SUMMARY
Polyphosphoinositides are lipid signaling molecules generated from phosphatidylinositol (PtdIns) with critical roles in vesicular trafficking and signaling. It is poorly understood where PtdIns is located within cells and how it moves around between membranes. Here we identify a hitherto unrecognized highly mobile membrane compartment as the site of PtdIns synthesis and a likely source of PtdIns of all membranes. We show that the PtdIns synthesizing enzyme, PIS associates with a rapidly moving compartment of ER origin that makes ample contacts with other membranes. In contrast, CDP-diacylglycerol synthases that provide PIS with its substrate reside in the tubular ER. Expression of a PtdIns-specific bacterial PLC generates diacylglycerol also in rapidly moving cytoplasmic objects. We propose a model in which PtdIns is synthesized in a highly mobile lipid distribution platform and is delivered to other membranes during multiple contacts by yet to be defined lipid transfer mechanisms.
INTRODUCTION
Polyphosphoinositides (PPIs) are phosphorylated forms of phosphatidylinositol (PtdIns) formed by a variety of PI- and PIP kinases in eukaryotic cells. These minute amounts of phospholipids have gained enormous interest because of their pivotal roles in regulating virtually every cellular process within eukaryotic cells. These lipids first rose to prominence as precursors of important second messengers, generated upon stimulation of certain groups of cell surface receptors (Michell, 1975). However, PPIs have proven to be more versatile in that they also regulate ion channels and transporters, they control membrane fusion and fission events and hence are master regulators of vesicular transport, secretion, and endocytosis and they also play key roles in lipid transport and disposition (Balla et al., 2009).
Significant progress has been made in identifying the enzymes that produce and eliminate PPIs and characterizing their biology (Sasaki et al., 2009). The distribution and dynamics of PPIs changes in different membrane compartments have been determined with antibodies or PPI binding protein modules used as GFP fusion proteins in live or fixed cells (Downes et al., 2005; Halet, 2005). Similar progress has not been made in understanding the localization, movements and importance of the PtdIns lipid pools. PtdIns is, of course, the precursor of all PPIs but also is a structural phospholipid. Our current knowledge on PtdIns synthesis and distribution originates from pioneering studies that used cell fractionation and metabolic labeling to identify the ER as the site of PtdIns synthesis (Agranoff et al., 1958) and the plasma membrane (PM) where PtdIns is sequentially phosphorylated to PtdIns 4-phosphate (PtdIns4P) and PtdIns 4,5-bisphosphate [PtdIns(4,5)P2] (Hokin and Hokin, 1964; Michell et al., 1967). Early studies by the Hokins (Hokin and Hokin, 1955) showed that receptor-mediated hydrolysis of phosphoinositides increases the metabolic labeling of PtdIns, and based on several subsequent studies, [reviewed by Michell (Michell, 1975)] it has always been assumed that there is a special fraction of PtdIns synthesis that responds to increased phosphoinositide hydrolysis and is dedicated to the replenishment of the PM signaling pools.
Attempts to identify a special compartment where the increased PtdIns resynthesis takes place have largely been unsuccessful (De Camilli and Meldolesi, 1974; Lapetina and Michell, 1972), leading to conclusions that PtdIns very rapidly equilibrates between various membranes possibly with the aid of PI-transfer proteins (PITPs) (Cockcroft and Carvou, 2007). However the functions of PITPs appear to be more complex than mere lipid distribution and membrane-specific roles assigned to specific molecular pathways such as presenting the PtdIns substrate to PI kinases have been emerging (Cockcroft and Garner, 2011; Ile et al., 2006). Also, because of the tight connection between phosphoinositide hydrolysis and PtdIns resyntheis during agonist stimulation, the question was repeatedly raised whether PtdIns is also synthesized in the PM (Imai and Gershengorn, 1987; Monaco et al., 2006) or some other membrane compartment such as endosomes (Sillence and Downes, 1993). However, no systematic study has attempted to map the PtdIns pools in unfractionated intact cells.
Similarly, few studies addressed the functional significance of PtdIns pools in the various membrane compartments, because of the difficulty to alter the overall PtdIns levels in living cells, let alone within various membrane compartments. Lithium ions in combination with strong phospholipase C (PLC) -coupled receptor stimulation have been shown to deplete PtdIns pools due to the blockade in inositol recycling (Balla et al., 1988; Batty and Downes, 1994; Jenkinson et al., 1994). Although this method has been successfully applied to invertebrates (Acharya et al., 1998), especially in the retina where the light induced PtdIns turnover is extremely robust (Wu et al., 1995), it has been difficult to drive down PtdIns pools with Li+ treatment in vertebrates. However, two recent studies identified disrupted or mutated PtdIns synthase (PIS) genes causing ER stress and steatosis in the liver and eye developmental defects in zebrafish (Murphy et al., 2011; Thakur et al., 2011). In the present study, we set out to perform a comprehensive analysis of the cellular PtdIns pools and their functional significance in living cells with a combination of approaches. First, we determined the localization of the PtdIns synthesizing machinery with GFP-tagged PIS and cytidine diphosphate-diacylglycerol (CDP-DAG) synthase (CDS) enzymes. We then employed a PtdIns-specific bacterial PLC enzyme that does not hydrolyze PPIs and targeted this enzyme to various membrane compartments. We monitored the produced DAG by a sensitive DAG sensor fused to GFP and also followed PtdIns3P, PtdIns4P and PtdIns(4,5)P2 by GFP-fused FYVE or PH domains. We determined the impact of PI-PLC expression on the sizes of the PtdIns pools using long-term labeling with myo-[3H]inositol and assessed their effects on the turnover rate by shorter 32P-phosphate incorporation experiments. These analyses identified a highly mobile special subcompartment of the ER as the site of PtdIns synthesis and distribution. These studies open new possibilities to better understand the process of PtdIns synthesis and its supply to other membrane compartments.
