Abstract
Epigenetic mechanisms have long been associated with the regulation of gene-expression changes accompanying normal neuronal development and cellular differentiation; however, until recently these mechanisms were believed to be statically quiet in the adult brain. Behavioral neuroscientists have now begun to investigate these epigenetic mechanisms as potential regulators of gene-transcription changes in the CNS subserving synaptic plasticity and long-term memory (LTM) formation. Experimental evidence from learning and memory animal models has demonstrated that active chromatin remodeling occurs in terminally differentiated postmitotic neurons, suggesting that these molecular processes are indeed intimately involved in several stages of LTM formation, including consolidation, reconsolidation and extinction. Such chromatin modifications include the phosphorylation, acetylation and methylation of histone proteins and the methylation of associated DNA to subsequently affect transcriptional gene readout triggered by learning. The present article examines how such learning-induced epigenetic changes contribute to LTM formation and influence behavior. In particular, this article is a survey of the specific epigenetic mechanisms that have been demonstrated to regulate gene expression for both transcription factors and growth factors in the CNS, which are critical for LTM formation and storage, as well as how aberrant epigenetic processing can contribute to psychological states such as schizophrenia and drug addiction. Together, the findings highlighted in this article support a novel role for epigenetic mechanisms in the adult CNS serving as potential key molecular regulators of gene-transcription changes necessary for LTM formation and adult behavior.
Keywords: acetylation, amygdala, behavior, DNA methylation, epigenetics, hippocampus, histone, histone deacetylases, histone methylation, long-term memory, prefrontal cortex
A plausible role for epigenetic mechanisms in human learning & remembering
Memory is the basis of the entire human psyche. It comprises the many processes by which an organism takes in information presented as environmental stimuli and converts that information to stored knowledge, capable of being recalled at any time to make executive decisions, form a narrative or generate a comprehensive picture of the world. The general classifications for memory include sensory memory (SM), short-term memory (STM) and long-term memory (LTM) [1]. Information from the surrounding environment is continuously presented first to the senses, and what each modality registers is defined as SM. The initial sensory port into the conscious mind acts as a buffering system for our focus, as only the percentage of SM which is attended to will be further translated into STM. Transient in nature, STM quickly decays if it is not consolidated and converted into LTM [2]. After LTM is formed, these memories can be recalled from long-term storage to act as perspective on additional environmental influences, participate in decision-making based on acquired knowledge, and subsequently influence behavioral responses. Recall of LTM to supply current thought processes is so fluid a phenomenon that, as humans, we experience this as a smooth stream of consciousness. Behavioral neuroscientists are interested in the biological basis of these processes, and one major goal in the learning and memory field has been to elucidate the molecular events that are triggered by learning to allow for the conversion of memory from the working short term into stored long term for later recall.
Epigenetic modifications represent an appealing candidate gene-transcription mechanism for accomplishing this task. Traditionally, the field of epigenetics has been explored primarily by developmental biologists interested in understanding how the regulation of gene-expression patterns influence cellular differentiation and mitotic maintenance. However, these epigenome-regulating mechanisms may have implications well beyond the process of early development. In fact, an emerging hypothesis is that such epigenetic modifications may continue to occur in the adult nervous system to mediate neuronal functions. As a consequence, researchers have begun to investigate whether chromatin structure regulation via DNA methylation and post-translational histone modifications could be responsible for gene transcriptional changes in the adult CNS that are necessary to consolidate and recall stored memories [3–6]. Indeed, inquiries into how chromatin structure is regulated in the CNS have now revealed that epigenetic modifications can be triggered by learning experiences to contribute to the process of LTM formation and to mediate synaptic long-term potentiation (LTP), the cellular correlate of LTM [3–6]. This newly hypothesized role for epigenetic mechanisms in the adult CNS has been characterized in the context of memories, which the brain retains long after the learning experience is first introduced.
The effects of histone and DNA modifications are dynamic and reversible, suggesting that the epigenome can be quickly altered in response to experiential stimuli to regulate the expression of memory-permissive genes, including immediate–early genes (IEGs) that are necessary for the synaptic plasticity accompanying active learning. Importantly, certain regulatory changes to the functioning epigenome can stably persist throughout the lifespan, a detail that might explain how patterns of stored memories can remain unchanged and dormant for extended periods of time until the conscious brain demands their recall. It follows, then, that certain behavioral patterns, emotional responses and even psychiatric disturbances can be attributed to the profound influence that experiential stimuli have on the functioning epigenome. In this article, we will focus on the role of epigenetic mechanisms in the process of converting STM to LTM, which requires gene-transcription activity in adult neurons during memory consolidation. In addition, we will highlight studies investigating how aberrant epigenetic processing can contribute to psychological states such as schizophrenia and drug addiction, and alter adult behavior.
A brief overview of epigenetic mechanisms in LTM formation
Epigenetic modifications typically encompass alterations to chromatin structure by the addition of functional groups either to histone proteins or directly onto the DNA. In turn, these epigenetic modifications affect the accessibility of transcription factors and enhancer element binding proteins to gene promoter sites. By convention, the restructuring of chromatin, or chromatin remodeling, impacts transcriptional regulation of genes without altering the DNA sequence [7]. Thus, regulation of chromatin structure represents the functional interface between the genome and transcription factors, which must bind consensus sequences within DNA in order to facilitate mRNA expression [8]. Epigenetic modifications are believed to be dynamic, and have defining properties, which make them differentially suited for acting as regulators of chromatin structure in memory.
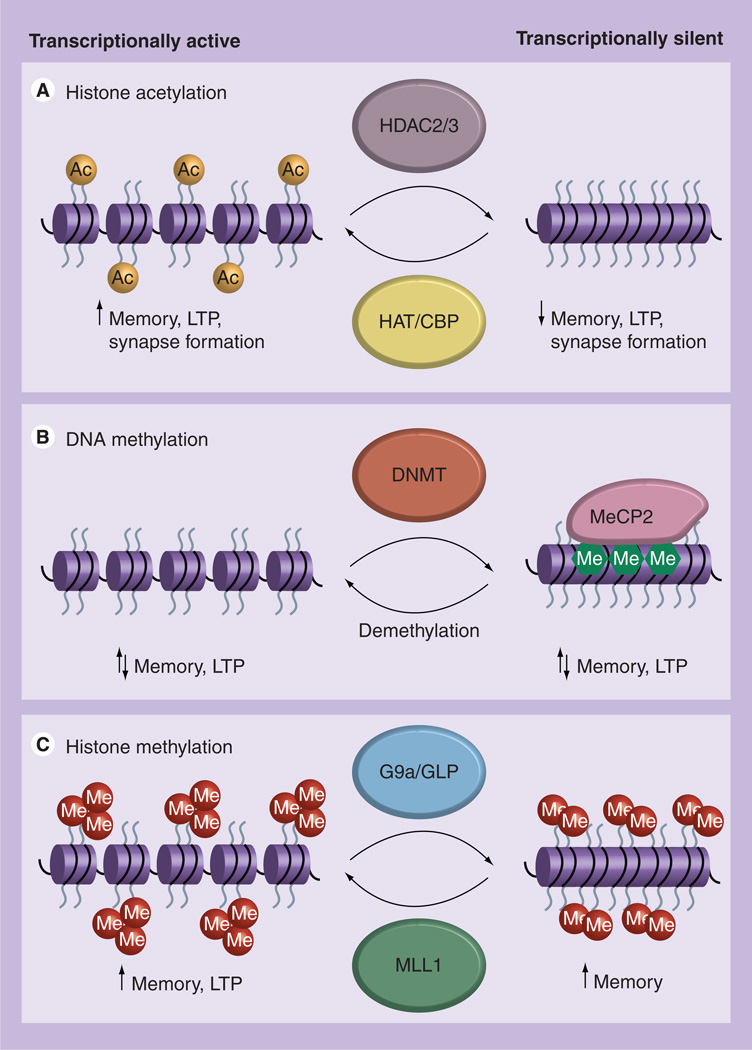
The unfolding and folding of chromatin results from enzymatic processes, which include the addition or removal of acetylation, phosphorylation, ubiquitination or sumoylation functional groups on amino acid residues at the unstructured N-terminus tails of histone proteins (H3 and H4) [8]. Histones H3 and H4 form a heterodimer, which combines with another heterodimer of histones H3 and H4 to form a tetramer. This tetramer combines with another tetramer composed of two histone H2A and H2B homodimers to form the compact octamer nucleosome core. In mature neurons, the addition of these functional groups to histone tails is generally transient and can be easily reversed and flexible in response to environmental stimuli [8]. As a reference, Figure 1 illustrates an overview of the specific epigenetic modifications that occur in mature neurons and are believed to play a role in LTM, LTP and synapse formation. Furthermore, the paragraphs below describe in brief detail the major epigenetic modifications in the adult CNS found to play a major role in the process of LTM formation and its cellular correlate, LTP.
