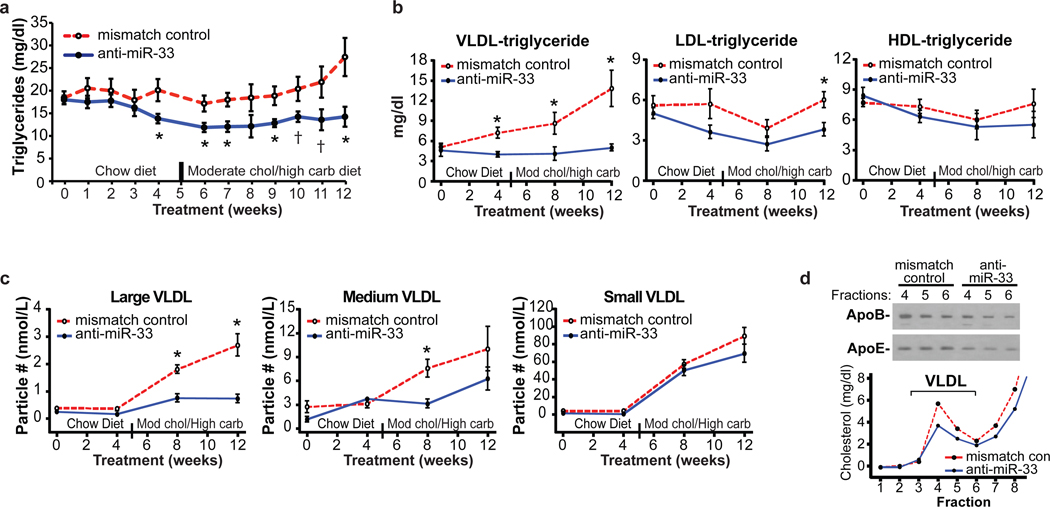
Given the reciprocal effects of anti-miR-33 treatment on the expression of genes involved in fatty acid oxidation and synthesis, we next measured plasma triglyceride levels. In anti-miR-33 treated monkeys, there was a striking reduction in plasma triglycerides (Fig. 4a). This decrease was apparent as early as 4 weeks, and reached a maximum reduction of 50% at the termination of the study. Fractionation of plasma lipoproteins revealed that this derived primarily from reduced VLDL-associated triglycerides throughout the study, and a reduction in LDL-triglyceride at 12 weeks (Fig. 4b). VLDL particle analysis by nuclear magnetic resonance (NMR) spectroscopy demonstrated decreased accumulation of large VLDL particles in anti-miR-33 treated monkeys (Fig. 4c), with a corresponding decrease in apoB and apoE in the VLDL fraction (Fig. 4d). Thus, by simultaneously increasing fatty acid oxidation via derepression of HADHB, CPT1A and CROT, and decreasing fatty acid synthesis via inhibition of the SREBP-1 pathway, anti-miR-33 treatment results in a pronounced reduction in plasma VLDLtriglyceride.
Figure 4. Triglyceride and VLDL Particle Analysis.
Levels of plasma (a) total triglyceride, (b) VLDL-triglyceride, LDL-triglyceride or HDL-triglyceride from anti-miR treated monkeys. *P ≤ 0.05; † P ≤ 0.1. (c) Quantification of small, medium and large VLDL particle number by NMR spectroscopy. *P ≤ 0.05. (d) Western blot of apoE and apoB in VLDL fractions obtained from FPLC fractionation of plasma.