RESULTS
Combination of a bacterial PtdIns-specific PLC and a sensitive DAG sensor reveals unexpected localization of PtdIns pools in living cells
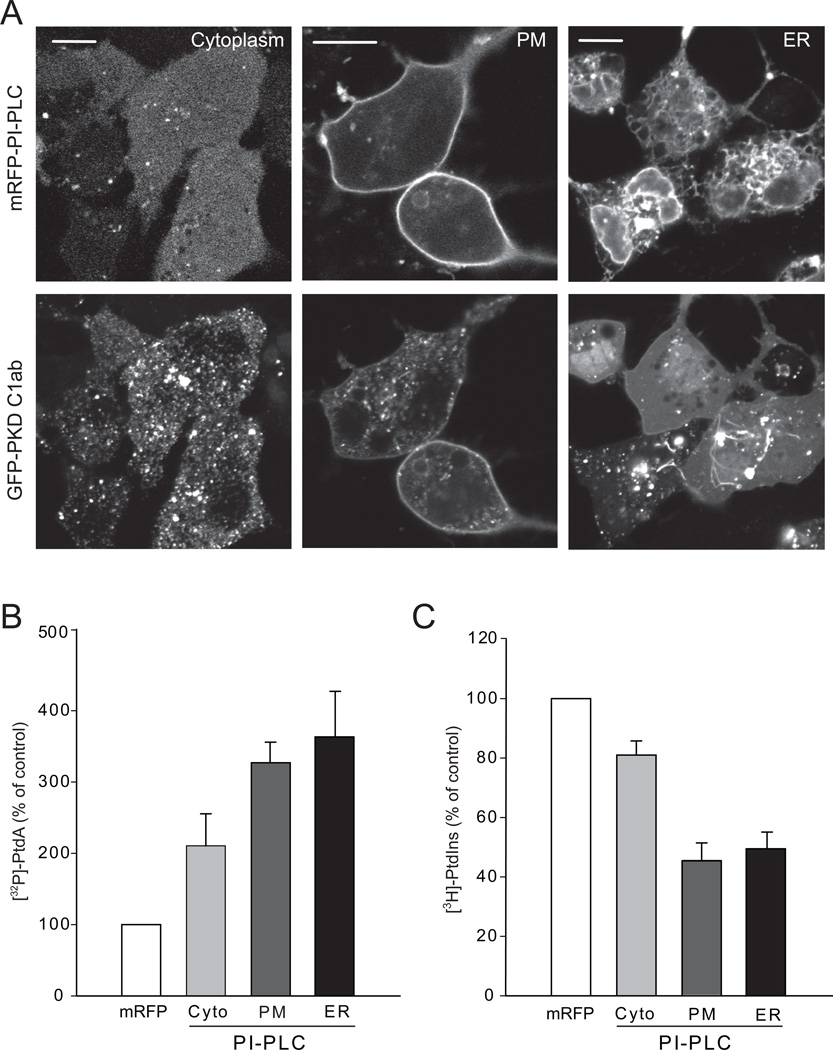
Phospholipase C enzymes isolated from a variety of bacteria specifically hydrolyze PtdIns or PtdIns-glycan (GPI) linkages without acting on PPIs, both in vitro and in vivo (Griffith and Ryan, 1999). To take advantage of this substrate restriction, we used a bacterial PI-PLC for expression in mammalian cells to specifically manipulate PtdIns levels. The Listeria monocytogenes PLC has substantial activity against PtdIns in addition to its natural substrate, GPI (Wei et al., 2005). We cloned this enzyme from Listeria monocytogenes (strain 10403S), removed its N-terminal hydrophobic signal sequence to prevent its targeting to the secretory pathway, and fused it to mRFP. A catalytically inactive mutant form (H86A) was also produced (using strain DP-L3430) to serve as a negative control (Bannam and Goldfine, 1999).