Figure 1. Epigenetic modifications in mature hippocampal neurons during memory formation and storage.
(A) Histone acetylation mediated by CBP HAT activity contributes to long-term memory storage, LTP and synapse formation. Blockade of histone deacetylation, as well as indications of nonhistone protein deacetylation, via pharmacological inhibition of HDAC2 and/or HDAC3 is likely to result in a hyperacetylated chromatin structure state, which leads to an enhancement in memory, LTP and increased synapse number. (B) Alterations in both DNA methylation and DNA demethylation have been demonstrated to occur at specific memory-related gene promoters in response to learning and synaptic activity. Studies involving loss of DNMT1 or DNMT3a activity indicate that both DNA methylation and DNA demethylation are important to the process of memory storage and LTP. MeCP2 has been demonstrated to bind methylated DNA to facilitate both active-gene transcription and gene silencing during memory formation. (C) Learning-induced changes in histone methylation represented by the active transcription mark, histone H3 lysine 4 trimethylation (H3K4me3), and the repressive transcription mark, histone H3 lysine 9 dimethylation (H3K9me2), support the idea that both gene activation and gene silencing are necessary for memory formation. Studies utilizing transgenic mice and pharmacological inhibitors indicate that alterations to H3K4me3, via MLL1, or H3K9me2 via G9a/GLP, lead to memory deficits as well as decreased LTP.
DNMT: DNA methyltransferase; HAT: Histone acetyltransferase; HDAC: Histone deacetylase; LTP: Long-term potentiation; MeCP2: Methyl-CpG binding protein 2.
Histone acetylation
In the context of memory formation and LTP, acetylation and deacetylation of histone tails have been primarily characterized in the H3 and H4 classes of histone proteins [9–25]. For example, the addition of acetyl groups to histone H3 and H4 tails by histone acetyltransferases (HATs) neutralizes the positive charge of the basic histone octamer and repels the negatively charged backbone of DNA, thus relieving chromatin compaction and promoting gene expression necessary for synaptic plasticity, LTP and memory formation. The interaction between DNA and the histone octamer is what keeps DNA tightly compacted. Therefore, the direct consequence of HAT activity is a functional conversion from heterochromatin, the closed form of chromatin, to euchromatin, the open form of chromatin, which indicates transcriptional activation of genes. A well-known example of a protein with HAT activity is the transcriptional coactivator p300 also known as cAMP response element binding (CREB) binding protein (CBP), which interacts with the co-p300/CBP-associated factor (PCAF). Interestingly, the coactivator p300/CBP/PCAF complex mediates acetylation of the amino terminal ends of all core histone proteins as well as other nonhistone transcription Y-related factors, which results in active transcription [26].
Conversely, removal of the negatively-charged acetyl group via histone deacetylase (HDAC) activity allows the DNA to re-establish a transcriptionally closed, or heterochromatic state. Interestingly, when global HDAC inhibitors such as suberoylanilide hydroxamic acid (SAHA; also known as Vorinostat) or sodium butyrate are infused into the hippocampus or amygdala during fear learning, LTM formation for this experience is improved and synaptic plasticity (LTP) at the cellular level is also enhanced. Moreover, while evidence supports that histone acetylation mediates learning-induced transcriptional regulation, the investigation of the specific HDAC isoforms involved in chromatin modifications in multiple memory systems are in the early stages of discovery.
Early investigations into the particular roles of HDAC isoforms at gene promoters in mature neurons have revealed that some HDACs (HDAC1 and HDAC5) may be positive regulators of neuronal responses whereas others (HDAC2 and HDAC3) are negative regulators [16,27–29]. For example, recent investigations by Koshibu et al. implicate protein phosphatase 1 (PP1) as a key molecule in orchestrating chromatin modifications through its interaction with HDAC1 at transcriptionally silent gene promoters [30]. In addition, HDAC5 has been implicated in the maintenance of the histone deacetylated state in brain regions such as the nucleus accumbens (NAc), the reward center of the adult brain [31]. Indeed, disruption of HDAC5 activity in the NAc results in a hypersensitive response to chronic drug abuse and stress in rodents [31].
In relation to memory formation, HDAC2 has been implicated in the regulation of histone acetylation levels in the hippocampus during fear memory formation. Guan et al. established that SAHA, a HDAC inhibitor specific to HDAC1, 2 and 6, improved memory and synaptic plasticity [16]. The authors were able to eliminate the role of HDAC6 in SAHA effects by synthesizing a novel HDAC6-specific inhibitor, WT-161. Administration of this new compound had no effect on fear–memory formation. In addition, the authors investigated the specific role of HDAC1 and 2 via genetic knockout and overexpressor mice models. Interestingly, they found that HDAC2 overexpression produced decreased dendritic spine density, synapse number, synaptic plasticity and attenuated fear–memory formation compared with HDAC1 overexpression. HDAC2 deficiency resulted in an increased synapse number and enhanced memory formation that was similar to effects observed with the HDAC inhibitor SAHA. Crucially, HDAC2 deficiency was not associated with gross changes in neuronal morphology or behavior, suggesting that HDAC2 can be safely targeted without broadly disrupting neuronal physiology.
In addition to HDAC2, HDAC3 has recently been defined as a negative regulator of LTM formation. McQuown et al. demonstrated that HDAC3 is part of a corepressor complex that has direct interactions with Class II HDACs that may be important for its molecular and behavioral consequences [29]. Intriguingly, increased acetylation levels by CBP HAT activity are often observed at the promoter regions of IEGs as a priming mechanism for the regulation of these genes, which are rapidly expressed in response to experience-driven stimuli [11,32]. Thus, pharmacologically manipulating histone acetylation levels via HDAC inhibitors represents a powerful tool for globally upregulating gene transcription in the adult CNS to promote memory formation and may potentially serve as a viable therapeutic target for treatment of cognitive impairments associated with neurological disorders.
DNA methylation
In addition to acetylation and deacetylation of histone proteins, DNA methylation is another epigenetic modification that has been implicated as a crucial mediator of LTM formation. DNA methylation is catalyzed by a group of DNA methyltransferase (DNMT) enzymes including DNMT1, DNMT3A and DNMT3B, which have been found to be highly expressed in the adult CNS [33–35] and are subject to activity-dependent regulation in the CNS [34]. In addition, in cortical neuronal cultures, altering DNA methylation levels using the DNMT inhibitor 5-azadeoxycytidine results in decreased expression of the memory-related gene, REELIN [34]. Thus, the presence of DNMT mRNA and protein, along with data demonstrating that DNMT activity is dynamically regulated within the CNS, supports DNA methylation changes in the normal functioning adult CNS.
