Abstract
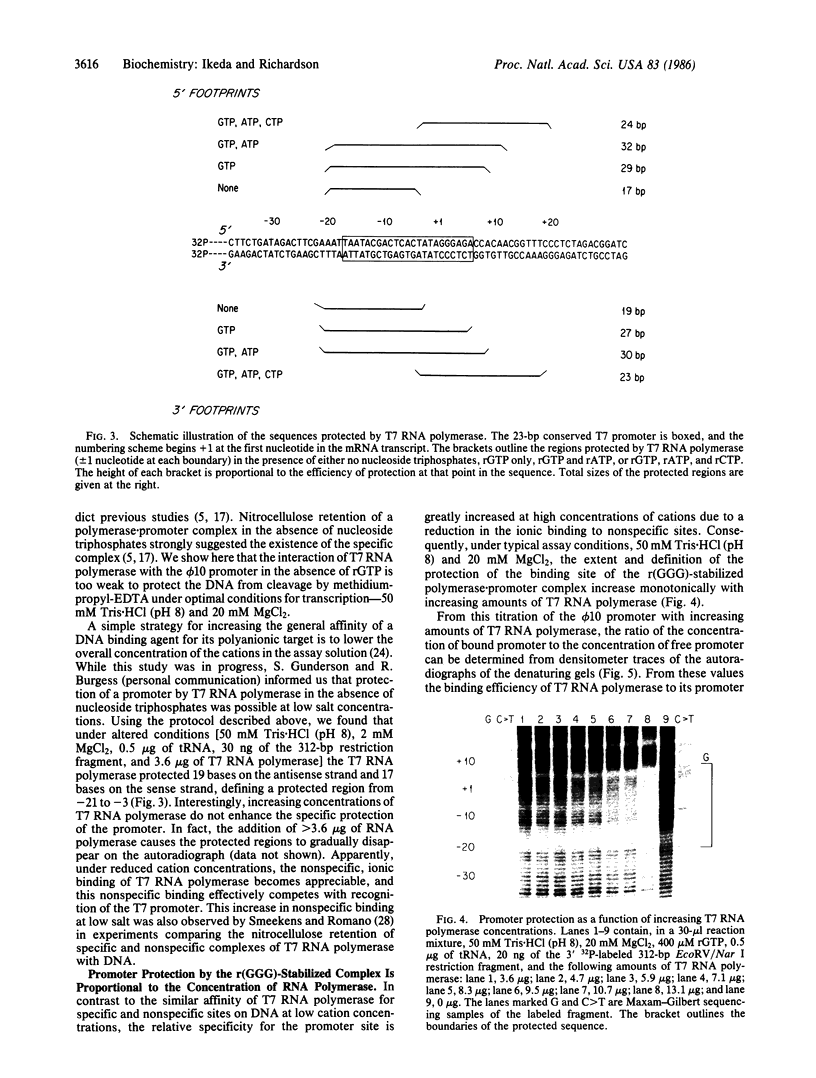
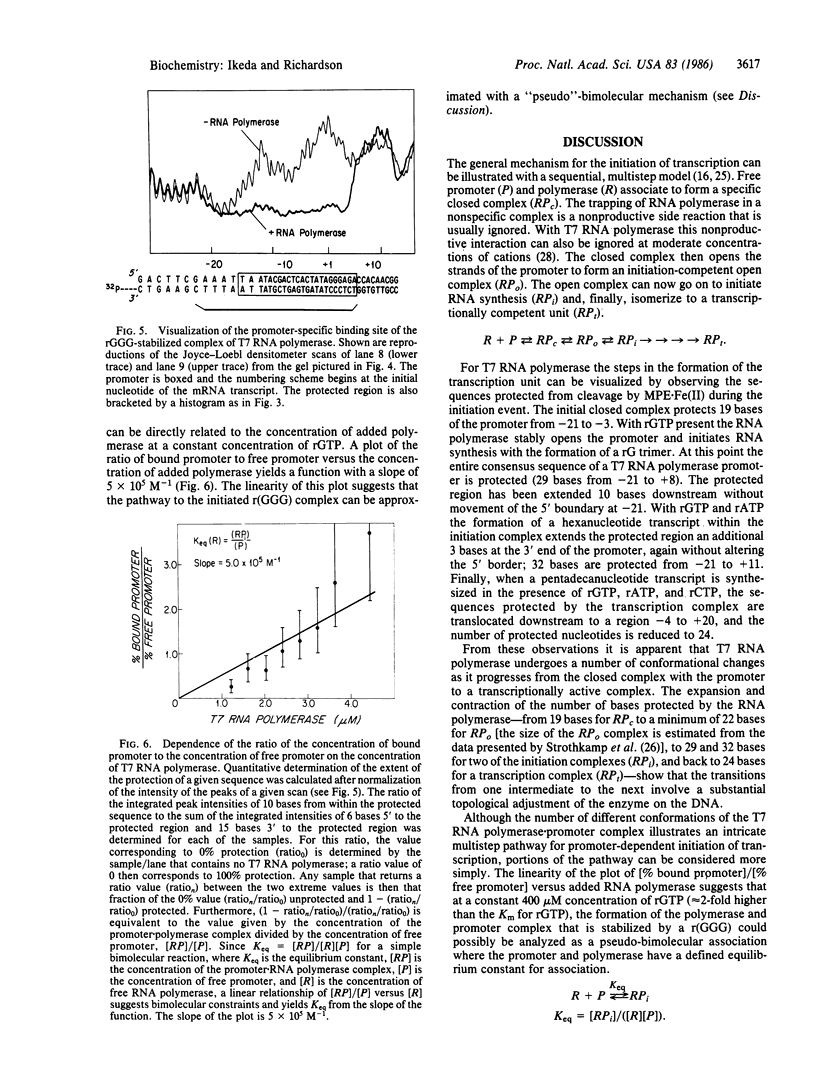
Promoters for T7 RNA polymerase have a highly conserved sequence of 23 continuous base pairs located at position -17 to +6 relative to the initiation site for the RNA. The complex of T7 RNA polymerase with the phage phi 10 promoter has been visualized indirectly by exploiting the ability of the polymerase to protect DNA sequences from cleavage by methidiumpropyl-EDTA X Fe(II). The DNA contacts made by T7 RNA polymerase have been mapped during binding and during the subsequent initiation of transcription. The RNA polymerase alone protects 19 bases in a region from -21 to -3. Synthesis of the trinucleotide r(GGG) expands the length of the sequence protected by the RNA polymerase and stabilizes the complex; 29 bases (-21 to +8) are protected, and the observed equilibrium association constant of the r(GGG) complex is 5 X 10(5) M-1. The formation of a hexanucleotide mRNA, r(GGGAGA), further extends the protected region; 32 bases (-21 to +11) are protected. Finally, the synthesis of a pentadecanucleotide mRNA leads to a translocation of the region protected by the protein; the sequence now protected is reduced to 24 bases (-4 to +20).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985 May 25;183(2):165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Fischer H., Hinkle D. C. Bacteriophage T7 DNA replication in vitro. Stimulation of DNA synthesis by T7 RNA polymerase. J Biol Chem. 1980 Aug 25;255(16):7956–7964. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hofer B., Müller D., Köster H. The pathway of E. coli RNA polymerase-promoter complex formation as visualized by footprinting. Nucleic Acids Res. 1985 Aug 26;13(16):5995–6013. doi: 10.1093/nar/13.