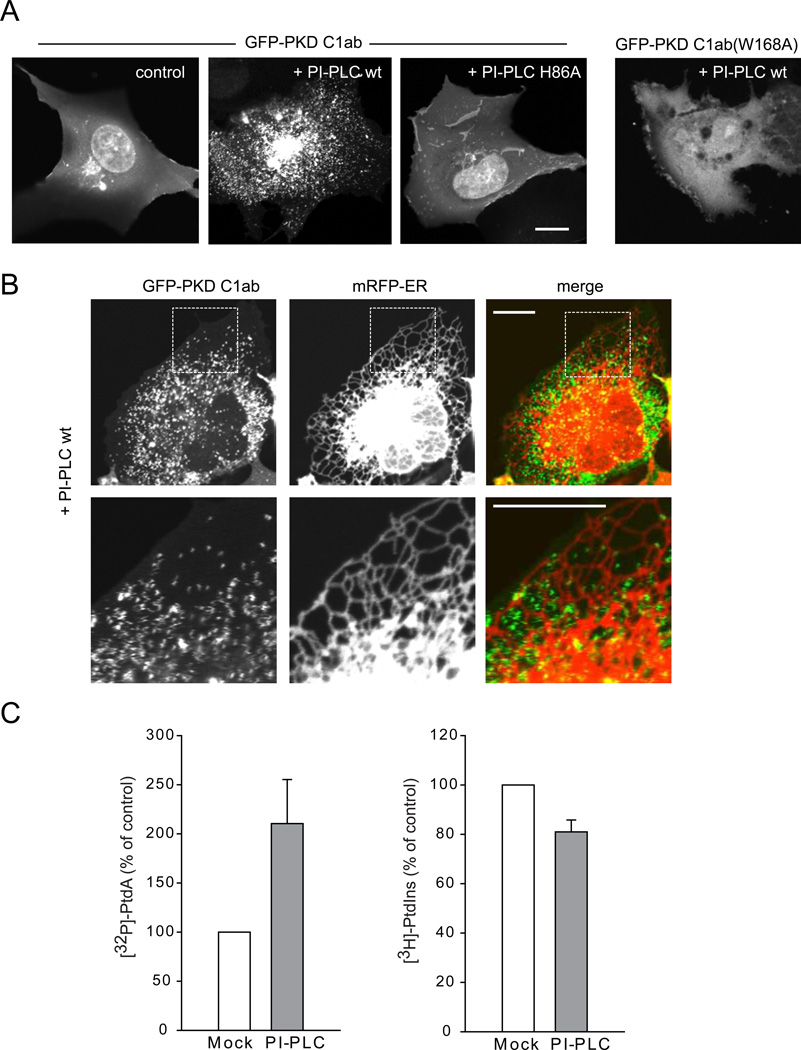
To capture the PtdIns hydrolytic product, DAG, we created a high affinity DAG sensor using the tandem C1 domains (C1ab) of PKD fused to the C-terminus of GFP. A nuclear export signal was added to the N-terminus of the probe to decrease its nuclear accumulation. This sensor was more sensitive than the C1a domain of PKCγ (Oancea et al., 1998) generally used for DAG detection. In quiescent cells it was mostly found in the cytoplasm with a faint signal in PM and Golgi membranes and some remaining signal in the nucleus (Figure 1A). However, it readily detected DAG after activation of endogenous PLC by angiotensin II (AngII) stimulation in HEK293 cells expressing the AT1a receptors (HEK293-AT1) (Movie S1) (Figure S1A). The sensor also responded to addition of recombinant PI-PLC externally (which cleaves GPI linkages on the outside and the DAG flips to appear in the inner leaflet), or to exogenous delivery of di-C8-DAG or PMA (Figures S1B–D). When the DAG sensor was expressed together with PI-PLC enzyme (both in HEK293-AT1 and COS-7 cells), a huge number of tiny and very rapidly moving particles appeared in the cytoplasm attracting the DAG sensor but not PLC itself (Figure 1A and Movie S2). The rapid movement of these DAG positive structures showed up as zigzagging traces mostly in areas between the tubular ER (labeled with an ER-targeted mRFP using the C-terminal Sac1 sequence) when using slower double scan mode in the confocal microscope (Figure 1B). Expression of the lipase defective mutant of PI-PLC (H86A) did not produce such particles (Figure 1A). Similarly, a mutant DAG sensor (W166A) [this highly conserved Trp has a major impact on DAG affinity of C1 domains (Dries et al., 2007)] that showed significantly reduced diC8-DAG sensitivity (Figure S1F) did not efficiently detect the rapidly moving particles (Figure 1A).
Figure 1. Association of the DAG sensor with small mobile structures after PI-PLC expression in the cytoplasm.
(A) COS-7 cells were transfected with wild-type or mutant GFP-PKD C1ab alone or together with either mRFP-PI-PLC or its lipase mutant H86L (also see Figure S1 and Movie S1). (B) Relationship of the DAG positive structures to the ER. COS-7 cells were co-transfected with CFP-PI-PLC, GFP-PKD C1ab and mRFP-ER using the Sac1 C-terminal ER localization signal. Enlarged images are shown in the lower panels. The zigzagging traces correspond to very rapid movements during scanning in confocal microscopy. Scale bars, 10 µm. (C) HEK293-AT1 cells were transfected with mRFP only or mRFP-PI-PLC and labeled with [32P]phosphate for the last 3 hrs of the one day transfection or myo-[3H]inositol for 24 hrs as described under Methods. Labeled lipids were extracted, separated by TLC, and quantified by a PhosphorImager for [32P] phosphate labeling or by densitometry of exposed films in case of myo-[3H]inositol labeled samples. Error bars indicate SEM (from 4 independent experiments, each performed in duplicates).
