Abstract
Alternative splicing is an important mechanism for increasing functional diversity from a limited set of genes. De-regulation of this process is common in diverse pathologic conditions. The androgen receptor (AR) is a steroid receptor transcription factor with functions critical for normal male development as well as the growth and survival of normal and cancerous prostate tissue. Studies of AR function in androgen insensitivity syndrome (AIS) and prostate cancer (PCa) have demonstrated loss-of-function AR alterations in AIS, and gain-of-function AR alterations in PCa. Over the past two decades, AR gene alterations have been identified in various individuals with AIS, which disrupt normal AR splicing patterns and yield dysfunctional AR protein variants. More recently, altered AR splicing patterns have been identified as a mechanism of PCa progression and resistance to androgen-depletion therapy. Several studies have described the synthesis of alternatively spliced transcripts encoding truncated AR isoforms that lack the ligand-binding domain, which is the ultimate target of androgen depletion. Many of these truncated AR isoforms function as constitutively active, ligand-independent transcription factors that can support androgen-independent expression of AR target genes, as well as the androgen-independent growth of PCa cells. In this review, we will summarize the various alternatively spliced AR variants that have been discovered, with a focus on their role and origin in the pathologic conditions of AIS and PCa.
The Androgen Receptor: Gene, mRNA, and Protein
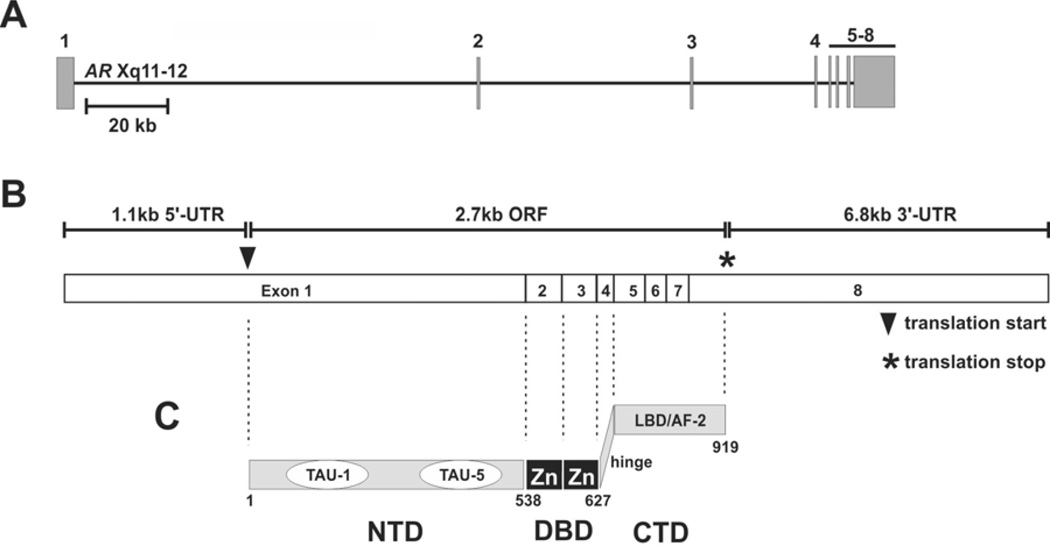
The AR is a 110kDa member of the steroid receptor transcription factor family that mediates the cellular actions of the androgens testosterone and dihydrotestosterone (DHT). The structural organization of the AR gene, mature spliced mRNA, and protein domains are similar to the family members estrogen receptor-α (ERα), estrogen receptor-β (ERβ), and progesterone receptor (PR) (Figure 1) (Lubahn et al. 1989). Males contain one copy of the AR gene located at chromosome position Xq11-12. AR exon 1 encodes the entire AR NH2-terminal domain (NTD), which represents nearly 60% of the AR protein but is variable in length by virtue of polymorphic (CAG)n and (GGN)n repeat units encoding polyglutamine and polyglycine tracts, respectively (Ding et al. 2004, 2005; Ferro et al. 2002). AR exon 2 encodes the first zinc-finger in the AR DNA binding domain (DBD), which is the DNA recognition helix that makes contact with major groove residues in an androgen-response element (ARE) half-site (Shaffer et al. 2004). AR exon 3 encodes the second zinc-finger in the AR DBD, which is a dimerization interface that mediates binding with a neighboring AR molecule engaged with an adjacent ARE half-site (Shaffer et al. 2004). Exons 4–8 encode a short flexible hinge region and 11 alpha-helices folded in an alpha-helical sandwich to form the well-characterized AR COOH-terminal domain (CTD), which harbors the AR ligand-binding domain (LBD) and transcriptional activation function-2 (AF-2) co-regulator binding interface (Bain et al. 2006; Estebanez-Perpina et al. 2005; He et al. 2004; Hur et al. 2004; Matias et al. 2000; Sack et al. 2001).
Figure 1.
Modular organization of the AR gene, mRNA, and protein. (A) Exon organization of the ~180kb AR gene located on chromosome Xq11-12. (B) Normal splicing of AR exons 1–8 yields a 10.6kb mRNA species with large 5’ and 3’ untranslated regions (UTR) and a 2.7kb open reading frame (ORF). (C) The AR protein consists of a large AR NH2-terminal domain (NTD) harboring transcriptional activation unit-1 (TAU-1) and TAU-5, a central DNA binding domain (DBD) consisting of two zinc fingers, and a COOH-terminal domain (CTD) harboring the AR ligand binding domain (LBD) and transcriptional activation function-2 (AF-2).
Each of these discrete protein domains plays an important functional role in the classical mode of AR activation by androgenic ligands. AR protein in the ligand-free state is localized in the cytoplasm, where it is engaged with Hsp90, other molecular chaperones, and high molecular-weight immunophilins by virtue of interaction with the AR CTD (Pratt and Toft 1997; Smith and Toft 2008). Androgen binding induces a change in the conformation and composition of this multi-protein complex, which exposes the bipartite nuclear localization signal in the hinge region (Zhou et al. 1994), thus allowing direct interaction with importin-α and subsequent nuclear translocation through the nuclear pore complex (Cutress et al. 2008). In the nucleus, AR binds as a dimer to AREs in promoter and enhancer elements of target genes, the best-characterized of which are PSA, TMPRSS2, and hK2 (Dehm and Tindall 2006). Transcriptional activation of these target genes is a complex, multi-step process that requires ordered-stepwise recruitment of a plethora of co-regulatory proteins (Heemers and Tindall 2007; Heinlein and Chang 2004). Although the AR AF-2 domain is able to recruit well-characterized co-activators such as SRC-1, SRC-2/TIF-2, and SRC-3/AIB1, its contribution to transcriptional activity is relatively weak (He et al. 2004). In contrast, the AR NTD is a potent transcriptional activator, and this activity has been mapped to two primary transactivation domains termed transactivation unit-1 (TAU1) and TAU5. These two domains have been shown to be necessary and sufficient for full AR transcriptional activity (Callewaert et al. 2006). TAU1 activity has been more precisely mapped to a discrete 5 amino-acid Leu-Lys-Asp-Ile-Leu (LKDIL) motif; however, the co-regulatory proteins that interact with this region have not been elucidated (Callewaert et al. 2006; Chamberlain et al. 1996). Our own work has mapped TAU5 activity to a discrete helical Trp-His-Thr-Leu-Phe (WHTLF) motif, and we have demonstrated that this region is important for AR transcriptional activity under conditions of no/low androgens (Dehm et al. 2007). Overall, the multi-protein complexes nucleated by active AR results in recruitment of the basal transcriptional machinery and a tightly-controlled level of target gene transcription.