The development of novel therapies to exploit the atheroprotective properties of HDL is an area of intense investigation1. In randomized clinical trials raising plasma HDL by augmenting apoAI levels or treating with niacin has shown direct benefits in patients with coronary artery disease, including reducing cardiovascular event rates and plaque volume1,12. However, the development of HDL-raising drugs has proven particularly challenging12. Previous studies by our group and others have shown that inhibiting miR-33a in mice is an effective strategy to raise HDL3–5, and to enhance reverse cholesterol transport and regress atherosclerotic plaques6. Although promising, these studies in mice are limited in their translational insight due to the lack of miR-33b expression, which may contribute substantially to miR-33 levels in humans. The current study in non-human primates is the first to show that inhibiting both miR-33a and miR-33b has a profound and sustained effect on circulating HDL levels. Importantly, this study also establishes that miR-33 antagonism markedly suppresses plasma VLDL triglyceride levels, attributable in part to regulation of key genes involved in fatty acid oxidation7,8 and synthesis. As low HDL and high VLDL triglycerides are commonly associated with metabolic syndrome13, miR-33 inhibitors may have clinical utility for the treatment of this growing health concern. Of note, as recently reported in mice7, inhibition of miR-33 in monkeys also increased hepatic expression of IRS2, a key component of insulin signaling which also becomes dysfunctional in metabolic syndrome13. While the monkeys used in this study were normoglycemic, future studies in primate models of obesity/diabetes will be important to test the effects of miR-33 inhibition on insulin signaling. Taken together, our study demonstrates that pharmacological inhibition of miR-33a and b leads to a sustained increase in plasma HDL cholesterol and a coincident decrease in VLDL triglycerides without evidence of adverse effects. These findings in non-human primates support the development of antagonists of miR-33 as potential therapeutics for dyslipidemia, atherosclerosis, and related metabolic diseases.
METHODS SUMMARY
All experiments were performed in accordance with National Institutes for Health guidelines for animal research and were approved by the Wake Forest University Health Science Institutional Animal Care and Use Committee. Male African green monkeys (n=6/group) were injected subcutaneously with 5 mg/kg of 2' fluoro/methoxyethyl modified anti-miR-33 or mismatch anti-miR (Regulus Therapeutics) twice weekly for 2 weeks and then weekly until termination of the study. Monkeys were fed a weighed amount of chow or a high carbohydrate, moderate cholesterol semisynthetic diet. Serum and whole blood samples were analyzed using Superchem and CBC tests (ANTECH Diagnostics), respectively. Plasma lipoprotein cholesterol and triglyceride distribution were determined by on-line, high performance gel filtration chromatography. Pooled plasma was separated by FPLC on a Superose 6 10/300 GL column (GE Healthcare). Plasma lipoprotein particle number and size were determined using nuclear magnetic resonance spectroscopy (LipoScience, Inc). Plasma apoAI, apoAII, and apoE were measured by ELISA. Detailed description of RNA and protein analyses, as well as HDL characterization assays can be found in the online version of the paper at www.nature.com/nature.
FULL METHODS
African Green Monkeys
Adult male African green monkeys (Chlorocebus aethiops) (n=12, age 5–10 years) were obtained from St Kitts Island. Monkeys were housed in an AAALAC accredited facility under the direct care of the Wake Forest University Health Sciences (WFUHS) Animal Resources Program and necropsied at the termination of the study. All experiments were approved by the WFUHS Institutional Animal Care and Use Committee (IACUC). Monkeys were singly housed in climate-controlled conditions with 12 hour light and dark cycles.