Isotope labeling experiments were then performed in HEK293-AT1 cells to verify the effects of expressed PI-PLC on PtdIns hydrolysis. Since PI-PLC activity generates DAG, which is rapidly converted to phosphatidic acid (PtdOH), DAG production is associated with an increase in 32P incorporation into PtdOH. As shown in Figure 1C, cells expressing PI-PLC showed significantly increased PtdOH radioactivity compared to controls expressing either mRFP or the mutant PI-PLC. Longer labeling (24 h) with myo-[3H]inositol, on the other hand, showed that PtdIns levels were only moderately reduced by the expressed bacterial enzyme (Figure 1C). These results suggested that in cells expressing PI-PLC, a significant fraction of the PtdIns pool is associated with highly mobile cytoplasmic particles.
PtdIns synthesis is localized to a unique compartment
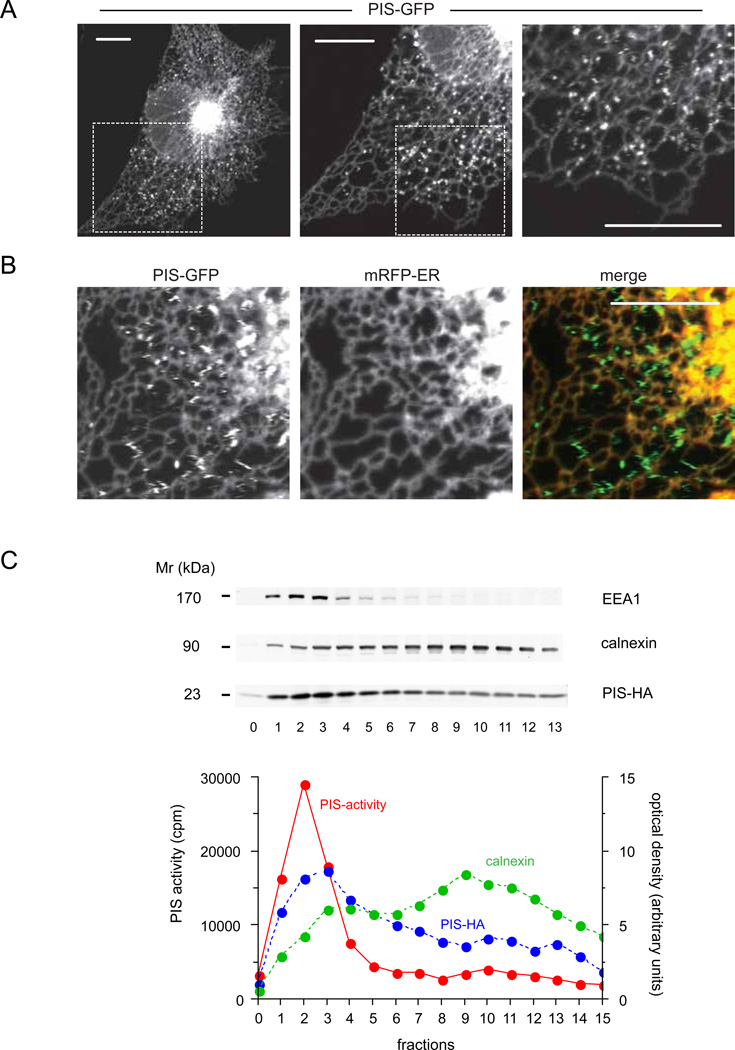
PtdIns synthesis involves the conjugation of myo-inositol with CDP-DAG by a single PIS enzyme that has been previously identified (Nikawa and Yamashita, 1997; Tanaka et al., 1996). To determine the intracellular distribution of PIS, the enzyme was tagged at its C-terminus with GFP and expressed in HEK293-AT1 or COS-7 cells. Strikingly, PIS was not only found in ER tubules and in the central perinuclear ER-Golgi region, but was particularly enriched in rapidly moving mobile structures reminiscent of vesicles (Figure 2A and Movie S3). These structures showed no co-localization with any of the known endomembrane markers, such as lysosomes, peroxisomes, early- or late endosomes or mitochondria, although they did make ample contacts with these organelles (Figure S2A). Also, these PIS vesicles showed no co-localization with lipid droplets induced by oleic acid treatment and visualized with mCherry-Tip47 (Figure S2A). Although PIS was also detectable in the tubular ER, an ER-targeted mRFP was not associated with the PIS positive moving objects (Figure 2B) and several other ER markers (type-I IP3R, STIM1, calreticulin signal-sequence with ER retention signal) also did not appear in the PIS positive mobile structures (not shown).
Figure 2. Cellular distribution of PIS-GFP.