Naturally Occurring AR Variants: AR Alternative Splicing In Normal Tissues
Given the modular structure of the AR protein, and the important functional roles for discrete AR protein domains, it is anticipated that alternative splicing events would have profound effects on activity of the AR signaling axis. AR45 is a naturally-occurring variant of the human AR, which was originally identified via 5’ Rapid Amplification of cDNA Ends (RACE) with RNA isolated from human placenta tissue (Ahrens-Fath et al. 2005). AR45 mRNA was found to arise from alternative splicing of a previously-unreported exon located within intron 1 of the AR gene (Figure 2). This alternative exon, termed exon 1B, is situated approximately 22.1 kb downstream of AR Exon 1 and can be spliced in place of AR exon 1 to yield a 45kDa receptor isoform containing the entire AR DBD, CTD, and a novel 7 amino acid Met-Ile-Leu-Trp-Leu-His-Ser sequence at the N-terminus in place of the wild-type AR NTD. RT-PCR analysis indicated that AR45 mRNA is expressed in diverse tissues including heart, muscle, uterus, prostate, lung, and breast, with no apparent expression in brain. However, these RT-PCR experiments were not quantitative, so it is unclear how AR45 expression levels compared with wild-type AR in these tissues. It is also unclear whether AR45 mRNA is translated into a protein in these tissues, although Western blot analysis of LNCaP lysates demonstrated the presence of a ~45kDa species that was immunoreactive with an antibody specific for the AR CTD. Functionally, ectopically expressed AR45 protein was shown to bind androgen, localize to the nucleus, interact with the full-length AR NTD, and inhibit full-length AR activity in a ligand- and DBD-dependent manner. Moreover, overexpression of AR45 in LNCaP cells inhibited proliferation. Together, these data indicate that AR45 is a negative regulator of AR signaling. However, AR45 was also demonstrated to harbor transcriptional activity under conditions of overexpression of the AR co-activators β-catenin or TIF2 (Ahrens-Fath et al. 2005), which is a phenomenon that has been demonstrated for DBD/CTD fragments derived from the AR in other studies (Alen et al. 1999; He et al. 1999). A more recent study revealed a potentially interesting role for AR45 in the increased risk of drug-induced cardiac arrhythmias in women. The Human Ether-a-go-go-Related Gene (HERG) K+ channel has been implicated in this gender bias, and DHT-mediated stabilization of HERG protein was observed in cells transfected with AR45 but not full-length AR (Wu et al. 2008).
Figure 2.
Alternative splicing of AR exon 1B gives rise to the AR45 mRNA isoform. (A) Schematic of the AR gene locus. The location of exon 1B is illustrated in red. (B) The AR45 mRNA is predicted to encode a NH2-terminally truncated protein with an intact DNA binding domain and COOH-terminal domain.
Loss-of-Function AR Variants: AR Alternative Splicing In Androgen Insensitivity Syndrome
The X-linked inheritance pattern of androgen insensitivity syndromes (AIS) is caused by AR genetic alterations in 46, XY males. These AR gene alterations result in AR proteins with impaired function that prevents normal androgen signaling and proper development of internal and external male phenotypes (Brinkmann 2001). The most severe AIS phenotype is complete androgen insensitivity syndrome (CAIS), where individuals have a complete female appearance. Partial androgen insensitivity syndrome (PAIS) includes a wide range of phenotypes including individuals with a primarily female appearance as well as individuals with a primarily male appearance. In general, the severity of AIS is proportional to the level of impaired AR activity caused by specific alterations in the gene. The best-characterized AR gene alterations in AIS are single point mutations that result in an amino acid substitution or a premature translation termination, insertions/deletions resulting in a reading frame shift, or a complete/partial gene deletion (Gottlieb et al. 2004). However, a number of alternatively spliced AR mRNAs have been identified in AIS syndrome, resulting in AR proteins with impaired function. Importantly, gene defects have been associated with these alternatively spliced variants, often within intronic splice donor or splice acceptor sites (Figure 3). Therefore, it would be more appropriate to refer to these events as disruptions in normal splicing as opposed to bona fide alternative splicing in order to reflect the fact that the pathologic transcripts are produced from an altered gene sequence.
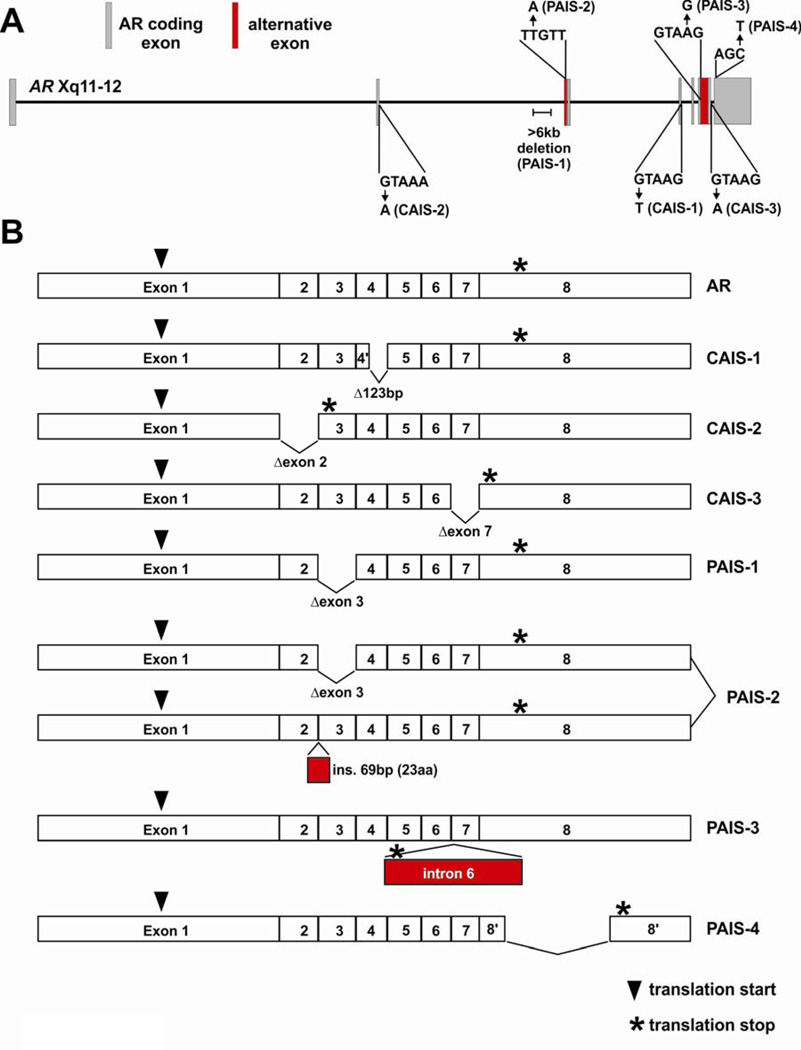
Figure 3.
Mutations and structural alterations in the AR gene disrupt mRNA splicing patterns in androgen insensitivity syndrome (AIS). (A) Schematic of the AR gene locus with intronic point mutations and structural alterations that have been identified in individuals with complete AIS (CAIS) or partial AIS (PAIS). Predominant AR mRNA isoforms expressed in individuals with these specific gene alterations are illustrated in (B).