Monkeys were provided water ad libitum and were initially fed twice a day a weighed amount of a chow diet (Monkey Diet 5038, Lab Diet), such that their daily caloric intake was 70 kcal/day/kg body weight. To induce dyslipidemia, the monkeys were fed twice a day a weighed amount of a high carbohydrate, moderate cholesterol semisynthetic diet (Table S3) prepared at the Wake Forest Primate Center Diet Laboratory, such that their daily caloric intake was 90 kcal/day/kg body weight. The use of a similar semi-synthetic high-fructose diet in cynomolgus monkeys has been shown to increase plasma triglyceride levels.14
Liver Biopsy
Liver samples were collected from chow fed monkeys prior to and following 4 weeks of anti-miR treatment. After an overnight fast, the monkeys were initially anesthetized with Ketamine (10mg/kg, IM) and pre-treated with Atropine (0.04 mg/kg, IM). Following intubation, anesthesia was maintained throughout the surgical procedure with Isofluorane (3.5–5.0% induction, 1–2% maintenance, inhalation). The monkeys were shaved and prepped for surgery in accordance with standard veterinary medical practice (3 sequential scrubs/rinses with Novolsan/isopropanol and final spray of betadine solution). Anesthesia was monitored at 15-minute intervals during the surgical procedure. If there was a change after the monkeys reached the desired plane of anesthesia, anesthesia was increased or decreased at the direction of the surgical veterinarian. Heart rate, oxygen saturation, end expired CO2, capillary refill time, respiration rate, and temperature were recorded at 15-minute intervals or more frequently if needed. Under sterile conditions an abdominal incision was made and a 1 gram wedge of liver was taken. The monkeys were administered Buprenorphine HCL (0.01mg/kg, IM) for pre-emptive analgesia. The laparotomy incision was closed in a standard 3-layer fashion with the abdominal wall closed using a synthetic absorbable suture in a simple interrupted pattern. The subcutis was similarly closed with a continuous suture pattern using a synthetic absorbable material, and the skin was opposed with a continuous pattern of intradermally placed suture. The monkeys were returned to their cages and monitored during anesthetic recovery in accordance with the WFUHS IACUC policy on survival surgery and post-surgical care. For 10 days following the surgery, monkeys were monitored for post-operative pain, which was alleviated with Buprenorphine (0.01 mg/kg, IM, BID) and Ketoprofen (2.0–3.0 mg/kg, IM, SID).
Anti-miR Treatment
Regulus Therapeutic provided 2' fluoro/methoxyethyl (2'F/MOE) modified, phosphorothioate backbone modified anti-miR-33 (TGCAATGCAACTACAATGCAC) and mismatch control (TCCAATCCAACTTCAATCATC) anti-miR. Monkeys were injected subcutaneously with 5 mg anti-miR/kg body weight twice weekly during the first 2 weeks and then once weekly during the remaining 10 weeks of the study.
Monitoring for Adverse Effects
To monitor for any adverse effects of anti-miR treatment, body weights were measured weekly. In addition, at week 0, 4, 8, and 12 of anti-miR treatment, ANTECH Diagnostics analyzed serum and whole blood samples using the Superchem and CBC tests, respectively.
RNA Extraction and Quantitative PCR
Liver tissue was homogenized in 300µl of Trizol (Invitrogen) with 1mm zirconium silicate beads using the Bullet Blender (Next Advance Inc.). Insoluble material and beads were removed by centrifugation, the volume of Trizol was brought up to 1ml and RNA was extracted according to the manufacturer’s protocol. RNA integrity was verified using the Agilent Bioanalyzer. Reverse transcription was carried out on 1µg of total RNA using the iScript cDNA Synthesis kit (Biorad). Quantitative PCR was performed using the Eppendorf Mastercycler with primers directed against the genes listed in Table S2.
miR-33a and miR-33b detection was performed using Panomics Quantigene 2.0 microRNA assay according to the manufacturer’s instructions. 1mg input total RNA was used for each liver sample. Copy number quantification was done by generating a standard curve using miR-33a and miR-33b oligonucleotides synthesized from Integrated DNA Technologies.
Affymetrix Gene Array Analysis
Microarray analysis of liver biopsies from monkeys treated with anti-miR-33 or a control anti-miR were performed in groups of 6 using Affymetrix human U133 plus 2.0 arrays. Treatments were performed at 3 time points: 0, 4, and 12 weeks. Data from the arrays were normalized, quality-controlled, and compared using two-way ANOVA tests. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE31177 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31177). Signature seed-matched heptamer genes to miR-33 were filtered using p-values with an alpha<0.05 and sorted by fold change. The miR-33 seed sequence was taken from MirBase (mirbase.org) and seed-matched genes were derived from genes with 3' UTR seed-sequence matches in the Ensembl Biomart database (biomart.org).