(A) COS-7 cells were transfected with human PIS-fused at its C-terminus with GFP and imaged after 24 h with confocal microscopy. PIS-GFP was localized to the central perinuclear ER and peripheral ER tubules. In addition, note the rapidly moving PIS positive structures that show up as zigzagging traces on the higher magnification scans (A, right panel, see also Figure S2 and Movie S3). (B) Relationship of the PIS positive structures to the ER. COS-7 cells were transfected with PIS-GFP and mRFP-ER. Confocal images show that the mobile structures containing PIS molecule do not contain the ER marker, mRFP-ER. Scale bars, 10 µm. (C) Density gradient separation of membranes. COS-7 cells were transfected with PIS-HA and broken cell membranes were separated on a 10 ml OPTIPREP 10–25% gradient by overnight ultracentrifugation. The distributions of selected markers are shown along with PIS activity measurements from the top 0.5 ml fractions (0–13 of 20). (Fraction “0” is the last 0.5 ml of the sample on top of the gradient proper).
Next, we performed density gradient centrifugation of membranes prepared from COS-7 cells expressing either PIS-GFP or PIS tagged with a HA-tag at the C-terminus on a 10–25% continuous OPTIPREP gradient. The endogenous ER marker, calnexin as well as the expressed PIS enzymes showed a wide distribution along this shallow gradient. However, there was an enrichment of PIS in the lighter membrane fractions relative to the ER marker and these fractions were where EEA1 was also detected (Figure 2C). Remarkably, the PIS enzymatic activity was mostly associated with the protein found in the lighter membranes. These results were in good agreement with previous fractionation data (Imai and Gershengorn, 1987; Sillence and Downes, 1993) showing high PIS activity in membrane fractions separable from ER membranes.
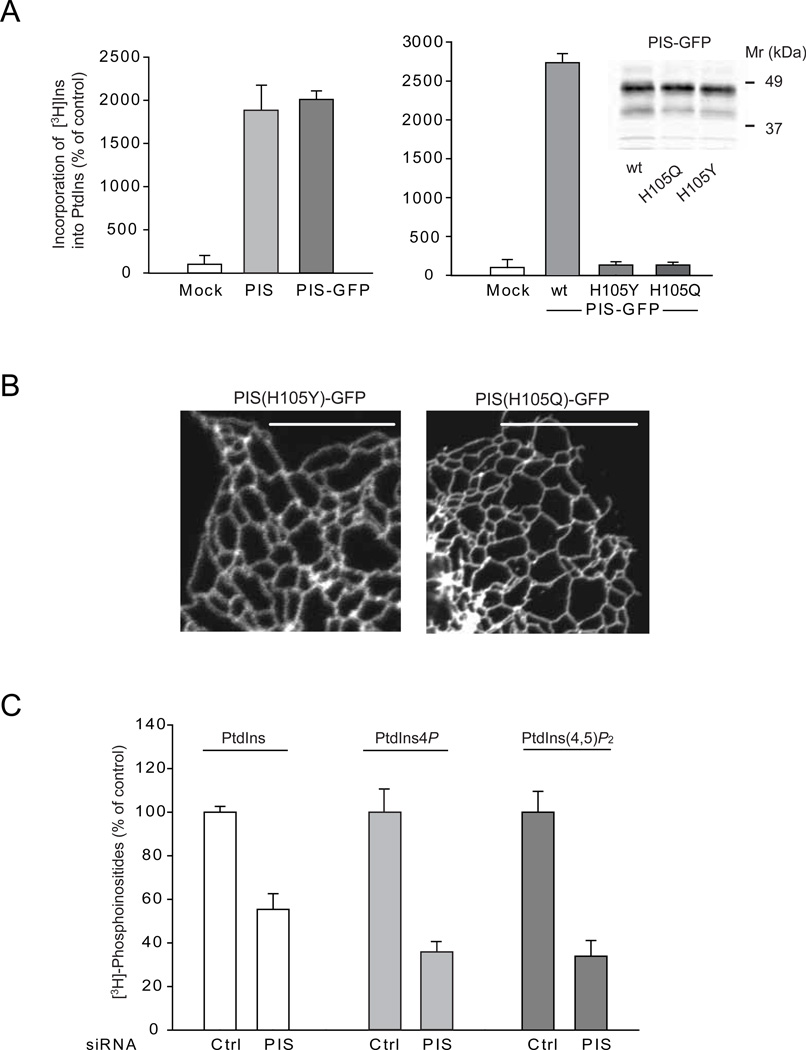
GFP-tagged PIS was also found catalytically active by in vitro PIS assays in membranes prepared from COS-7 cells transfected with untagged PIS or the PIS-GFP (Figure 3A). To determine whether a catalytically inactive mutant PIS would change its cellular localization, we mutated residue His105 to either Gln or Tyr. The corresponding His mutations of the yeast PIS enzyme rendered the enzyme catalytically inactive (Kumano and Nikawa, 1995). Interestingly, the Gln- but not the Tyr mutation allowed the yeast to grow in the presence of high inositol (Kumano and Nikawa, 1995; Nikawa and Yamashita, 1982). As shown in Figure 3A, both of these mutant PIS enzymes was catalytically inactive in our PIS assay. Importantly, neither of these mutant forms could be found in mobile vesicles (Figure 3B). Occasional small clusters of the mutant enzymes could be observed along the ER tubules in a few cells but they did not move or separated from the ER tubules. Although it is not known how these mutations affect the enzyme structurally, these data suggest a connection between the activity and sorting of the enzyme to the mobile compartment. RNAi-mediated down-regulation of endogenous PIS significantly reduced the level of myo-[3H]-inositol labeled PtdIns with concomitant decreases in PtdIns4P and PtdIns(4,5)P2 (Figure 3C). However, overexpression of PIS or CDS2 did not enhance the labeling of PtdIns with either [3H]-inositol or 32P-phosphate (not shown) in agreement with earlier reports (Lykidis et al., 1997).