AR Splicing Disruptions in CAIS
In 1990, AR gene sequencing in a patient with CAIS yielded the observation that the sequences of coding exons were unaltered, but a G→T mutation occurred within the splice donor site of intron 4 (Ris-Stalpers et al. 1990). This mutation was shown to lead to the use of an alternative cryptic splice donor in exon 4, resulting in an in-frame 123bp deletion from AR mRNA and a concomitant 41aa internal deletion of the AR protein (CAIS-1 in Fig. 3). This variant displayed no ligand-binding activity and no transcriptional activity on an androgen-responsive promoter-reporter gene. Since this initial observation, similar mutations in splice donor sites of introns 2 and 7 have been described in two other individuals with CAIS, leading to skipping of exons 2 and 7 from AR mRNAs, respectively (Hellwinkel et al. 1999; Lim et al. 1997). The exon 2-deleted AR protein (CAIS-2 in Fig. 3) was prematurely truncated by virtue of a premature stop codon encoded by out-of-frame exon 3, and the exon 7-deleted AR protein (CAIS-3 in Fig. 3) was ~98kDa as opposed to the 110kDa wild-type AR. As expected, these alternatively-spliced AR variants were unable to bind androgens (Hellwinkel et al. 1999; Lim et al. 1997).
AR Splicing Disruptions in PAIS
Alternatively-spliced AR variants have also been described in individuals with PAIS, indicating that cells with altered AR splicing patterns can retain some level of residual AR activity. This could result from AR variants maintaining some degree of transcriptional activity, or a residual level of splicing that permits synthesis of wild-type AR mRNA and protein. A >6kb deletion has been described in AR intron 2, which resulted in skipping of exon 3 and synthesis of an AR variant protein (PAIS-1 in Fig. 3) that lacks the 39aa zinc finger encoded by this exon (Ris-Stalpers et al. 1994). This variant displayed normal ligand binding, but no transcriptional activity on an AR-responsive promoter-reporter construct. PAIS was attributed to some residual splicing of AR exons 1–8 and a low-level of expression of wild-type AR protein. Similarly, a T→A mutation 11bp upstream of exon 3 was shown to be associated with two alternatively-spliced AR transcripts (PAIS-2 in Fig. 3), one of which arose through skipping of exon 3, and one of which resulted from use of a cryptic splice acceptor site 71bp upstream of exon 3 (Bruggenwirth et al. 1997). The protein encoded by use of this alternative splice acceptor site contained an additional 23aa in-frame insert in the middle of the AR DBD directly between the two AR zinc fingers. This receptor was shown to bind androgens with normal affinity, but had no DNA binding activity in a bandshift assay. Interestingly, this same AR variant, named AR23, was detected in a metastatic prostate cancer specimen via a yeast functional assay (Jagla et al. 2007). However it is not clear whether this patient harbored a T→A mutation at position −11. In another individual, an A→G mutation in the splice donor site of intron 6 resulted in expression of larger AR mRNA species (PAIS-3 in Fig. 3) with retention of this intron (Sammarco et al. 2000). An in-frame stop codon 79bp into this intron resulted in a smaller, truncated AR protein. As expected, androgen binding was impaired for this receptor variant, but no additional functional assays were performed. Finally, an additional PAIS splicing mutation that has been identified was a C→G mutation in the last position of codon 886 and concomitant replacement of the final 33aa of the wild-type AR protein with 8 novel amino acids (PAIS-4 in Fig. 3) as a result of a cryptic splice-donor site (Hellwinkel et al. 2001). In this case, androgen binding capacity was impaired and no androgen-induced transcriptional activity was observed on an androgen responsive promoter-reporter gene.
Role of the AR in Prostate Cancer Development and Progression
The prostate is an androgen- and AR-dependent tissue. Prostate cancer (PCa) cells retain this androgen- and AR-dependent property. Androgen depletion therapy (ADT) is a collective term used to refer to surgical castration, pharmacologic castration via leutenizing hormone releasing hormone analogs, or administration of antiandrogens such as bicalutamide. These modes of ADT are the standard systemic treatments for locally advanced, relapsed, or metastatic PCa. Initially, these therapies are able to effectively inhibit AR transcriptional activity in PCa cells, leading to reduced expression of AR target genes such as prostate specific antigen (PSA) and tumor regression. However, changes eventually occur in the biology of tumors during ADT that allows resumption of tumor growth, and metastatic spread. This stage of the disease, termed castration-resistant prostate cancer (CRPCa), is a major cause of PCa morbidity and mortality. Therefore, this transition from a treatable, androgen-dependent disease to a lethal, castration-resistant disease has been an area of intense interest. Studies on the mechanisms of PCa progression during ADT have demonstrated that in nearly all cases resumed AR transcriptional activity is a critical event (Knudsen and Scher 2009). Therefore, as appears to be the case with other targeted cancer therapies, resistance of PCa to ADT usually occurs through changes in the drug target. Several important mechanisms of AR reactivation during ADT have been identified including AR point mutations, intratumoral androgen production, and AR protein expression/gene amplification. These mechanisms of AR reactivation have been reviewed in detail elsewhere (Agoulnik and Weigel 2006; Chen et al. 2009; Culig and Bartsch 2006; Heemers and Tindall 2007; Knudsen and Scher 2009; Pienta and Bradley 2006). Investigation of intratumoral androgen production mechanisms and modes of AR activation have led to the development of abiraterone acetate and MDV3100 as new therapies for men with CRPCa. Abiraterone is an inhibitor of the CYP17 enzyme, which catalyzes two steps in androgen biosynthesis. This drug blocks intra-tumor conversion of progesterone and adrenal androgens to DHT (Attard et al. 2009) and a recent Phase III trial demonstrated an overall survival benefit for abiraterone in men with metastatic CRPCa (de Bono et al. 2011). MDV3100 is a non-steroidal antiandrogen that can inhibit AR even under conditions of protein overexpression (Tran et al. 2009). Phase I/II clinical trials have demonstrated efficacy for MDV3100 in a subset of patients with CRPCa, and this drug is currently being tested in Phase III trials (Payton 2010; Scher et al. 2010). Overall, these clinical advances have served as confirmation that targeting AR signaling in CRPCa remains an important option for treatment of this stage of the disease.
Gain-of-Function AR Variants Discovered in the CWR22 PCa Model
It was recently discovered that AR alternative splicing can give rise to COOH-terminally truncated AR protein isoforms that lack the AR LBD. These AR isoforms are missing the region of the receptor that would be predicted to mediate responses to traditional ADT, as well as new AR-targeted therapies such as abiraterone and MDV3100. Key observations leading to this discovery were initially reported in 2002, when it was demonstrated that 22Rv1 cells expressed two separate AR protein species of approximately 112 kDa and 75–80 kDa (Tepper et al. 2002). By antibody mapping, it was demonstrated that the smaller 75–80kDa AR species was composed of the AR NTD and DBD, but lacked the AR LBD. Further biochemical characterization revealed that the AR ΔLBD isoform was constitutively nuclear and could bind DNA independent of androgens. Co-immunoprecipitation experiments indicated that the AR ΔLBD isoform did not interact with full-length AR, suggesting that this species functioned as an independent factor. In another study, Western blots with benign and cancerous prostate tissue demonstrated that AR immunoreactive species with a similar mobility to the AR ΔLBD isoform were frequently expressed in clinical PCa (Libertini et al. 2007). Together, these studies provided the first evidence that AR protein species lacking the LBD could play an important role in modulating PCa therapy resistance.