HPLC-ES/MS Quantification of Anti-miR
Anti-miR was quantified in liver samples by ion-pairing (IP) HPLC-ES/MS. Separation was accomplished using 1200 HPLC-MS system consisting of binary pump, diode array UV detector, a column oven, an autosampler, and model 6100 single quadrupole mass spectrometer (Agilent Technologies). Typically, 50 mg of each sample was extracted using a phenol/chloroform/isoamyl alcohol (25:24:1) extraction method followed by a two-step solid phase extraction method (strong anion exchange followed by reverse phase C18). The extracted material was reconstituted in water and injected directly on XBridge™ OST C18 column (50× 2.1mm; 2.5mm particles; Waters). The column was maintained at 55°C, and the flow rate on the column was 0.1 ml/min. The column was equilibrated with 25% acetonitrile in 5 mM tributylammonium acetate, pH 7.0. A gradient from 30 to 60% acetonitrile over 10min was used to separately elute compound of interest and internal standard (27-mer F/MOE compound). Peak areas were quantified online using single ion monitoring mode (SIM) with m/z = 1868 for RG428651 and m/z = 1843 for RG522293. Mass spectra were obtained using drying gas flow rate of 12 l/min at 350°C, nebulizer pressure of 35 psig and capillary voltage of 4000V. Chromatograms were analyzed using Chemstation software. Compounds levels were back-calculated using quadratic fit and calibration curve range of 15.6–500 mg/g tissue. Low limit of quantitation (LLOQ) was equal to 31.3 mg/g tissue.
Protein Extraction and Western Blot Analysis of Liver
Liver tissue was homogenized in 500µl RIPA buffer using the Bullet Blender, as described above. Lysates were cleared by centrifugation and a total of 50µg of protein was separated using SDS-PAGE and transferred to nitrocellulose or PVDF. Membranes were blotted with antibodies against ABCA1 (rabbit, gift from M. Fitzgerald 15), Cpt1a (goat, Novus Biologicals), CROT (rabbit, Abcam) and tubulin (mouse, Sigma). Secondary antibodies labeled with IRDye800 or IRDye700 (Rockland Immunochemicals Inc.) were visualized using the Li-cor Odyssey imaging system.
Cholesterol Efflux Assays
THP-1 cells (1×106 per well) were differentiated in 10nM PMA for 72 hours in RPMI supplemented with 10% FBS. Cells were loaded by incubation with 37.5µg/ml acetylated LDL (Biomedical Technologies Inc) and labeled with 0.5µCi/ml of 3H-cholesterol (PerkinElmer) for 24 hours. Excessive label was removed by extensive washing with PBS before cells were equilibrated in 2mg/ml fatty-acid free BSA in RPMI. To use as an acceptor in efflux studies, HDL was isolated by combining pooled serum samples (from n=6 monkeys treated with either mismatch control or anti-miR33) with 20% polyethyleneglycol (PEG; Sigma) followed by centrifugation, to precipitate the apoB-containing lipoproteins as previously described.16 The cholesterol content of this PEG-HDL was determined to be 526 ug/ml for control anti-miR and 863 ug/ml for anti-miR-33 treated monkeys. Media containing equal volumes (25µl) of this PEG-isolated HDL was added to the labelled cells for 6 hours. Alternatively, 2.5% pooled serum from each group of monkeys was added to the cells as an efflux acceptor for 6 hours. Supernatants were collected and 3H counted and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol+cell-associated 3H-cholesterol). Data are expressed as mean ± SD of triplicate wells, and represents an experimental n=3.
Luciferase Assays
HEK293 cells were seeded 24 hours prior to transfection in 24-well plates. A plasmid containing the full-length 3’UTR of ABCA1 downstream of firefly luciferase (Genecopoeia Inc) was transfected into cells in the presence or absence of the following vectors: pre-miR-33a, pre-miR-33b or a control miRNA (System Biosciences Inc) along with 20nM of either the mismatch anti-miR or anti-miR-33. Twenty-four hours after transfection, cells were harvested and luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase activity was normalized to renilla luciferase activity as a control for transfection efficiency. Data are expressed relative to ABCA1 3’UTR activity in the presence of a control miRNA, and are mean ± SD of triplicate samples of an experimental n=3.