DNMT1, DNMT3A and DNMT3B are involved in both the maintenance and de novo methylation of DNA [33–35] and mediate DNA methylation via the transfer of a methyl group from the donor S-adenosylmethionine to the 5′-position of the pyrimidine ring of a cytosine residue that is covalently linked to an adjacent guanine residue (CpG site) in the backbone of DNA [36]. CpG-rich regions of DNA are termed ‘CpG islands’, which are found within inactive promoter regions [7,37–40]. The methylated CpG residues are docking sites for proteins such as methyl CpG binding protein-2 (MeCP2), which contains the methyl-binding domain [41]. MeCP2 binds to methylated DNA and this interaction is typically associated with recruitment of HDAC proteins to further facilitate suppression of gene transcription [42–44].
Although DNA methylation at CpG islands is thermodynamically very stable and occurs in inactive gene promoter regions, there exist several lines of evidence to suggest that this process is reversible in terminally differentiated neurons. In this regard, several DNA demethylating mechanisms in mature neurons have been proposed. One possibility, which still remains unclear, is that the DNMT enzymes themselves contribute to the DNA demethylation process [35]. Loss of DNMT activity in the hippocampi of DNMT1 or DNMT3A knockout mice results in the animals’ inability to form new memories, indicating that DNMT activity is of crucial importance in the process of LTM formation, either through the active methylation of memory-repressive genes via DNMT or DNMT participation in the demethylation of key memory-permissive genes.
Another potential mechanism for the active demethylation of DNA in mature neurons is the deamination of 5-methylcytosine to thymine, which triggers normal base-excision repair mechanisms. Indeed, research findings have also proposed that members of the Gadd45 family, known to be involved in DNA repair, are crucial regulators of DNA demethylation in the CNS [45,46]. Moreover, 5-methylcytosine can be hydroxylated by the TET enzymes to the 5-hydroxymethylcytosine form; base-excision repair mechanisms might again participate in restoring the original unmethylated cytosine [47–50]. However, the key players involved in dynamic DNA demethylation changes in mature neurons in the context of memory formation and storage are unexplored at present. Nonetheless, active DNA methylation and DNA demethylation support the idea that these epigenetic markings are not intractably set in place in the adult CNS by early-life experiences and can be further manipulated and reversed by learning events to influence an organism’s behavior and adaptation to future stimuli.
Histone methylation
Histone methylation is another well-characterized post-translational modification of histones that was discovered over 40 years ago [51]. Unlike histone acetylation and phosphorylation, histone methylation can have differing effects on gene-transcription activity depending on the lysine residue of the histone tail being modified within the cell. An additional level of complexity is present because each lysine residue can add up to three methyl residues, which can exist in three states: mono-, di-, and tri-methylated mediated by histone methyltransferases (HMTs). These lysine methylated states can have disparate effects on the transcriptional machinery and are differentially distributed across the chromatin fibers. For example, the trimethylated form of the lysine 4 residue of histone H3 (H3K4me3) is associated with transcriptional activation sites, whereas the monomethylated form at the same lysine 4 site is associated with enhancer regions that are at a distance from the transcriptional start sites of the genes [52,53]. Another noteworthy example of methylation disparity is observed with the monomethylated marks at lysine 9 and 27 of histone H3 and lysine 20 of histone H4 that are associated with active gene transcription, whereas the di- and tri-methylated forms at lysine 9 (H3K9me2 and H3K9me3) are associated with gene repression [45]. The specific pattern of these modifications affects the assembly of the transcriptional machinery, differentially working in both the recruitment and inhibition of transcription factors or DNMTs at the gene promoter sites [8,54,55]. The recruitment of DNMT enzymes to methylated histone lysine sites is a mechanism by which DNA methylation and histone methylation operate in tandem to regulate chromatin structure [8,54,55].
Histone methylation was once considered to be as persistent of an epigenetic mark as DNA methylation [37]. However, emerging information suggests that histone methylation patterns bring a certain stable yet dynamic complexity to the epigenome that can signal a vast number of subsequent regulatory marks necessary for transcriptional control, notably in the adult CNS during LTM formation [45,56,57]. For example, Gupta et al. found that certain histone methylation patterns, including the histone H3 methylation marks H3K4me3 and H3K9me2, serve to activate and repress gene transcription, respectively, in the hippocampus during fear–memory consolidation [58]. These marks have been demonstrated to be catalyzed by different HMT enzymes in the hippocampus during memory formation. Specifically, H3K4me3 corresponds to an increase in the activity of mixed-lineage leukemia 1 (MLL1), while H3K9me2 is due to the enzymatic action of the G9a dimethyltransferase [16,59–61]. Indeed, the addition of methyl groups to histone H3 proteins is as important to the process of memory formation as the methylation marks on DNA [58]. Thus, both histone and DNA methylation are likely to be involved in the evolution of the adult CNS to respond to experience-induced LTM [62].
Coordinated chromatin-structure regulation of genes in memory formation
The timing and rate of gene transcription is strongly influenced by the microenvironment surrounding a given gene promoter. In this respect, there are several factors to consider in the context of epigenetic regulation of gene transcription such as the accessory proteins that are recruited to the promoter region and whether the steric nature of the chromatin allows for transcription factors to bind and function, all in accordance with the epigenetic state of the genome. How epigenetic modifications work in concert to coordinate gene-transcription changes in mature neurons during memory formation is at an early stage of discovery and may prove to be complex. As summarized in Table 1, a number of epigenetic modifications in mature neurons have been demonstrated to be triggered by various learning paradigms to mediate gene-expression changes in several memory-related brain regions. In the next few paragraphs, we will discuss current studies that describe the coordinated chromatin remodeling events involved in the regulation of genes in mature neurons across several brain regions in the context of memory formation and storage.
Table 1.
Types of learning and memory paradigms, brain regions, epigenetic modifications and associated gene-expression changes.