The mechanism proposed for synthesis of the AR ΔLBD protein species in PCa cells was via proteolytic degradation of full-length AR by calpain-2 (Libertini et al. 2007). This was based on the presence of a consensus calpain cleavage site in the AR hinge region and the observation that calpain-2 was able to cleave full-length AR protein in cell extracts derived from LNCaP and 22Rv1 cells, as well as the CRPCa CWR-R1 cell line which was also derived from the CWR22 xenograft model (Chen et al. 2010; Libertini et al. 2007). In addition, the full-length AR mRNA in 22Rv1 cells had been shown to be larger due to tandem duplication of AR exon 3, and this larger form of the AR appeared to have enhanced susceptibility to this mechanism of cleavage (Libertini et al. 2007). However, several recent studies have suggested that alternative splicing could also be an important contributor to the synthesis of truncated AR species lacking the AR LBD in these and other cells. This new concept was initially based on the observation of differential siRNA targeting of the various AR species in 22Rv1 cells (Dehm et al. 2008). For example, siRNA targeted to AR exon 7 abolished expression of full-length AR in 22Rv1 cells, but had no effect on the smaller 75–80kDa species. Conversely, siRNA targeted to AR exon 1 knocked down expression of all AR immunoreactive species in 22Rv1 cells. Functionally, androgen-dependent expression of AR target genes and androgen-dependent cell growth was shown to be attributable to full-length AR, whereas androgen-independent expression of AR target genes and constitutive, androgen-independent growth was shown to be supported by the smaller 75–80kDa species. These observations strongly suggested that different mRNA species encoded the different AR protein species observed in these cells, and provided the foundation for 3’-RACE and other approaches to identify their origin (Dehm et al. 2008; Guo et al. 2009; Hu et al. 2009; Hu et al. 2011; Marcias et al. 2010). These efforts led to the identification of a series of alternatively spliced AR mRNAs expressed in 22Rv1 cells that resulted from cryptic exons located within AR introns 2 and 3. The AR isoforms or variants encoded by these alternatively spliced mRNAs have been shown to function as constitutively active, ligand-independent transcription factors that can support the CRPCa phenotype in various model systems. Specific details of the individual AR isoforms that have been identified in models derived from the CWR22 xenograft are discussed in the following section. A major barrier to clarity regarding this emerging topic is the nomenclature that has been employed by different investigators, leading to instances where different names have been given to the same isoform (e.g. AR 1/2/3/2b, AR-V4, AR5, and ARV6 are all encoded by contiguously-spliced AR exons 1/2/3/2b; AR-V7 and AR3 are both encoded by contiguously-spliced AR exons 1/2/3/CE3; and ARv567es and AR-V12 are both encoded by skipping of exons 5–7). Also, there are instances where different isoforms have been given the same name (e.g. AR-V7 encoded by contiguously spliced AR exons 1/2/3/CE3 and ARV7 encoded by contiguously spliced AR exons 1/2/3/3/3’/4/5/6/7/8). Therefore, we will discuss these isoforms in the context of their exon constituency, and acknowledge all of the various names that have been given to these isoforms to date (Figure 4).
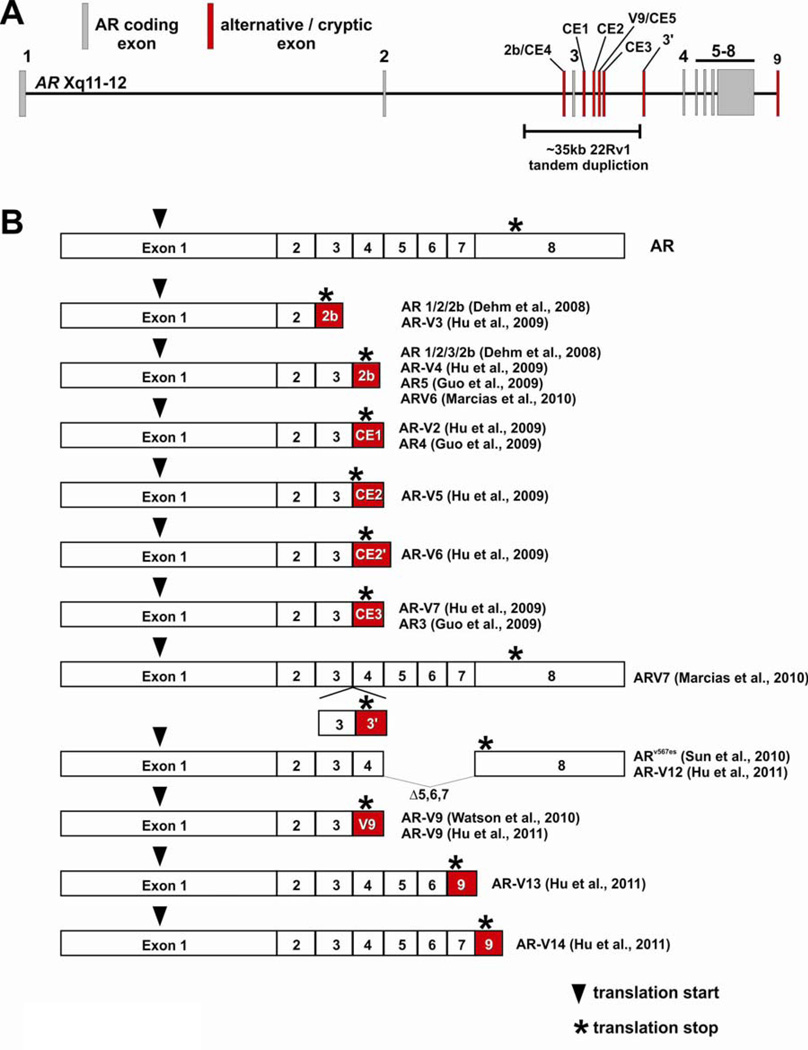
Figure 4.
Alternatively spliced AR isoforms identified in prostate cancer (PCa). (A) Schematic of the AR gene locus with locations of alternatively spliced, cryptic exons illustrated in red. The location of an intragenic tandem duplication identified in 22Rv1 cells is indicated. (B) Alternative splicing of cryptic exons in the AR locus or exon skipping gives rise to COOH-terminally truncated AR mRNA isoforms that encode constitutively active, ligand-independent transcription factors. CE2’ denotes the use of an alternative splice acceptor site for splicing of exon CE2 downstream from exon 3. Additional alternatively-spliced mRNA isoforms which have not been cloned or characterized are not illustrated in this figure, but are discussed in the text.
Splicing of AR exons 1/2/2b and 1/2/3/2b
AR exon 2b was discovered following 5’-RACE experiments with RNA derived from 22Rv1 cells (Dehm et al. 2008). AR exon 2b was shown to splice downstream of either AR exon 2 or 3, giving rise to truncated AR proteins containing the AR NTD, one or both zinc fingers of the AR DBD, and a short 11 amino acid COOH-terminal extension encoded by AR exon 2b. Expression of the 75–80kDa immunoreactive species in 22Rv1 cells was reduced upon transfection with exon 2b-specific siRNA, which corresponded with an inhibition of androgen-independent cell proliferation. These data confirmed that transcripts containing AR exon 2b were translated to functional proteins. Importantly, there was no effect of exon 2b siRNA on androgen-stimulated proliferation, further supporting the concept that truncated AR isoforms could support features of the CRPCa phenotype independent of full-length AR. Functionally, AR 1/2/2b and AR 1/2/3/2b were shown to function as constitutively active transcription factors in promoter-reporter assays. The AR 1/2/2b AR mRNA was also shown to be expressed in VCaP cells as well as the LuCaP 23.1 and 35 models of PCa progression.