Plasma Lipid, Apolipoprotein, Lipoprotein Concentrations
Monkeys were sedated with Ketamine (10 mg/kg, IM) and blood was collected into EDTA-containing vacutainers. Plasma was isolated by centrifugation at 1,500×g for 30 min at 4°C. Plasma total cholesterol and triglyceride were measured using the Cholesterol Reagent Set (Pointe Scientific) and Triglyceride Reagent and Free Glycerol Reagent (Sigma), respectively. Plasma lipoprotein cholesterol and triglyceride distribution was determined by on-line, high performance gel filtration chromatography17,18 using Infinity Cholesterol or Infinity Triglyceride reagent (Thermo). Plasma lipoprotein particle number and size were determined by LipoScience, Inc using nuclear magnetic resonance spectroscopy as described previously.19 Plasma apoAI, apoAII, and apoE levels measured by ELISA.20
Plasma Cholesterol and Apolipoprotein Distribution
An equal volume of plasma from each monkey within a treatment group was pooled. The pooled plasma was separated on a Superose 6 10/300 GL column (GE Healthcare) at a flow rate of 0.4 ml/min. From 20 to 60 minutes post-injection, fractions were collected at 1 min intervals. Total cholesterol content of the fractions was determined using the Cholesterol Reagent Set (Pointe Scientific). An equal volume of each fraction was mixed with 5X SDS sample buffer and separated on a NuPAGE Novex 4–12% Bis Tris Midi Gel using 1X NuPAGE MOPS SDS running buffer (Invitrogen). The proteins were transferred to a nitrocellulose membrane, which was subsequently blocked with 5% (w/v) non-fat dried milk dissolved in wash buffer. The membrane was incubated overnight at 4°C in one or more of the following goat anti-monkey affinity purified antibodies: anti-apoAI, anti-apoAII, anti-apoB or anti-apoE. All anti-monkey apolipoprotein antibodies were created and tested for specificity at WFUHS. Secondary antibodies against goat IgG were conjugated to HRP (Sigma), and proteins were visualized with ECL reagent (Perkin Elmer) and exposure to blue X-ray film (Phenix).
Plasma Cytokine Analysis
The levels of cytokines in plasma samples were measured by Human Proinflammatory 7-plex Cytokine Ultrasensitive kit (MESO Scale Discovery). The assay was done following manufacturer’s assay procedure. Briefly, 25 µl of monkey plasma samples or kit calibrators were added to assay diluent in a 96-well 7-plex assay plate. After incubation for 2 hours, the plate was washed then incubated with detection antibody solution for 2 hours. Read Buffer was added to each well in the assay plate, and the signal was analyzed by the SECTOR Imager (MESO Scale Discovery). Seven cytokines including IL-1β, IL-12p70, IFNγ, IL-6, IL-8, IL-10 and TNF-α were analyzed simultaneously by the imager. The data was quantified using a standard curve generated from kit calibrators for each of the cytokines.
Flow cytometric analysis of endothelial VCAM-1 and E-selectin
Human umbilical vein endothelial cells (HUVEC, Lonza) were cultured in EGM-2 according to the manufacturer’s protocol. HUVEC (passage 3–5) were plated in 24-well plates with 1×105 cells/400 µl per well in EGM-2 plus 20% FBS. After a 5 hour re-attachment period, cells were pre-incubated for 16 hours with HDL isolated from pooled serum samples (n=6), after which TNF-α (0.5 ng/ml, Peprotech) was added to the culture medium for an additional 4 hours. Cell-surface expression of VCAM-1 and E-selectin were then measured by flow cytometry, using a VCAM-1 monoclonal antibody (BD Biosciences) followed by an anti-mouse FITC conjugated antibody (Sigma), or an E-selectin PE conjugated antibody (Chemicon). Cells were detached using 5 mM EDTA in PBS. Cellular FITC and PE were then analyzed by flow cytometry (C6 Flow Cytometer, Accuri Cytometers, BD Biosciences). Controls included an isotype-matched control antibody and no primary antibody. Native HDL3 (isolated by sequential ultracentrifugation of human plasma) at concentration of 0.5 mg/ml protein was used as a positive control.