Figure 3. Relationship between PIS activity and localization and effects of PIS knock-down on cellular phosphoinositide levels.
(A) Crude membranes prepared from COS-7 cells expressing either an empty vector, the untagged PIS or PIS-GFP or its mutants (H105Y and H105Q) were assayed for PtdIns synthase activity as described under Methods. The incorporation of myo-[3H]inositol into PtdIns using CDP-DAG as a substrate was significantly increased in cells transfected with either forms of wild-type PIS but not the mutant enzymes. The results of a representative experiments are shown performed in duplicates. The inset on the right shows equal expression of the wild-type and mutant PIS-GFP proteins. (B) Cellular localization of mutant PIS-GFP enzymes (H105Y and H105Q) expressed in COS-7 cells. Note the lack of the mobile PIS positive structures. (C) HEK293-AT1 cells were treated with control siRNA or PIS siRNA for 3 days and labeled with myo-[3H]inositol for 24 hrs as described under Methods. Labeled lipids were extracted from the cell pellets, separated by TLC, and analyzed by a densitometry of exposed films. Error bars indicate SEM (from 3 independent experiments, each performed in duplicates). In other experiments where the separated lipids were eluted and counted with a scintillation counter, the labeling of PtdIns4P and PtdIns(4,5)P2 were 6 and 4 %, respectively, of that of PtdIns.
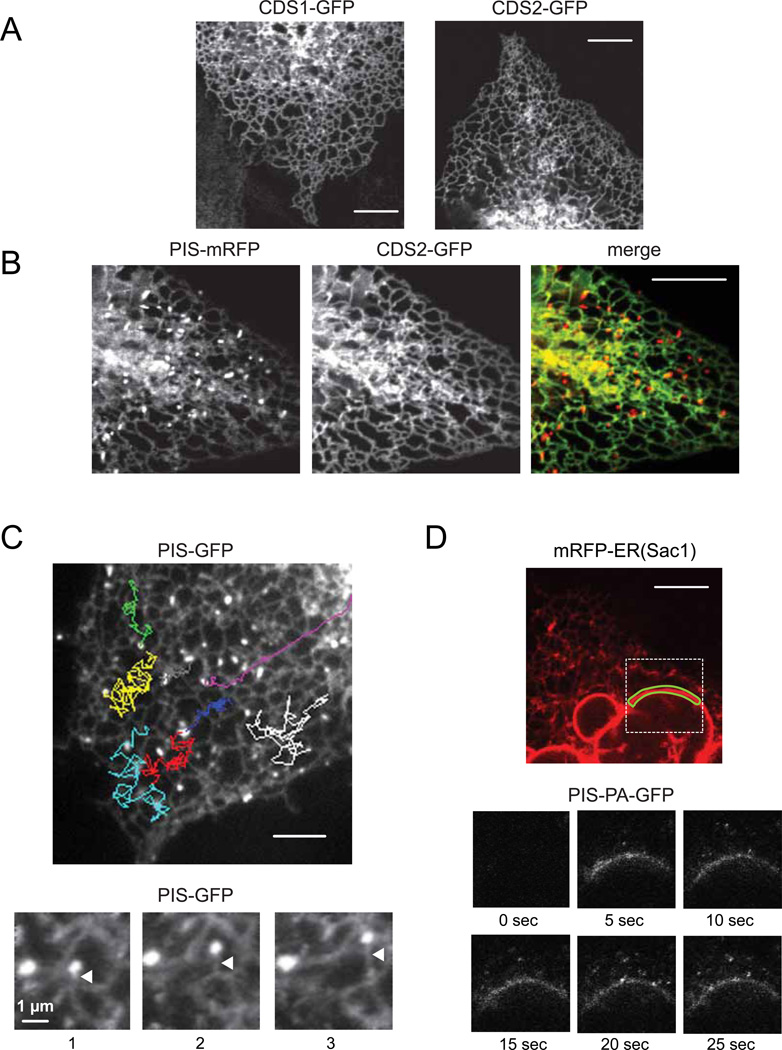
The unique distribution of PIS prompted us to investigate the localization of the two mammalian CDS enzymes (Halford et al., 1998; Heacock et al., 1996) that supply the substrate for PIS. Both of these proteins were tagged with GFP at their C-termini and were found associated with the ER, mostly with the nuclear envelope and the peripheral ER tubules (Figure 4A). Importantly, we could not detect any of the two CDS enzymes on the PIS positive dynamic structures (Figure 4B). These data confirmed that the ER is an important part of CDP-DAG production but also indicated a higher complexity of PtdIns synthesis with prominent differences between the localization of the CDS and PIS enzymes. However, we could not completely rule out that some CDS enzyme, undetectable by our methods, can be present in the mobile compartment.