Additional studies by other groups have confirmed the existence of mRNA species containing exon 2b in the 22Rv1 model system, but with variability in the precise 3’ composition of the transcript. For example, one transcript was reported with exon 2b (referred to as exon CE4 in the study) spliced between exons 2 and 3, with exon CE1 at the 3’ terminus (Hu et al. 2009). This isoform was termed AR-V3 in this study, and was proposed to encode a 53 aa COOH-terminal extension due to a TAA trinucleotide deletion in the sequence of CE4 compared with the sequence of exon 2b. The reason for this discrepancy is not clear, but could be due to procedural differences or the fact that 22Rv1 cells harbor 2 genomic copies of AR exon 2b (Li et al. 2011). Another transcript was also identified with exon 2b spliced between the duplicated copies of exon 3, with exon CE1 at the 3’ terminus (Hu et al. 2009). This configuration, termed AR-V4, matched the splicing pattern for the AR5 mRNA reported in a separate study (Guo et al. 2009). Importantly, AR5 mRNA expression was detected by RT-PCR at a low level in benign prostate tissue as well as PCa tissue. In yet another study, AR exon 2b (referred to as exon 2’ in that study) was shown to be spliced between the duplicated copies of exon 3 in the context of an mRNA species, termed V6, with contiguous splicing of AR exons 1–8 (Marcias et al. 2010). Together, these data indicate that many AR mRNAs in the 22Rv1 cell line contain exon 2b spliced after exon 2 or 3, with possible variation in the composition of the 3’ untranslated region of these transcripts.
Splicing of AR exons 1/2/3/CE1 and 1/2/3/3/CE1
Two AR mRNA species composed of contiguously spliced AR exons 1/2/3/CE1 (termed AR-V1) and AR exons 1/2/3/3/CE1 (termed AR-V2) were demonstrated to be expressed in 22Rv1 cells following the bioinformatic identification of AR exon CE1 as an expressed sequence tag mapping to AR intron 3 (Hu et al. 2009). The AR-V1 transcript composed of AR exons 1/2/3/CE1 was detected by quantitative RT-PCR at a higher level in RNA isolated from CRPCa tissue specimens vs. hormone naïve PCa. In another study, the AR 1/2/3/CE1 isoform was also detected in 22Rv1 cells, and termed AR4 (Guo et al. 2009). No studies have been performed to demonstrate translation of either of these mRNAs into endogenous proteins.
Splicing of AR exons 1/2/3/CE2
Two separate AR mRNA species composed of contiguously spliced AR exons 1/2/3/CE2 have been identified, with different locations for the splice acceptor site used for incorporation of exon CE2 into these mRNAs (Hu et al. 2009). As with the identification of AR exon CE1, exon CE2 was originally discovered by bioinformatic identification of an expressed sequence tag mapping to AR intron 3. The isoform generated from utilization of the most 5’ splice acceptor site in exon CE2, termed AR-V5, results in a protein predicted to encode a single aspartate residue following exon 3 coding sequence. The isoform generated from utilization of the most 3’ splice acceptor site in exon CE2, termed AR-V6, results in a protein predicted to encode a 6 amino acid COOH-terminal extension following exon 3 coding sequence. No studies have been performed to demonstrate translation of this mRNAs into an endogenous protein.
Splicing of AR exons 1/2/3/CE3
The best-characterized alternatively spliced AR isoform identified to date is encoded by contiguously splicing of AR exons 1/2/3/CE3. In one study, this isoform was named AR-V7 (Hu et al. 2009), and in another study this same isoform was named AR3 (Guo et al. 2009). As with AR exons CE1 and CE2, exon CE3 was originally discovered based on bioinformatic identification of an expressed sequence tag mapping to AR intron 3 (Hu et al. 2009). Importantly, AR 1/2/3/CE3 mRNA was detected in various clinical PCa specimens as well as normal prostate tissue (Hu et al. 2009). Quantitative RT-PCR demonstrated that the RNA levels of this isoform could predict biochemical recurrence following surgery (Hu et al. 2009). The 16aa COOH-terminal extension encoded by AR exon CE3 has been amenable to antibody development (Guo et al. 2009; Hu et al. 2009), and the utilization of these isoform-specific antibodies has revealed widespread protein expression of this isoform in cell lines, xenografts, and clinical specimens. Importantly, protein expression of this isoform was shown to be increased in CRPCa vs. hormone naïve PCa (Guo et al. 2009). Also, cytoplasmic levels of this isoform could predict biochemical recurrence following surgery (Guo et al. 2009). It is interesting to note that the AR 1/2/3/CE3 isoform is also expressed at the mRNA and protein level in benign prostate tissue, indicating that there may also be a role for this splicing event in normal cells (Guo et al. 2009). Functionally, AR 1/2/3/CE3 was demonstrated to function as a constitutively active, ligand-independent transcription factor that could induce a CRPCa growth phenotype in LNCaP cells grown in vitro and as xenografts in vivo (Guo et al. 2009). Interestingly, in one study, gene expression profiling of LNCaP cells transfected with an expression vector encoding AR 1/2/3/CE3 demonstrated that the ligand-independent transcriptional activity of this isoform could largely recapitulate the gene expression program activated by ligand-induced full-length AR, including the PSA gene (Hu et al. 2009). However, another study that used shRNAs to selectively knock-down the AR 1/2/3/CE3 or full-length AR isoforms in 22Rv1 and CWR-R1 cells demonstrated that AR 1/2/3/CE3 activated a significantly different gene expression program than ligand-activated AR (Guo et al. 2009). One differentially-regulated gene that was identified was AKT1 (Guo et al. 2009). The AR 1/2/3/CE3 isoform was shown to activate AKT1 expression at the mRNA and protein level, whereas full-length AR had no effect on AKT1 expression. Mechanistically, this could be attributed to the finding that AR 1/2/3/CE3 but not full-length AR could bind to two AREs located in the AKT1 promoter region. Overall, this body of work demonstrates that an AR variant derived from contiguous splicing of AR exons 1/2/3/CE3, termed AR-V7 or AR3, is a clinically validated AR isoform that could play an important role in supporting the CRPCa phenotype.
Insertion of an extra copy of exon 3 and a novel exon 3’ between AR exons 3 and 4
Additional alternatively-spliced AR isoforms have recently been cloned from 22Rv1 cells using a yeast functional assay to identify AR cDNAs with constitutive or ligand-induced transcriptional activity (Marcias et al. 2010). This approach led to the identification of an additional cryptic exon in AR intron 3 termed 3’. Sequencing of the isolated cDNA, which was named ARV7 in this study, demonstrated that exon 3’ spliced between an extra copy of AR exon 3 and exon 4, giving rise to a truncated AR isoform containing the entire AR NTD, the entire DBD, an extra zinc finger encoded by the extra copy of exon 3, and a novel 23 aa COOH-terminal extension encoded by exon 3’.
Gain-of-Function AR Variants Discovered in Other PCa Models
The identification of alternatively-spliced, truncated AR isoforms lacking the AR LBD in cell lines derived from the CWR22 PCa xenograft model, and extension of the findings to other models of PCa progression as well as human tissues demonstrated that this phenomenon may be a widespread mechanism of resistance to ADT therapies targeting the AR LBD. Indeed, examination of AR mRNA expression profiles in various other PCa models has led to the identification of additional AR mRNA species that are predicted to encode proteins that display constitutive, ligand-independent transcriptional activity. These isoforms and key details of their discovery are outlined below.