Statistical Analyses
All comparisons were made using a student’s t-test (p≤0.05) and data are expressed as mean ± SEM, unless otherwise noted. Data from the arrays were normalized, quality-controlled, and compared using two-way ANOVA tests.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health to KJM (R01AG02055, R01HL108182); EAF (P01HL098055, R01HL084312, R01HL58541); C.F.-H. (1P30HL101270, R01HL107953); RET (R00HL088528) and the Canadian Institutes of Health Research (KJR).
Footnotes
Conflicts of interest: E.A.F. is a Merck Advisory board member and receives honoraria for speaking engagements. C.C.E., X.L., O.G.K., V.K. are employees of Regulus Therapeutics. K.J.R., C.F-H. and K.J.M. have a pending patent on the use of miR-33 inhibitors.
AUTHOR CONTRIBUTIONS
K.J.M. and R.E.T. contributed equally to this study. K.J.M, R.E.T, C.C.E. and K.J.R. designed the study. C.J.L, R.E.T, A.L.M, S.M.M and K.J.R. assisted in the necropsy. K.J.R, R.E.T, F.N.H, J.M.vG, F.J.S. and T.D.R. performed the biological assays. C.C.E, X.L, O.G.K. and V.K. conducted the microRNA and microarray analyses, E.A.F and C.F-H assisted with experimental design and data interpretation. K.J.M and K.J.R wrote the first draft of the manuscript, which was commented on by all authors.
AUTHOR INFORMATION
The microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE31177 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31177). Reprints and permission information is available at www.nature.com/reprints.
REFERENCES
- 1.Degoma EM, Rader DJ. Novel HDL-directed pharmacotherapeutic strategies. Nature reviews. Cardiology. 2011;8:266–277. doi: 10.1038/nrcardio.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends in endocrinology and metabolism: TEM. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. The Journal of clinical investigation. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerin I, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010 doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of Clinical Investigation. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geary RS. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin Drug Metab Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 12.Chyu KY, Peter A, Shah PK. Progress in HDL-Based Therapies for Atherosclerosis. Current atherosclerosis reports. 2011 doi: 10.1007/s11883-011-0189-9. [DOI] [PubMed] [Google Scholar]
- 13.Alberti KG, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 14.Wagner JE, et al. Old world nonhuman primate models of type 2 diabetes mellitus. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2006;47:259–271. doi: 10.1093/ilar.47.3.259. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald ML, et al. ATP-binding cassettee transporter A1 contains an NH2-terminal singal anchor sequence that translocates in protein's first hydrophilic domain to the expo;asmic space. J. Biol. Chem. 2001;276:15137–15145. doi: 10.1074/jbc.m100474200. [DOI] [PubMed] [Google Scholar]
- 16.Yvan-Charvet L, et al. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieft KA, Bocan TMA, Krause BR. Rapid on-line determination of cholesterol distribution among plasma lipoproteins after high-performance gel filtration chromatography. J Lipid Res. 1991;32:859–866. [PubMed] [Google Scholar]
- 18.Garber DW, Kulkarni KR, Anantharamaiah GM. A sensitive and convenient method for lipoprotein profile analysis of individual mouse plasma samples. J Lipid Res. 2000;41:1020–1026. [PubMed] [Google Scholar]
- 19.Jeyarajah EJ, Cromwell WC, Otvos JD. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin Lab Med. 2006;26:847–870. doi: 10.1016/j.cll.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Koritnik DL, Rudel LL. Measurement of apolipoprotein A-I concentration in nonhuman primate serum by enzyme-linked immunosorbent assay (ELISA) J.Lipid Res. 1983;24:1639–1645. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.