Figure 4. Cellular localization of CDS enzymes and the origin and movements of PIS positive mobile structures.
(A) COS-7 cells were transfected with human CDS1 or CDS2-fused at their C-termini with GFP and imaged after 24 hrs with confocal microscopy. Both CDS-GFPs were distributed at perinuclear and tubular regions of the ER. (B) PIS-mRFP and CDS2-GFP were co-expressed in COS-7 cells and their images showed that the PIS positive mobile structures did not contain CDS2 molecule. (C) Tracking the movements of selected PIS positive mobile objects from a time-lapse recording of COS-7 cells expressing PIS-GFP. The MetaMorph software was used to follow the movements of individual objects shown by different colors. (Movie S4). Selected frames from a series of time-lapse images from PIS-GFP expressing COS-7 cells (lower panels). Here, the PIS mobile objects appear to remain attached to ER tubules as the latter were extending. These images were processed using the basic filters function of the MetaMorph software (low pass, 10 pixels) to decrease noise. Arrowheads point to the growing ER tubule. (D) COS-7 cells were co-transfected with mRFP-ER and PIS-tagged with photoactivable GFP (PIS-PA-GFP). The small region of the nuclear envelope, outlined with green line, was repeatedly photoactivated by 405 nm laser and the images were taken at the indicated times (see also Movie S5). Note that PIS positive mobile objects became visible after photoactivating only the small region of the nuclear envelope (lower panels). See also Figure S3 and Movie S7 for localization of PIS relative to PM-ER contact zones and Movies S6 and S8 for analysis of the relationship between PIS membranes and Golgi and PM, respectively.
The PIS positive mobile pool is derived from the ER
Tracking of the rapidly moving small PIS objects showed movements along and between the tubular ER with multiple contacts with the latter (Figure 4C and Movie S4). Some of the time-lapse images showed that these small PIS positive objects were tethered to growing ER-tubules (Figure 4C lower). To investigate whether they originated from the ER, we generated a PIS protein tagged with the photoactivable green fluorescent protein (PA-GFP) (Patterson and Lippincott-Schwartz, 2002). Due to the highly mobile and dynamic nature of the tubular ER network, we chose the relatively more static and well-defined nuclear envelope as a site of photoactivation. As shown in Figure 4D, small rapidly moving PIS positive objects became visible very rapidly after repeated photoactivation of a small segment of the nuclear envelope. At the same time, the signal was also diffusing along the nuclear membrane (see also Movie S5).
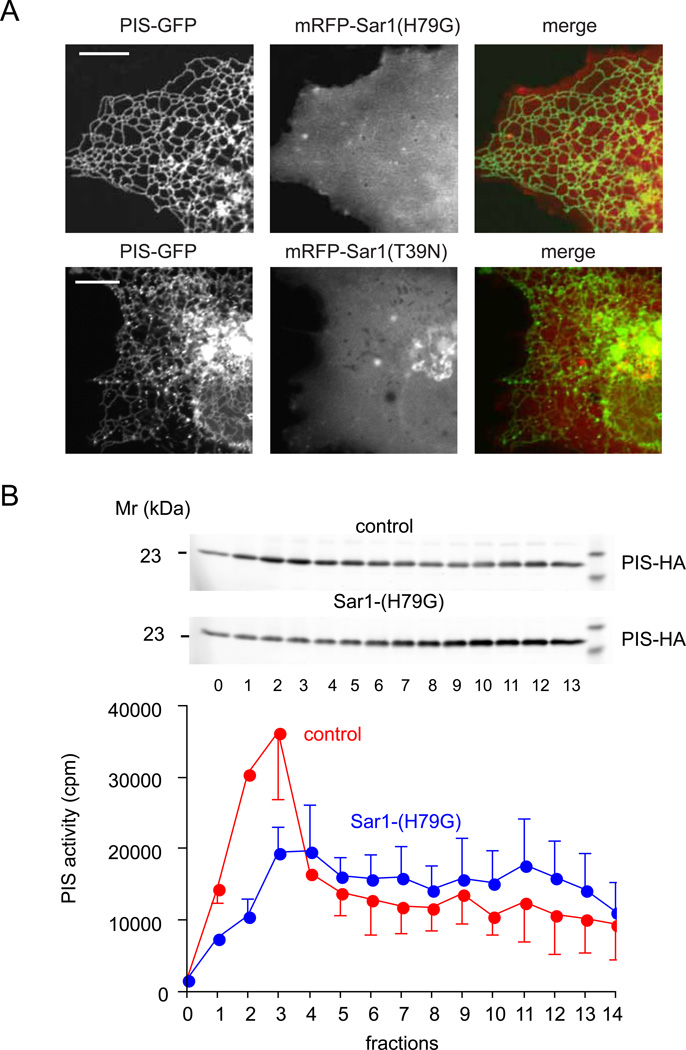
To determine whether the emergence of these structures from the ER is linked to ER exit sites, we performed co-localization experiments with Sec31, which marks this compartment. However, we found no significant co-localization between PIS-GFP and endogenous Sec31 (Figure S2B). Next we determined whether interfering with the small GTPase protein, Sar1, a known regulator of numerous ER-originated membrane remodeling events (Donaldson and Jackson, 2011) affects the formation of the PIS positive structures. As shown in Figure 5A, expression of the GTP-locked mutant form of Sar1 (H79G), tagged with mRFP and HA-tags at the N-terminus, completely eliminated the PIS-positive mobile compartment and kept PIS associated only with the tubular ER (Figure 5A). Interestingly, the GDP-locked form of Sar1 (T39N) was less effective in preventing the formation of the moving vesicles, although it also greatly reduced the number of such objects and the cells showing them (Figure 5A). Gradient fractionation performed on cells expressing PIS-HA with Sar1(H79G) showed that the PIS activity of light fractions was greatly reduced and the protein was mostly present in the ER positive heavier fractions (Figure 5B). These data indicated that the light membrane fractions showing the highest PIS activity corresponded to the mobile compartment and that these objects emerged from the ER membranous network with a Sar1-dependent mechanism.