Splicing of AR exons 1/2/3/4/8 in LuCaP 86.2 and LuCaP 136 Xenografts
An AR mRNA isoform arising through skipping of exons 5–7, termed ARv567es, was discovered in the LuCap 86.2 and LuCaP 136 xenografts following examination of AR mRNA profiles via RT-PCR (Sun et al. 2010). Interestingly, in the LuCaP 86.2 xenograft, full-length AR mRNA did not appear to be expressed at either the mRNA or protein level, which is in contrast to 22Rv1 and other models where full-length AR expression levels remain high. No experiments have been performed to demonstrate that the ARv567es mRNA is translated to an endogenous protein in these xenograft models, although the xenografts do display high-level expression of a ~80kDa species that is immunoreactive with an AR NTD-directed antibody (Sun et al. 2010). Skipping of AR exons 5–7 was confirmed via RT-PCR in several additional xenograft models of PCa progression, patient samples of metastatic CRPCa, primary PCa, as well as benign prostate epithelium. These findings indicate that skipping of exons 5–7 by the splicing machinery is frequent, and can even occur in non-cancerous tissue. Functionally ARv567es was shown to act as a constitutively active, ligand-independent transcription factor that could support CRPCa cell growth in vitro and in vivo. Interestingly, ARv567es was also shown to interact with full-length AR and enhance its ligand-dependent and ligand-independent activity, perhaps through a mechanism of protein stabilization or even induction of full-length AR translocation to the nucleus in the absence of androgens.
Deep Sequencing Identifies Additional Alternative AR Splicing Patterns in VCaP Cells
The VCaP cell line has been an additional cell-based PCa model of interest for studying AR alternative splicing in PCa progression because these cells express a smaller 75–80kDa AR species that is recognized by antibodies directed to the AR NTD, but not the LBD (Dehm et al. 2008). An adaptation of the 3’-RACE method was recently described wherein 3’-RACE products produced using a forward primer spanning the AR exon 2/3 junction were directly sequenced using 454 and SOLiD next-generation sequencing platforms (Watson et al. 2010). This approach confirmed splicing of AR exons CE1 and CE3 downstream of exon 3 in VCaP cells. However, this approach also identified 4 additional isoforms, named AR-V8 through AR-V11, that resulted from exon 3 read-through, splicing of novel cryptic exons in AR intron 3, or use of a different splice acceptor site in exon CE3. Similar to previously-identified AR variants, these novel mRNAs would be predicted to encode variable-length COOH-terminal extensions following exon 3 sequence. Additional RNA species were identified in VCaP cells that appeared to arise through exon skipping, and splicing of a novel cryptic exon within AR intron 5. No full-length cDNAs were isolated from VCaP cells that corresponded to these splicing events detected by deep sequencing, so it is unclear whether these putative new AR isoforms represent splicing intermediates, splicing errors targeted for nonsense-mediated RNA decay, or bona fide alternatively spliced AR mRNAs that encode functional proteins. However, this work demonstrated the power of using next-generation sequencing technology for studying AR splicing events in PCa samples.
Deep Sequencing Identifies Alternative AR Splicing Patterns in Myc-CaP Cells
The development of the 3’-RACE/next-generation sequencing workflow using VCaP cells led to deployment of this approach in the Myc-CaP model of PCa. Myc-CaP cells were derived from prostate tumors that arose in mice expressing a prostate-specific myc-transgene (Watson et al. 2005). A recent study demonstrated the synthesis of four novel alternatively-spliced AR isoforms in these cells, which were named mAR-V1-4 (Watson et al. 2010). mARV1 was composed of contiguously spliced mouse exons 1/2/3, followed by read-through into intron 3. Remarkably, mAR-V2 and mAR-V4 were found to harbor novel exons located outside of the AR gene. One exon, located ~250 kb downstream of the AR locus, was shown to be spliced after AR exon 3 to yield the mAR-V2 isoform. The other exon, located ~1 Mb upstream of the AR locus, was shown to be spliced after AR exon 4 to yield mAR-V3 and mAR-V4. mAR-V3 differed from mAR-V4 by virtue of skipping of AR exon 3. Importantly, expression of mAR-V2 and mAR-V4 was confirmed at the protein level following isoform-specific knock-down of these species in Myc-CaP cells. Functional study of the mAR-V2 and mAR-V4 isoforms demonstrated that mAR-V4 was a constitutively active, ligand-independent transcription factor, and was constitutively localized to the nucleus. Conversely, mAR-V2 did not display transcriptional activity and was found to be localized predominantly to the cytoplasm. Consistent with these findings, ectopic expression of mAR-V4, but not mAR-V2, could support the growth of LNCaP xenografts in castrated mice. Interestingly, studies with the AR antagonist MDV3100 and siRNA directed to the full-length AR mRNA demonstrated that the CRPCa-promoting effects of mAR-V4 required functional full-length AR. Similar observations were made with the AR 1/2/3/CE3 isoform, indicating that truncated AR variants may not be able to take over the complete function of full-length AR under castrate conditions.
Genomic Tiling Arrays Identifies Alternative AR Splicing Patterns in Clinical CRPCa
Another novel workflow that has been developed to identify additional alternatively-spliced AR isoforms is based on selective linear amplification of sense RNA (SLASR) using an Exon 3-anchored forward primer, followed by detection of amplified products using an AR gene tiling array (Hu et al. 2011). This approach was able to detect synthesis of mRNAs containing AR Exons 2b/CE4, CE1, CE2, and CE3 in 22Rv1 cells, but also detected expression of a novel isoform termed AR-V9, which was produced by contiguous splicing of AR exons 1/2/3 and a novel cryptic exon termed CE5, located within AR intron 3. This exon configuration matched the AR-V9 isoform previously reported in VCaP cells (Watson et al. 2010). This approach also led to the identification of an additional novel AR exon expressed in RNA derived from CRPCa tissue specimens. This exon, termed “exon 9” was located downstream of AR exon 8, and was shown to be the most 3’ exon in three discrete AR mRNA species, termed AR-V12 (AR exons 1/2/3/4/8/9, which would encode the ARv567es variant), AR-V13 (AR exons 1/2/3/4/5/6/9), and AR-V14 (AR exons 1/2/3/4/5/6/7/9) (Hu et al. 2011). Functional evaluation demonstrated that AR-V9 and AR-V12 (identical to ARv567es) displayed varying degrees of constitutive, ligand-independent transcriptional activity depending on the cell line studied, and AR-V9 was shown to function independently of full-length AR in LNCaP cells.