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt B. A., Dunn J. J., Studier F. W. Nucleotide sequence of the gene for bacteriophage T7 RNA polymerase. J Mol Biol. 1984 Feb 25;173(2):265–269. doi: 10.1016/0022-2836(84)90194-3. [DOI] [PubMed] [Google Scholar]
- Oakley J. L., Strothkamp R. E., Sarris A. H., Coleman J. E. T7 RNA polymerase: promoter structure and polymerase binding. Biochemistry. 1979 Feb 6;18(3):528–537. doi: 10.1021/bi00570a023. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Recognition and initiation site for four late promoters of phage T7 is a 22-base pair DNA sequence. Nature. 1979 Jul 5;280(5717):35–39. doi: 10.1038/280035a0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Romano L. J., Tamanoi F., Richardson C. C. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins: requirement for T7 RNA polymerase. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4107–4111. doi: 10.1073/pnas.78.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. D. Four T7 RNA polymerase promoters contain an identical 23 bp sequence. Cell. 1979 Apr;16(4):815–825. doi: 10.1016/0092-8674(79)90097-7. [DOI] [PubMed] [Google Scholar]
- Smeekens S. P., Romano L. J. Promoter and nonspecific DNA binding by the T7 RNA polymerase. Nucleic Acids Res. 1986 Mar 25;14(6):2811–2827. doi: 10.1093/nar/14.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Strothkamp R. E., Oakley J. L., Coleman J. E. Promoter melting by T7 ribonucleic acid polymerase as detected by single-stranded endonuclease digestion. Biochemistry. 1980 Mar 18;19(6):1074–1080. doi: 10.1021/bi00547a005. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]