| Study | Learning paradigm |
Brain Regions involved |
Epigenetic modifications | Affected gene expression | Ref. |
|---|---|---|---|---|---|
| Guan et al. (2009) | FC (cued) | Amygdala | ↑ H3 acetylation | ↑ CREB | [16] |
| Koshibu et al. (2009) | ↑ DNMT3A | ↑ c-Rel | [30] | ||
| Monsey et al. (2011) | ↓ HDAC2 | ↑ BDNF | [64] | ||
| Ahn et al. (2008) | ↑ H4 acetylation | [77] | |||
| Gupta et al. (2010) | ↑ H3K4me3 | [58] | |||
| ↑ MLL1 | |||||
| Oliveira et al. (2007) | FC (context) | Amygdala, hippocampus, mPFC |
↑ H3 acetylation, phosphorylation | ↑ BDNF | [109] |
| Guan et al. (2009) | ↑ H2B, H4 acetylation | ↑ NF-κB/c-Rel | [16] | ||
| Koshibu et al. (2010) | ↓/↑ DNA methylation | ↑ Egr-1/Zif-268/Krox24/NGF1-A | [75] | ||
| Miller et al. (2008) | ↑ CBP/PFAC | ↑ NF-κB/p65 | [65] | ||
| Levenson et al. (2004) | ↑ p300 | ↑ ERK, MAPK | [20] | ||
| Ahn et al. (2008) | ↓ HDAC2 | ↑ reelin | [77] | ||
| Lubin et al. (2008) | ↑/↓ MeCP1/2 | ↓ PP1 | [22] | ||
| Koshibu et al. (2009) | ↑ H3K4me3 | ↑ c-fos | [30] | ||
| Fleischmann et al. (2003) | ↑ H3K9me2 | [110] | |||
| Gupta et al. (2010) | ↑ MLL1 | [58] | |||
| Schaefer et al. (2009) | ↑ G9a | [61] | |||
| Oliveria et al. (2007) | NOR | Hippocampus | ↑ p300 | ↑ CREB | [109] |
| Ahn et al. (2008) | ↓ HDAC1 | ↑ c-Rel | [77] | ||
| Koshibu et al. (2009) | ↑ CBP | ↓ PP1 | [30] | ||
| Jones et al. (2001) | ↑ Egr-1/Zif-268/Krox24/NGF1-A | [111] | |||
| Guan et al. (2009) | MWM | Hippocampus | ↓ HDAC2 | ↑ CREB | [16] |
| Koshibu et al. (2009) | ↓ HDAC1 | ↓ PP1 | [30] | ||
| Barrett et al. (2008) | ↑ PCAF | ↑ Zif-268 | [10] | ||
| Jones et al. (2001) | ↑ c-fos | [111] | |||
| Fleischmann et al. (2003) | [110] | ||||
| McQuown et al. (2011) | NOL | Hippocampus | ↓ HDAC3 | ↑ Nr4a2 | [29] |
| Haettig et al. (2011) | ↑ CBP | [112] | |||
BDNF: Brain-derived neurotrophic factor; CBP: CRE-binding protein; CREB: cAMP response element; DNMT: DNA methyltransferase; Egr-1: Early-growth response-1; ERK: Extracellular signal-regulated kinase; FC: Fear conditioning; HDAC: Histone deacetylase; MeCP1/2: Methyl-CpG binding protein 1/2; MLL1: Mixed-lineage leukemia 1; mPFC: Medial prefrontal cortex; MWM: Morris water maze; NOL: Novel object location; NOR: Novel object recognition; Nr4a2: Nuclear receptor subfamily 4, group A, member 2; PFAC: co-p300/CBP-associated factor; PP1: Protein phosphatase 1.
During the process of LTM formation, upregulation of several mRNA transcripts for a diverse range of genes have been demonstrated to be modulated. These genes encode a number of proteins including growth factors, structural proteins, channels/transporters, transcription and signaling transduction molecules, all for the purpose of supporting synaptic strengthening and the maintenance of the memory trace [63]. Importantly, experience-driven expression of these genes does not only occur in one brain region, but in several memory-related brain regions, including the hippocampus and the amygdala, depending on the environmental experience presented. Whether learning induces gene-expression patterns across different brain regions to work independently or in conjunction with each other is currently unknown.
In relation to brain region-specific epigenetic modifications, several learning paradigms have demonstrated histone acetylation changes to occur in concert with DNA methylation in the adult brain (see Table 1). For example, cued-fear conditioning paradigms that recruit emotional fear responses from the amygdala correlate with increased levels of H3 acetylation and an upregulation of DNMT3A activity, and the same epigenetic trends have been demonstrated in the hippocampus during contextual-fear conditioning, which necessitates the integration of spatial recognition [20,64,65]. High-density microarray assays of brain tissue from these animals demonstrate significant gene-transcription activity in both the hippocampus and amygdala regions during fear conditioning with very little overlap, indicating that the functional genome manifests differently in specific brain areas; moreover, eight clusters of similar genes from the amygdala and six from the hippocampus demonstrate time-based variance [66]. These data suggest that the expression patterns of certain memory-related genes are altered during memory formation. Using several learning and behavior tasks, behavioral neuroscientists have been able to identify important IEGs whose protein expression in the hippocampus or amygdala is altered in response to learning, including BDNF, CREB and NF-κB.
DNA methylation events measured in the promoter region of BDNF during memory consolidation reflect a changing epigenetic landscape triggered by learning, and the subsequent expression pattern of the BDNF protein has a profound effect on the establishment of new memories [58,67]. Bisulfite sequencing, a technique for quantifying the percent of DNA methylation changes, revealed altered DNA methylation at the BDNF gene promoters assayed shortly after fear conditioning that corresponds to an increase in exon-specific BDNF mRNA transcripts in the hippocampus, potentially impacting synaptic plasticity, spine morphology and neuronal circuitry changes subserving memory formation [22,68]. Such activity in neurons in response to learning coincides with the phosphorylation and subsequent dissociation of the MeCP2 repressive complex from active BDNF promoters [69]. Furthermore, a recent animal study focusing on chromatin remodeling in cortical neurons from the medial prefrontal cortex (mPFC), another important brain region involved in LTM storage, found that DNA methylation changes induced by fear conditioning endured for at least 30 days after training [70]. Whether these persistent DNA methylation changes in the mPFC promote or contribute to memory storage remains unclear. Nevertheless, persistent changes in DNA methylation 30 days post-learning are surprising and further studies are necessary to better elucidate how DNA methylation in the mPFC contributes to the maintenance of these long-term memories.
Together, the studies described above indicate that memory processing is related to the methylation status of DNA in specific memory-related brain regions. These include the hippocampus and amygdala, areas that function in primary LTM formation and can store memories for short durations. The expression of growth factors such as BDNF demonstrated to be methylation sensitive is likely to contribute to the necessary plasticity in these processes; however, chromatin changes observed in response to learning are not retained long-term in these regions. On the other hand, DNA methylation changes observed in the mPFC that can store memories for long durations do persist long after the initial learning event, indicating that DNA methylation is differentially utilized throughout the brain to mediate the process of LTM formation and maintenance.
In response to learning, intracellular signaling cascades involving cAMP production are triggered by synaptic depolarization via NMDA receptor activity [71,72]. These events result in active regulation of transcription factors, including CREB. This transcription factor is phosphorylated in an activity-dependent manner to bind CRE consensus sequences within DNA, consequently recruiting the coactivator CBP which has HAT activity [71,72]. CBP HAT activity, as previously mentioned, results in increased histone acetylation levels around gene promoters, which subsequently promotes active gene transcription [73]. For example, spatial memory coincides with CREB1 activation in association with at least two HAT enzymes in the hippocampus, CBP and PCAF, to modulate histone acetylation near the promoter regions of plasticity-related genes [74]. Conversely, binding of the CREB2 isoform, also known as activating transcription factor 4 (ATF4), to DNA subsequently engages HDAC5 to gene promoter sites, which has the effect of repressing transcription [15]. Thus, the CREB family of transcription factors can serve as regulators of histone acetylation levels by recruiting either HAT or HDAC enzymes to gene promoters during memory formation [22,63].
In response to fear memory, histone modifications can also occur in the amygdala to regulate changes to IEGs such as BDNF and CREB [75]. The amygdala mediates fear-related emotional memories and incorporates several epigenetic modifications during this process. These epigenetic modifications have been demonstrated to be regulated by PP1 activity. The PP1 enzyme acts as a negative regulator of memory formation by mediating the removal of phosphate groups from a number of cellular proteins including histones, resulting in transcriptional silencing. Furthermore, PP1 associates with HDAC1 and the histone demethylase jumonji domain-containing protein 2A (JMJD2A) to repress gene-expression changes during memory formation [30]. However, in hippocampus-dependent tasks such as novel-object recognition and Morris water maze learning paradigms, PP1 activity is associated with increased CREB gene expression that corresponds to an enhancement in LTM formation [30]. These studies further underscore the idea that epigenetic modifications are differentially regulated in brain regions, yet integrated to ultimately alter the morphology of synapses and the protein expression of synaptic receptors within these brain regions to facilitate the process of memory formation and storage.