Regulation of AR Splicing and Synthesis of Truncated AR Isoforms
Truncated AR mRNA and/or protein expression, in particular the AR-V7/AR3 isoform resulting from splicing of AR exons 1/2/3/CE3 and the ARv567es isoform, resulting from skipping of AR exons 5–7, has been shown to occur in normal prostate tissues, indicating that the mere presence of these factors is unlikely to be pathogenic. This observation also suggests that there may be normal functions attributable to AR alternative splicing and synthesis of truncated AR isoforms. This possibility is also supported by studies of the AR45 isoform, which is synthesized at the mRNA level in normal tissues of various origin. However, a consistent finding has been an increased level of expression of alternatively-spliced, truncated AR isoforms in CRPCa cells vs. androgen-dependent PCa cells (Dehm et al. 2008; Guo et al. 2009; Hu et al. 2009; Sun et al. 2010). This suggests that ADT exerts a selective pressure favoring the expression of constitutively active, truncated AR isoforms. One possible mechanism for this increase in truncated AR isoform expression could be changes in expression or activity of factors that regulate AR splicing patterns. In this case, plasticity in these splicing patterns would be expected, which represents an opportunity to develop therapies to modify aberrant splicing patterns. However, studies of alternatively-spliced AR variants synthesized in individuals with AIS has demonstrated that alterations in AR splicing patterns can also be stable due to sequence or structural changes in the genomic DNA template, which represents a “new normal” for that cell. With these concepts in mind, our laboratory recently reported that the disruptions in AR splicing patterns in 22Rv1 cells was linked to a ~35 kb intragenic tandem duplication of a genomic segment encompassing AR exon 3 and many of the cryptic AR exons that are expressed in these and other cells (Li et al. 2011). These include exons 2b/CE4, CE1, CE2, CE3, CE5, as well as the 4 additional exons that were recently identified in the AR-V8-V11 isoforms expressed in VCaP cells. This rearrangement was not observed in the CWR22Pc cell line, which is an androgen-dependent sub-line developed from the original CWR22 xenograft (Li et al. 2011). However during long term castration, CWR22Pc cells progressed to a CRPCa growth phenotype that was marked by emergence of a rare sub-population of cells harboring this gene rearrangement, and emergence of high-level truncated AR expression (Li et al. 2011). This finding demonstrates that alterations in the AR gene may underlie disruptions in mRNA splicing patterns observed in CRPCa cells such as 22Rv1. This concept is also supported by the observation that the mAR-V4 isoform results from contiguous splicing of mouse AR exons 1/2/3/4, and a cryptic exon located ~1Mb upstream of the AR locus, which, similar to the AR 1/2/3/2b isoform, likely requires alterations in gene architecture to be synthesized (Watson et al. 2010). Remarkably, the AR gene in Myc-CaP cells, where mAR-V4 was discovered, is frequently amplified, indicating that amplification may also be associated with alterations in gene architecture (Watson et al. 2005). By examining high-resolution gene copy number data derived from clinical CRPCa specimens, we observed frequent copy number imbalances along the length of amplified AR genes, which is a finding consistent with complex gene rearrangements (Li et al. 2011). Together, these data suggest that changes in the genomic DNA template may underlie disrupted AR splicing patterns and pathologic synthesis of truncated, constitutively active AR isoforms in CRPCa cells. Further studies are required for a more complete understanding of the mechanisms regulating synthesis of alternatively-spliced, truncated AR isoforms in PCa.
Perspectives and Future Directions
Various alternatively-spliced AR isoforms have been identified in pathologic conditions including AIS and CRPCa. Alternatively spliced AR isoforms have also been observed in non-cancerous tissues; however, it remains to be determined whether these have a role in normal cellular processes. In CRPCa, a common theme is the synthesis of AR isoforms with a modular organization consisting of the AR NTD, the AR DBD, and short COOH-terminal extensions of variable length and sequence. Constitutive, ligand-independent transcriptional activity of these isoforms represents a conceptually simple mechanism for the development of resistance to PCa therapies that target the LBD. However, several studies have clearly demonstrated that additional understanding is required in this area. For example, there is evidence to suggest that the exons encoding the COOH-terminal extensions of truncated AR isoforms may provide important regulatory information in addition to harboring in-frame translation stop codons. This is particularly important because replacement of AR exon 4 with an alternative cryptic exon would disrupt the well-characterized AR nuclear localization signal (NLS) encoded by correctly-spliced exons 3 and 4. Future studies are required to resolve this issue, and determine the molecular basis for the differences in nuclear localization of the different AR variants that have been reported (Sun et al. 2010; Watson et al. 2010). Similarly, differences in transcriptional activity of different AR variants have been reported (Dehm et al. 2008; Guo et al. 2009; Hu et al. 2011; Watson et al. 2010). It will also be important to understand whether these different COOH-terminal extensions adjacent to the AR DBD can influence DNA-binding activity. The mechanisms by which full-length AR could influence AR isoform function (and vice versa) will also require further investigation, particularly in light of the findings that the ARv567es isoform interacts with wild-type AR (Sun et al. 2010) and that interfering with full-length AR expression or activity affects the function of the ectopically expressed AR-V7 and mAR-V4 isoforms (Watson et al. 2010). However, these findings contrast with studies in models that naturally express truncated AR isoforms. For example, the LuCaP 86.2 xenograft model does not appear to express full-length AR, indicating that normal AR activity is not required for ARv567es function (Sun et al. 2010). Similarly, knock-down of full-length AR in 22Rv1 cells does not affect androgen-independent proliferation or constitutive activation of AR target genes supported by truncated AR isoforms (Dehm et al. 2008). Finally, it will be critical in future studies to evaluate quantitatively the levels of alternatively-spliced AR isoforms relative to wild-type AR to differentiate between background splicing byproducts and significant alterations in AR splicing patterns that could affect response to PCa therapies targeting the AR LBD.
Acknowledgements
S.M.D. is supported by a Prostate Cancer Foundation Young Investigator Award, a Department of Defense Prostate Cancer Research Program New Investigator Award (PC094384), and the National Cancer Institute (CA141011 and Cancer Center Support Grant P30 077598). S.M.D. is a Masonic Scholar of the Masonic Cancer Center, University of Minnesota. D.J.T. is supported by the National Cancer Institute (CA121277, CA125747, and CA91956) and the T.J. Martell Foundation.