NF-κB is another transcription factor with many regulatory functions in the nervous system, which has been demonstrated to be involved in the modulation of epigenetic mechanisms in mature neurons during memory formation. Originally believed to be involved only in immune responses, NF-κB transcription subunits have been demonstrated more recently to have additional roles in controlling gene-expression changes in the CNS [76]. In particular, regulation of gene transcription by members of the NF-κB superfamily, p65/RelA and c-Rel, is necessary for neuronal activity and consolidating LTM. For example, c-Rel knockout mice exhibit deficits in both contextual and cued-fear conditioning as well as in novel object recognition memory tasks [77]. The NF-κB DNA binding complex, which can contain combinations of the p65/RelA or c-Rel sub-units, is activated when the inhibitor κB (IκB) protein is phosphorylated, an event that leads to its further ubiquitination and ensuing degradation. This phosphorylation is mediated by the IκB kinase (IKK). The active retrieval of fear memory requires the specific isoform, IKKα, to indirectly activate the NF-κB DNA binding complex, as well as to regulate histone H3 phosphorylation and acetylation at the IκB and the IEG Zif-268 gene promoters in the hippocampus [23]. These results suggest that specific signaling components of the NF-κB pathway serve to regulate histone modifications (phosphorylation and acetylation) during the storage of fear memories. Inhibition of the NF-κB signaling pathway, either at the IKK or NF-κB DNA binding complex level, results in disruption of fear–memory formation [23]. Moreover, infusion of the HDAC inhibitor, sodium butyrate, rescues the observed memory deficits either by increasing histone H3 acetylation levels, or potentially by enhancing p65/RelA acetylation levels within the NF-κB complex [23]. Indeed, CBP HAT activity and certain HDAC isoforms have been demonstrated to mediate acetylation or deacetylation of both histone and nonhistone proteins in the process of memory formation [78,79]. For example, the p65/RelA subunit can be acetylated at lysine 310 and lysine 314/315 by CBP, and both modification sites are associated with transcriptional activity [80–82]. Thus, HAT and HDAC proteins are now generally referred to as protein lysine-modifying enzymes rather than histone-modifying enzymes. Together, these findings are exciting, as they not only implicate epigenetic mechanisms in memory formation, but also demonstrate the ability of HDAC inhibitors to rescue memory via acetylation of both histone (H3) and nonhistone (NF-κB) proteins.
Epigenetic patterns established by early-life experiences create behavioral phenotypes
Memories which are processed in the hippocampus and amygdala learning centers of the brain and transferred into long-term storage in the mPFC profoundly influence the behavior of the individual. The lasting influence of epigenetic patterning on memory and behavior is illustrated in how early-life experiences biologically dictate an individual’s reaction to present-day stressors encountered in the environment. An eloquent series of experiments by Weaver et al. demonstrates how epigenetic responses to experiential stimuli work to regulate expression patterns of the glucocorticoid receptor (GR), involved in the stress response, within the hippocampi of rat offsprings as a function of maternal care and ensuing stress levels [83]. For these studies, the authors separated mothers with their pups into two groups, each classified by maternal behavior. In one group the mothers readily licked and groomed their newborn offspring, providing an early nurturing environment. Mothers in the other group often withheld this fostering stimulation from the neonates, thus representing a more adverse early atmosphere for the pups [83].
Upon maturation, the offspring of these high-licking and low-licking mothers were observed for behavioral responses to stress-inducing events and categorized, in order to expose any differential consequences of their early-life situation. The researchers found that pups of the high-licking mothers had lower cortisol plasma levels in response to acute stress. They also had higher expression levels of GR mRNA in the hippocampus along with lower cortisol-releasing hormone mRNA levels in the hypothalamus [83]. These responses in GR and cortisol-releasing hormone mRNA levels correlated with dampened output from the hypothalamic–pituitary– adrenal (HPA) axis, the cascade responsible for cortisol release in response to stress, which is negatively regulated by the very hormone it produces to control for biological sensitivity to stress. Pups of the high-licking mothers were more sensitive to the stifling negative feedback of cortisol release than pups of low-licking mothers, suggesting that the former group was more equipped than the latter to respond moderately to anxiety-inducing circumstances.
Clearly, those early-life experiences intimately modulated the biology of the HPA axis, conceivably by transforming the receptor–ligand response to a stressful environment. The investigators further proposed a stimulus-driven epigenetic cascade to explain how physiological memory influences the future behavior of the HPA axis, providing a basic molecular model for environmental gene programming [83]. During prenatal development in the rat hippocampus, there is no 5-methylcytosine present at exon 17 of the GR gene; however, parturition generates hypermethylation of the DNA in this region, corresponding with tightly compacted chromatin and repression of the hormone receptor’s transcription [83]. It is the stimulation of the grooming that the pups received from their mothers that promotes the DNA demethylation of genes in the hippocampus of newborns. Specifically, the experience of being groomed induces the activity-dependent transcription factor, nerve growth factor 1-A (NGF1-A, also known as Zif-268), to target the methylated GR promoter. NGF1-A further recruits HAT enzymes to acetylate histone H3 at several sites along exon 17 of the GR promoter, and the association of HAT and NGF1-A promotes stable transcription of the GR-protein receptor in the hippocampus that is maintained past the neonate stage [83]. This model is encouraging, as the sustainability of the altered psychological state brought about by these observed epigenetic changes indicates the persistence of such behavior-modifying molecular effects into adulthood and throughout the lifespan. It remains to be determined how this dynamic yet stable reprogramming of the epigenome is maintained, since one of the central questions remaining unanswered in the field of epigenetics revolves around the details of the active demethylation process.
The work by Meaney and colleagues supports the theory that comprehensive behavioral traits can be produced through an established epigenome responding, for example, to continual adversity in an individual’s early environment to manifest later as a predisposition to anxiety. The changes encoded by these epigenetic modifications include tuning the output of the HPA axis, a well-characterized signaling pathway, and this alteration leads to enhanced sensitivity of the individual to a stressful environment. The heightened basal and evoked activity of the HPA axis in these model animals correlates with higher levels of cortisol release upon experience-induced stimulation, which corresponds to a very real behavioral phenotype. In humans, such programming can result from moderate to severe forms of trauma, such as childhood abuse, where increased methylation in the promoter of a neuronal glucocorticoid receptor called nuclear receptor subfamily 3, group C, member 1 (NR3C1) leads to decreased transcription of hippocampal GR; the translational study referenced here was performed using postmortem tissue from suicide victims either with or without a history of childhood abuse [84]. Higher concentrations of cortisol can even explain some of the memory deficits observed in chronically stressed animals, since cortisol accumulations in the hippocampus can have devastating effects on the anatomy and function of this brain region. In autopsy studies of rodents that have experienced either chronic stress or acute stress, extensive necrosis and apoptotic-related cell death have been detected in the hippocampus [85]. Furthermore, animals that experienced frequent stress, even at low levels, had region-specific increases in atrophied apical dendrites within the hippocampal formation, including areas CA1 and CA3, and, to a lesser extent, the dentate gyrus [86]. These problems, which can lead to various cognitive deficiencies, arise because the hippocampus is so vulnerable to the toxic effects of cortisol – a recurrent observation also noted in the postmortem brains of patients with schizophrenia, a psychiatric condition where stress and inappropriate epigenetic processing are known to contribute to the emergence of the disease.
Epigenetic processing in psychiatric diseases, including schizophrenia & addiction
The field of behavioral epigenetics finds strong footing in its attempts to explain the origins of psychological disturbances. Schizophrenia is a disease particularly well-suited for investigating how environmental conditions early in an organism’s lifespan are translated into faulty epigenetic regulation that continues to shape behavior well into adulthood [87]. It has been categorized as a neurodevelopmental disorder, in which prenatal stress or abnormally adverse family life during the childhood of a person at genetic risk for schizophrenia can be strong predictors of the psychosis to emerge when the patient is in his or her twenties [88]. Epigenetic changes can explain this neural-diathesis model of schizophrenia, since the pathology advances in the brain under the influence of environmental insults to the neurobiological vulnerability [89]. This abnormal stress response is believed to be connected to the aberrant transmission of dopamine observable in schizophrenia [90]. It also points to the possibility that the same epigenetic mechanisms are becoming dysregulated by early stress in patients, and then again in response to some acutely stressful incident that usually precedes the shift from a normally functioning brain to the pathological condition, so that these processes underlie the environmentally induced changes in both latent biological susceptibility and disease emergence.
Such epigenetic etiology for the disease has been considered for decades since it was observed that administering antidepressants comprised of the methyl donor molecule l-methionine actually worsened symptoms in the schizophrenic population [91]. This led researchers to investigate how the DNA methylation status of the genome might be responsible for a diagnosis of schizophrenia, and the important discovery that the reelin protein, known to be involved in synaptic plasticity and specific to GABA-ergic cortical interneurons, has a hypermethylated promoter when analyzed in postmortem brain tissue from schizophrenia patients that accounts for its dramatically reduced expression levels [92,93]. Indeed, it is a defining characteristic of the disease that patients suffer from diminished inhibitory GABA-ergic cortical activity, leading to hypofunctionality in the frontal cortex and the inability to smoothly switch focus between tasks, behavior which can be elicited in humans with psychiatric tests including prepulse inhibition and the Stroop task [94,95]. Later human autopsy studies confirmed that the methylated REELIN promoter does in fact correlate with increased activity of DNMT1 and decreased levels of reelin mRNA in schizophrenia patients [96–98].
The epigenetic status of the promoter region of GABA-related glutamic acid decarboxylase (GAD) has also been investigated, since its expression is severely decreased with schizophrenia [99,100]. Histone methylation plays a role in sustainably programming the expression of the important GAD67 isoform with schizophrenia, as the downregulation of this protein noted in postmortem human brain tissue is accompanied in the schizophrenic condition by low levels of the activating H3K4me3 mark and high levels of the repressive H3K27me3 mark around its promoter [100]. GAD67 expression is also reduced as a result of promoter hypermethylation, which may be the consequence of overactive DNMTs in the frontal cortex of schizophrenia patients, introducing another layer of epigenetic complexity [101]. In mice developed to model schizophrenia by mimicking the hypermethylated states of the REELIN and GAD67 promoters, the infusion of a nonspecific HDAC inhibitor, sodium valproate (VPA), normalized mRNA expression of these genes [101,102]. Furthermore, the drug had an effect on some of the behavioral hallmarks of the disease exhibited by these model animals. By increasing acetylation along histone H3 and surmounting hypermethylation of the REELIN promoter, treatment with VPA was able to rescue deficiencies previously noted in prepulse inhibition and social interaction [102]. These findings add to an epigenetic understanding of schizophrenia and demonstrate how tweaking one epigenetic modification (e.g., histone acetylation) may work to overcome the effects of another (e.g., DNA methylation).
Epigenetic modification may also influence the development and maintenance of addiction, another instance where such lasting changes appear to help define a psychiatric state. Addiction arises when the natural reward mechanisms of the NAc, a central brain region that deals with cultivating motivation, are hijacked by drugs and refocused on obtaining more and more of the substance [103]. In theory, this may be regarded as a strange misappropriation of the mechanisms of memory recall, since drug cravings represent a kind of constant physiological nostalgia for the levels of euphoria experienced when the user first took the drug. Lately, a greater emphasis has been placed on the potential of epigenetic mechanisms to dictate how neural substrates and comprehensive behavioral patterns are influenced by drugs of abuse, and addiction research indicates that neuronal plasticity is critical and that transcriptional alterations accompany such changes. For example, in defining the cellular events that transpire with nicotine addiction, Penton and Lester noted that nicotinic acetylcholine receptors containing the α4 and β2 subunits have the highest affinity for the ligand. With chronic administration of nicotine, binding sites with this composition are upregulated, implying that transcription and expression of these two subunits may also be increased [104]. Furthermore, Kenny and colleagues discovered that, while acute exposure to nicotine effectively decreased the mRNA levels of BDNF in the hippocampus, more chronic exposure served to upregulate these transcript levels, suggesting that the epigenetic regulation of the growth factor plays a role in the reinforcing properties of the addiction [105].
Investigations into the mechanisms controlling addiction, craving and withdrawal in cocaine users have provided evidence of several plasticity-related gene-expression changes. Cocaine abuse has been demonstrated to alter the dendritic morphology and spine distribution in neurons of the NAc, processes that require the structural plasticity obtainable by a fine tuning of epigenetic modifications [106]. To that end, it has been demonstrated that, in response to chronic-cocaine exposure in animals, increased HAT activity encourages acetylation of H3K9 and H3K14 associated with the promoter region of the BDNF gene, sustainably upregulating its expression for at least a week after the last experience with the drug and helping to explain craving and withdrawal [18]. Furthermore, Renthal et al., working with animal models of addiction, established that administration of SAHA, a HDAC inhibitor, strengthens the hold that cocaine has on brain reward mechanisms by indirectly increasing acetylation, as gauged by the SAHA-induced boost in cocaine-conditioned place preference tests [31]. This group later determined that drug-related changes in the functional activity of HDAC5, one of the main histone deacetylases native to the NAc, may arbitrate when and how the physiological attraction to cocaine and other drugs of abuse is translated into a long-term dependence on these substances. Experiments with HDAC5 knockout mice revealed a more intense regimen of cocaine-seeking behavior in these animals, where acetylation levels are naturally higher as compared with controls, and the altered phenotype is rescued by overexpressing HDAC5 in the NAc of the knockouts [31].
Epigenetically regulated plasticity changes may explain how exposure to cocaine and other drugs of abuse can facilitate the compulsion to consume more of that drug, and these changes can be different depending on whether administration is acute or chronic. Some of the earliest work on chromatin regulation in animal models of addiction uncovered that an immediate consequence of introducing cocaine into the nervous system is acetylation of histone H4 near the promoters of the IEGs c-fos and fos-B, with subsequent expression of the transcription factor ΔFosB in the NAc [107]. In addition, phosphorylation of histone H3 at serine 10 by the kinase MSK1 near certain genes in the striatum following acute cocaine exposure, including c-fos, may account for the cultivation of some drug-related behavioral adaptations, especially as the drug itself is known to inhibit the action of PP1, thus keeping the chromatin populated with negatively charged phosphate groups and in a transcriptionally permissive state [108]. In the same way, histone methylation patterns change in response to cocaine, again in a way that activates gene expression to allow for enhanced plasticity. The HMT G9a is the enzyme responsible for inserting the repressive H3K9me2 mark into promoters of genes, but G9a was found to be downregulated following chronic cocaine administration by ΔFosB activity, so that key addiction-related genes could be actively transcribed. Research on cocaine abuse has revealed that many of the structural alterations and plasticity changes in the NAc and other brain reward regions require active transcription, which necessitates a permissive epigenetic environment in the chromatin of critical gene promoters.