References
- Agoulnik IU, Weigel NL. Androgen receptor action in hormone-dependent and recurrent prostate cancer. J Cell Biochem. 2006;99:362–372. [Google Scholar]
- Ahrens-Fath I, Politz O, Geserick C, Haendler B. Androgen receptor function is modulated by the tissue-specific AR45 variant. FEBS J. 2005;272:74–84. doi: 10.1111/j.1742-4658.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol Cell Biol. 2009;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard G, Reid AH, Olmos D, de Bono JS. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–4940. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear Receptor Structure: Implications for Function. Annu Rev Physiol. 2006;69 doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Brinkmann AO. Molecular basis of androgen insensitivity. Mol Cell Endocrinol. 2001;179:105–109. doi: 10.1016/s0303-7207(01)00466-x. [DOI] [PubMed] [Google Scholar]
- Bruggenwirth HT, Boehmer AL, Ramnarain S, Verleun-Mooijman MC, Satijn DP, Trapman J, Grootegoed JA, Brinkmann AO. Molecular analysis of the androgen-receptor gene in a family with receptor-positive partial androgen insensitivity: an unusual type of intronic mutation. Am J Hum Genet. 1997;61:1067–1077. doi: 10.1086/301605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert L, Van Tilborgh N, Claessens F. Interplay between two hormone-independent activation domains in the androgen receptor. Cancer Res. 2006;66:543–553. doi: 10.1158/0008-5472.CAN-05-2389. [DOI] [PubMed] [Google Scholar]
- Chamberlain NL, Whitacre DC, Miesfeld RL. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem. 1996;271:26772–26778. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- Chen H, Libertini SJ, Wang Y, Kung HJ, Ghosh P, Mudryj M. ERK regulates calpain 2-induced androgen receptor proteolysis in CWR22 relapsed prostate tumor cell lines. J Biol Chem. 2010;285:2368–2374. doi: 10.1074/jbc.M109.049379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10:981–991. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culig Z, Bartsch G. Androgen axis in prostate cancer. J Cell Biochem. 2006;99:373–381. doi: 10.1002/jcb.20898. [DOI] [PubMed] [Google Scholar]
- Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci. 2008;121:957–968. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr, Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Regan KM, Schmidt LJ, Tindall DJ. Selective role of an NH2-terminal WxxLF motif for aberrant androgen receptor activation in androgen depletion independent prostate cancer cells. Cancer Res. 2007;67:10067–10077. doi: 10.1158/0008-5472.CAN-07-1267. [DOI] [PubMed] [Google Scholar]
- Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ. Molecular regulation of androgen action in prostate cancer. J Cell Biochem. 2006;99:333–344. doi: 10.1002/jcb.20794. [DOI] [PubMed] [Google Scholar]
- Ding D, Xu L, Menon M, Reddy GP, Barrack ER. Effect of a short CAG (glutamine) repeat on human androgen receptor function. Prostate. 2004;58:23–32. doi: 10.1002/pros.10316. [DOI] [PubMed] [Google Scholar]
- Ding D, Xu L, Menon M, Reddy GP, Barrack ER. Effect of GGC (glycine) repeat length polymorphism in the human androgen receptor on androgen action. Prostate. 2005;62:133–139. doi: 10.1002/pros.20128. [DOI] [PubMed] [Google Scholar]
- Estebanez-Perpina E, Moore JM, Mar E, Delgado-Rodrigues E, Nguyen P, Baxter JD, Buehrer BM, Webb P, Fletterick RJ, Guy RK. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J Biol Chem. 2005;280:8060–8068. doi: 10.1074/jbc.M407046200. Epub 2004 Nov 8024. [DOI] [PubMed] [Google Scholar]
- Ferro P, Catalano MG, Dell'Eva R, Fortunati N, Pfeffer U. The androgen receptor CAG repeat: a modifier of carcinogenesis? Mol Cell Endocrinol. 2002;193:109–120. doi: 10.1016/s0303-7207(02)00104-1. [DOI] [PubMed] [Google Scholar]
- Gottlieb B, Beitel LK, Wu JH, Trifiro M. The androgen receptor gene mutations database (ARDB): 2004 update. Hum Mutat. 2004;23:527–533. doi: 10.1002/humu.20044. [DOI] [PubMed] [Google Scholar]
- Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gampe RT, Jr, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell. 2004;16:425–438. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Hellwinkel OJ, Bull K, Holterhus PM, Homburg N, Struve D, Hiort O. Complete androgen insensitivity caused by a splice donor site mutation in intron 2 of the human androgen receptor gene resulting in an exon 2-lacking transcript with premature stop-codon and reduced expression. J Steroid Biochem Mol Biol. 1999;68:1–9. doi: 10.1016/s0960-0760(98)00157-5. [DOI] [PubMed] [Google Scholar]
- Hellwinkel OJ, Holterhus PM, Struve D, Marschke C, Homburg N, Hiort O. A unique exonic splicing mutation in the human androgen receptor gene indicates a physiologic relevance of regular androgen receptor transcript variants. J Clin Endocrinol Metab. 2001;86:2569–2575. doi: 10.1210/jcem.86.6.7543. [DOI] [PubMed] [Google Scholar]
- Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011 doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla M, Feve M, Kessler P, Lapouge G, Erdmann E, Serra S, Bergerat JP, Ceraline J. A splicing variant of the androgen receptor detected in a metastatic prostate cancer exhibits exclusively cytoplasmic actions. Endocrinology. 2007;148:4334–4343. doi: 10.1210/en.2007-0446. [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117.. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertini SJ, Tepper CG, Rodriguez V, Asmuth DM, Kung HJ, Mudryj M. Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res. 2007;67:9001–9005. doi: 10.1158/0008-5472.CAN-07-1072. [DOI] [PubMed] [Google Scholar]
- Lim J, Ghadessy FJ, Yong EL. A novel splice site mutation in the androgen receptor gene results in exon skipping and a non-functional truncated protein. Mol Cell Endocrinol. 1997;131:205–210. doi: 10.1016/s0303-7207(97)00109-3. [DOI] [PubMed] [Google Scholar]
- Lubahn DB, Brown TR, Simental JA, Higgs HN, Migeon CJ, Wilson EM, French FS. Sequence of the intron/exon junctions of the coding region of the human androgen receptor gene and identification of a point mutation in a family with complete androgen insensitivity. Proc Natl Acad Sci U S A. 1989;86:9534–9538. doi: 10.1073/pnas.86.23.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcias G, Erdmann E, Lapouge G, Siebert C, Barthelemy P, Duclos B, Bergerat JP, Ceraline J, Kurtz JE. Identification of novel truncated androgen receptor (AR) mutants including unreported pre-mRNA splicing variants in the 22Rv1 hormone-refractory prostate cancer (PCa) cell line. Hum Mutat. 2010;31:74–80. doi: 10.1002/humu.21138. [DOI] [PubMed] [Google Scholar]
- Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, et al. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- Payton S. Prostate cancer: MDV3100 has antitumor activity in castration-resistant disease. Nat Rev Urol. 2010;7:300. doi: 10.1038/nrurol.2010.69. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–1671. doi: 10.1158/1078-0432.CCR-06-0067. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Ris-Stalpers C, Kuiper GG, Faber PW, Schweikert HU, van Rooij HC, Zegers ND, Hodgins MB, Degenhart HJ, Trapman J, Brinkmann AO. Aberrant splicing of androgen receptor mRNA results in synthesis of a nonfunctional receptor protein in a patient with androgen insensitivity. Proc Natl Acad Sci U S A. 1990;87:7866–7870. doi: 10.1073/pnas.87.20.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris-Stalpers C, Verleun-Mooijman MC, de Blaeij TJ, Degenhart HJ, Trapman J, Brinkmann AO. Differential splicing of human androgen receptor pre-mRNA in X-linked Reifenstein syndrome, because of a deletion involving a putative branch site. Am J Hum Genet. 1994;54:609–617. [PMC free article] [PubMed] [Google Scholar]
- Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek SR, Jr, et al. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc Natl Acad Sci U S A. 2001;98:4904–4909. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammarco I, Grimaldi P, Rossi P, Cappa M, Moretti C, Frajese G, Geremia R. Novel point mutation in the splice donor site of exon-intron junction 6 of the androgen receptor gene in a patient with partial androgen insensitivity syndrome. J Clin Endocrinol Metab. 2000;85:3256–3261. doi: 10.1210/jcem.85.9.6815. [DOI] [PubMed] [Google Scholar]
- Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, Rathkopf D, Shelkey J, Yu EY, Alumkal J, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Toft DO. Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol Endocrinol. 2008;22:2229–2240. doi: 10.1210/me.2008-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper CG, Boucher DL, Ryan PE, Ma AH, Xia L, Lee LF, Pretlow TG, Kung HJ. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62:6606–6614. [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Ellwood-Yen K, King JC, Wongvipat J, Lebeau MM, Sawyers CL. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res. 2005;65:11565–11571. doi: 10.1158/0008-5472.CAN-05-3441. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Chen K, Haendler B, McDonald TV, Bian JS. Stimulation of N-terminal truncated isoform of androgen receptor stabilizes human ether-a-go-go-related gene-encoded potassium channel protein via activation of extracellular signal regulated kinase 1/2. Endocrinology. 2008;149:5061–5069. doi: 10.1210/en.2007-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]