Conclusion
The accumulation of evidence in the past few decades leaves little doubt that dynamic regulation of gene expression is a necessary and critical process in the formation, storage and recall of memory and subsequent behavior. Both structural and synaptic plasticity require the activation of various transcription factors, IEGs, and growth factors, including BDNF, CREB and NF-κB. This is accomplished through the coordinated modification of epigenetic marks at the promoter regions of these genes. DNA methylation changes are essential for the formation of LTM, and inhibiting DNMT activity diminishes the capability of an organism to consolidate and recall memory. Likewise, histone acetylation and phosphorylation are methods by which portions of the genome can be rapidly opened up to the transcriptional machinery, and reversing this process through the activity of HDAC and protein phosphatases, respectively, can transcriptionally compromise DNA and have huge effects on memory and behavior. Furthermore, different histone methylation patterns, including the activating H3K4me3 marks and the repressive H3K9me2 and H3K27me3 marks, can recruit or prohibit transcription by either facilitating the tighter binding of transcription factors or providing steric hindrance against them at gene promoter sites. Stable gene expression influenced by experiences early on in life work to shape aspects of behavior that have very real biological underpinnings, as illustrated by the epigenetic programming of GR expression and HPA-axis sensitivity by differential levels of stress generated through maternal care in rodents and humans. Such behavioral phenotypes are the focus of psychiatric diseases encompassing schizophrenia and drug addiction that involve the continued dysregulation of epigenetic processing, evidenced by hypermethylation of the REELIN and GAD67 gene promoters with schizophrenia, and the hyperacetylation of several histone sites in the development of cocaine addiction. Epigenetic modifications in the CNS will continue to be an area of intense research for those who hope to elucidate the full trajectory of molecular events in the transduction of experiential stimuli into consolidated memory, and the profound way that such stored memory is recalled to fuel the psyche and dictate behavior.
Future perspective
Although exceptional progress has been made in the field of epigenetics since these regulatory chromatin-modifying mechanisms were first described in development, much is still unclear regarding their consequence in the adult CNS and in memory and behavior. For example, even as it is understood that memory formation is associated with increased epigenetic activity in the CNS, not all of the gene targets affected have been validated. It will be an arduous task; however, if the entire pathway of memory consolidation and recall is to be characterized, then the identity of important memory-related genes and the time course of epigenetic alterations and expression patterns will have to be recorded. This work up may well change depending on the kind of memory being studied, be it hippocampus-dependent spatial memory, fear-related cued memory, or even memory associated with motor skills that could shed light on the complex mechanisms triggering drug-seeking behavior in addicts. Furthermore, the particular relationship between DNA methylation and histone methylation must be further explored, as these modifications represent a more subtle and long-term adjustment to transcriptional competency than the transient and overtly electrostatic effects of histone phosphorylation and acetylation.
The concept of DNA methylation itself, though the most ancient known regulator of cellular memory, is still not fully understood. Specifically, the dynamic nature of DNA demethylation has yet to be satisfactorily elucidated. The new discovery of 5-hydroxymethylcytosine is exciting, since it represents a potential transition between the 5-methylcytosine produced by DNMT activity and the return to an unmethylated cytosine residue. The bisulfite treatment used by many epigenetic researchers to quantify methylation levels within particular promoters does not functionally account for the presence of 5-hydroxymethylcytosine, only 5-methylcytosine. The chromatin immunoprecipitation technique, which uses antibodies aimed at these differently methylated marks, might be employed in order to parse out the changes contributed by each of these DNA methylation transition molecules. Finally, the field of behavioral epigenetics is at an early stage of discovery, whose impact will be further explored with future studies. Indeed, debate over the quality and extent of the epigenome’s influence on the adult brain continues to grow at an exciting pace, as new information surfaces regarding the powerful and dynamic consequences of environmental influences on behavior.
Executive summary.
A plausible role for epigenetic mechanisms in human learning & remembering
-
▪
Epigenetic modifications are triggered in the processes of memory formation.
-
▪
Short-term memory is consolidated into long-term memory (LTM), and epigenetic upregulation of transcription factors and immediate–early genes accompany this conversion.
A brief overview of epigenetic mechanisms in LTM formation
-
▪
Epigenetic regulation of the working genome is accomplished through changes made to chromatin structure that do not affect the DNA sequence.
-
▪
The activity of epigenetic enzymes including DNA methyltransferases, histone acetyltransferases/histone deacetylases and histone methyltransferases/histone demethylases within a promoter region control how accessible the DNA of a specific gene is to transcription factors, and these epigenetic marks have been observed to change with learning and experience.
Epigenetic regulation of immediate–early genes in memory formation
-
▪
The consolidation and recall of LTM requires plasticity, which is influenced by epigenetic mechanisms.
-
▪
Histone modifications and DNA methylation changes are observed in the hippocampus, amygdala and cortex in response to learning, corresponding with the expression of key memory-related immediate–early genes and transcription factors.
Epigenetic patterns established by early-life experiences create behavioral phenotypes
-
▪
Early-life adversity is tightly associated with regulation of the hypothalamic–pituitary–adrenal axis, the governing mechanism of the stress response.
-
▪
Chronic stress acts through epigenetic mechanisms to reprogram expression patterns of the glucocorticoid receptor, influencing the sensitivity of an individual’s response to environmental stressors.
Epigenetic processing in psychiatric diseases, including schizophrenia & addiction
-
▪
Hypermethylation of the REELIN promoter may account for the significant decrease in GABA-ergic plasticity observed in schizophrenia.
-
▪
The cultivation of addiction requires epigenetically regulated plasticity changes within neurons in the nucleus accumbens, a central reward region in the brain.
Conclusion
-
▪
The transduction of experiential stimuli into LTM necessitates the molecular mechanisms of epigenetics to regulate gene expression.
-
▪
Experience modifies the epigenome, and epigenetic modifications have been detected at several memory-related gene promoters, regulating their expression, which is critical for the plasticity that occurs in response to learning.
Future perspective
-
▪
In the coming years, molecular memory scientists will continue to work on mapping the complete and timely expression patterns of genes identified to be essential to the process of LTM formation. These future studies will correlate gene-expression changes with the causal epigenetic modifications.
Acknowledgements
The authors wish to thank the Federation of American Societies for Experimental Biology, the National Institute of Mental Health (MH082106), and the Evelyn F McKnight Brain Institute at the University of Alabama at Birmingham, USA.
Footnotes
Financial & competing interests disclosure
Funding was recieved from the National Institute of Mental Health (MH082106). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Craik FI. Levels of processing: past, present. and future? Memory. 2002;10:305–318. doi: 10.1080/09658210244000135. [DOI] [PubMed] [Google Scholar]
- 2.Monfils MH, Cowansage KK, LeDoux JE. Brain-derived neurotrophic factor. Linking fear learning to memory consolidation. Mol. Pharmacol. 2007;72:235–237. doi: 10.1124/mol.107.038232. [DOI] [PubMed] [Google Scholar]
- 3.Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- 4.Guzowski JF. Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus. 2002;12:86–104. doi: 10.1002/hipo.10010. [DOI] [PubMed] [Google Scholar]
- 5.Schafe GE, Nader K, Blair HT, LeDoux JE. Memory consolidation of Pavlovian fear conditioning: a cellular and molecular perspective. Trends Neurosci. 2001;24:540–546. doi: 10.1016/s0166-2236(00)01969-x. [DOI] [PubMed] [Google Scholar]
- 6.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagot RC, Meaney MJ. Epigenetics and the biological basis of gene × environment interactions. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:752–771. doi: 10.1016/j.jaac.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Alarcon JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colvis CM, Pollock JD, Goodman RH, et al. Epigenetic mechanisms and gene networks in the nervous system. J Neurosci. 2005;25:10379–10389. doi: 10.1523/JNEUROSCI.4119-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 15.Guan Z, Giustetto M, Lomvardas S, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111(4):483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 16.Guan JS, Haggarty SJ, Giacometti E, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Langley B, Lubin FD, et al. Epigenetics in the nervous system. J. Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Choi KH, Renthal W, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 20.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 21.Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate, invertebrate memory formation. Cell Mol. Life Sci. 2006;63:1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. ▪ Highlights the important role of the NF-κB signaling pathway in the regulation of epigenetic mechanisms during memory re consolidation and determining consequential behavioral effects in a fear memory paradigm.
- 23.Lubin FD, Sweatt JD. The IκB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J. Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory? Learn Mem. 2006;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutsch SI, Rosse RB, Mastropaolo J, Long KD, Gaskins BL. Epigenetic therapeutic strategies for the treatment of neuropsychiatric disorders: ready for prime time? Clin. Neuropharmacol. 2008;31:104–119. doi: 10.1097/WNF.0b013e318067e255. [DOI] [PubMed] [Google Scholar]
- 27.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 28.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McQuown SC, Barrett RM, Matheos DP, et al. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. ▪ Seeks to understand the contribution of the histone deacetylase 3 isoform to plasticity and memory processes, and considers how selectively blocking this negative regulator of memory, histone deacetylase 3, affects memory consolidation.
- 30.Koshibu K, Graff J, Beullens M, et al. Protein phosphatase 1 regulates the histone code for long-term memory. J. Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 32.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J. Cell Sci. 2003;116:4905–4914. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 33.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 37.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 38.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 39.Chen WG, Chang Q, Lin Y, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 41.Amir RE, van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 42.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 45.Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol. Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 47.Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 52.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 53.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum. Mol. Genet. 2009;18:3375–3383. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 55.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters AH, Schubeler D. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 2005;17:230–238. doi: 10.1016/j.ceb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 57.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol. Life Sci. 2009;66(3):407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gupta S, Kim SY, Artis S, et al. Histone methylation regulates memory formation. J. Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. ▪ Successfully demonstrates dynamic regulation of gene transcription during long-term memory formation by distinct patterns of histone methylation modifications. Specifically, it identifes both active and repressive histone methylation marks in the hippocampus during memory consolidation.
- 59.Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maze I, Covington HE, 3rd, Dietz DM, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaefer A, Sampath SC, Intrator A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y, Jakovcevski M, Bharadwaj R, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J. Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 64.Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS ONE. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 67. Levenson JM, Roth TL, Lubin FD, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. ▪ Among the first studies to illustrate alterations in DNA methylation in the hippocampus in response to long-term potentiation, the cellular correlate of long-term memory formation.
- 68.Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 70.Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walton M, Henderson C, Mason-Parker S, et al. Immediate early gene transcription and synaptic modulation. J. Neurosci. Res. 1999;58:96–106. [PubMed] [Google Scholar]
- 72.Mitterdorfer J, Froschmayr M, Grabner M, Moebius FF, Glossmann H, Striessnig J. Identification of PKA phosphorylation sites in the carboxyl terminus of L-type calcium channel α 1 subunits. Biochemistry. 1996;35:9400–9406. doi: 10.1021/bi960683o. [DOI] [PubMed] [Google Scholar]
- 73.Vecsey CG, Hawk JD, Lattal KM, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. J. Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bousiges O, de Vasconcelos AP, Neidl R, et al. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koshibu K, Graff J, Mansuy IM. Nuclear protein phosphatase-1: an epigenetic regulator of fear memory and amygdala long-term potentiation. Neuroscience. 2010;173:30–36. doi: 10.1016/j.neuroscience.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 76.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-κB functions in synaptic signaling and behavior. Nat. Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 77.Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou HC, Sweatt JD. c-Rel, an NF-κB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learn Mem. 2008;15:539–549. doi: 10.1101/lm.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomson S, Clayton AL, Mahadevan LC. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell. 2001;8:1231–1241. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- 79.Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-κB in rat amygdala improves long-term but not short-term retention of fear memory. Mol. Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 80.Yang XD, Tajkhorshid E, Chen LF. Functional interplay between acetylation and methylation of the RelA subunit of NF-κB. Mol. Cell Biol. 2010;30:2170–2180. doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 82.Rothgiesser KM, Fey M, Hottiger MO. Acetylation of p65 at lysine 314 is important for late NF-κB-dependent gene expression. BMC Genomics. 2010;11:22. doi: 10.1186/1471-2164-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. ▪ The authors present a clear consequence of epigenetic mechanisms and their relationship to early-life experiences and how effects are transmitted via epigenetic programming into a comprehensive behavioral phenotype into adulthood.
- 84.McGowan PO, Sasaki A, D’ Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lucassen PJ, Muller MB, Holsboer F, et al. Hippocampal apoptosis in major depression is a minor event and absent from subareas at risk for glucocorticoid overexposure. Am. J. Pathol. 2001;158:453–468. doi: 10.1016/S0002-9440(10)63988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watanabe Y, Gould E, McEwen BS. Stress induces atrophy of apical dendrites of hippocampal CA3 pyramidal neurons. Brain Res. 1992;588:341–345. doi: 10.1016/0006-8993(92)91597-8. [DOI] [PubMed] [Google Scholar]
- 87.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol. Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 89.Rudge JS, Mather PE, Pasnikowski EM, et al. Endogenous BDNF protein is increased in adult rat hippocampus after a kainic acid induced excitotoxic insult but exogenous BDNF is not neuroprotective. Exp. Neurol. 1998;149:398–410. doi: 10.1006/exnr.1997.6737. [DOI] [PubMed] [Google Scholar]
- 90.Jones SR, Fernyhough C. A new look at the neural diathesis – stress model of schizophrenia: the primacy of social-evaluative and uncontrollable situations. Schizophr. Bull. 2007;33:1171–1177. doi: 10.1093/schbul/sbl058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pollin W, Cardon PV, Jr, Kety SS. Effects of amino acid feedings in schizophrenic patients treated with iproniazid. Science. 1961;133:104–105. doi: 10.1126/science.133.3446.104. [DOI] [PubMed] [Google Scholar]
- 92.Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch. Gen. Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 93.Grayson DR, Chen Y, Costa E, et al. The human reelin gene: transcription factors (+), repressors (−) and the methylation switch (+/−) in schizophrenia. Pharmacol. Ther. 2006;111:272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 94.Reid MA, Stoeckel LE, White DM, et al. Assessments of function and biochemistry of the anterior cingulate cortex in schizophrenia. Biol. Psychiatry. 2010;68:625–633. doi: 10.1016/j.biopsych.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewis DA. GABAergic local circuit neurons and prefrontal cortical dysfunction in schizophrenia. Brain Res. Rev. 2000;31:270–276. doi: 10.1016/s0165-0173(99)00042-9. [DOI] [PubMed] [Google Scholar]
- 96.Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 97.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol. Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- 98.Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol. Psychiatry. 2007;12(6):593–600. doi: 10.1038/sj.mp.4002014. 519. [DOI] [PubMed] [Google Scholar]
- 99.Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim. Biophys. Acta. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong H, Csernansky CA, Goico B, Csernansky JG. Hippocampal neurogenesis follows kainic acid-induced apoptosis in neonatal rats. J. Neurosci. 2003;23:1742–1749. doi: 10.1523/JNEUROSCI.23-05-01742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tremolizzo L, Doueiri MS, Dong E, et al. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol. Psychiatry. 2005;57:500–509. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 103.Kelley AE. Memory and addiction: shared neural circuitry and molecular mechanisms. Neuron. 2004;44:161–179. doi: 10.1016/j.neuron.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 104.Penton RE, Lester RA. Cellular events in nicotine addiction. Semin. Cell Dev. Biol. 2009;20:418–431. doi: 10.1016/j.semcdb.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kenny PJ, File SE, Rattray M. Acute nicotine decreases, and chronic nicotine increases the expression of brain-derived neurotrophic factor mRNA in rat hippocampus. Brain Res. Mol. 2000;85:234–238. doi: 10.1016/s0169-328x(00)00246-1. [DOI] [PubMed] [Google Scholar]
- 106.Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33:267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berton O, McClung CA, Dileone RJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 108.Brami-Cherrier K, Valjent E, Herve D, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J. Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fleischmann A, Hvalby O, Jensen V, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J. Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones MW, Errington ML, French PJ, et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 112.Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem. 2011;18:71–79. doi: 10.1101/lm.1986911. [DOI] [PMC free article] [PubMed] [Google Scholar]