Figure 5. Sar1 activity is required for the generation of the dynamic PIS organelle.
(A) COS-7 cells were co-transfected with mRFP-HA-Sar1(H79G) or mRFP-HA-Sar1(T39N) and PIS-GFP. Simultaneous presence of the Sar1 GTP-locked mutant (H79G) completely eliminated the mobile PIS positive structures. Sar1-GDP (T39N) had a similar inhibitory effect but it was not as effective. (B) Density gradient separation of membranes from COS-7 cells expressing PIS-HA enzyme with or without Sar1-H79G. Broken cell membranes were separated on a 10 ml OPTIPREP 10–25% gradient by overnight ultracentrifugation and PIS distribution and PIS enzymatic activity were determined by Western Blotting and an enzymatic assay, respectively. The average and range of two separate experiments are shown.
To investigate whether the dynamic PIS membranes might represent transport vesicles between the ER and the Golgi, we performed additional photoactivation experiments with PIS-PA-GFP and a red Golgi marker (mKO-GalT). As shown in Movie S6, photoactivation of PIS outside the Golgi region highlighted the highly mobile PIS structures but these did not fuse with Golgi membranes or increased the fluorescence associated with the Golgi compartment. This finding indicated that the dynamic PIS pool was not a transport intermediate between the ER and the Golgi.
Since PtdIns is present in the PM and the question of whether PtdIns synthesis also takes place in the PM has been repeatedly raised, we also examined the relationship between the PIS positive structures and the ER-PM contact zones. These junctional zones were visualized with the STIM1 protein in COS-7 cells (Lewis, 2007). As shown in Figure S3, the PIS positive particles did make repeated contacts with the STIM1 positive junctional zones (also see Movie S7), but they were not particularly associated with or enriched in these areas (examples of both smaller and larger junctional areas are shown in Figure S3 upper and lower panels, respectively).
It was also important to decide whether the mobile PIS positive structures fused with the PM. This was analyzed by TIRF microscopy. The static picture shown in Figure S3B only demonstrates close contact between the PIS-positive objects and the PM, but Movie S8 shows the kinetics of these movements. Fusion events can be detected in TIRF microscopy as the fluorescence of the fusing object diffuses away from the site of contacts. This is in contrast to the appearance and disappearance of objects as they move to and from the PM without fusion (Ma et al., 2004). Thorough analysis of several TIRF recordings failed to detect any fusion events while the PIS positive objects made ample contacts with the PM (Movie S8). However, we cannot rule out the existence of “kiss and run” events that could easily mediate lipid transfer from this organelle to the PM.
Depleting PtdIns pools with targeted PI-PLC enzymes
Next we investigated the effects of targeted PtdIns depletion on the levels of PtdIns and the other phosphoinositides. We targeted PI-PLC to the ER outer surface by adding the short ER targeting sequence of Sac1 (Varnai et al., 2007). To target the enzyme to the PM, we added the Lyn N-terminal PM targeting sequence (Inoue et al., 2005) to its N-terminus (Figure 6A). We co-expressed the DAG sensor to determine its distribution during these manipulations. Cells were also labeled with myo-[3H]inositol (24 hrs) or with 32P-phosphate (3 hrs) to analyze the size and turnover of the PI pools. These experiments showed that PtdIns pools can be efficiently depleted and turned over by targeting PI-PLC to either the ER or the PM as indicated by the large increase in 32P-labeling of PtdOH (Figure 6B). Membrane targeted PI-PLC also evoked a substantial decrease in myo-[3H]inositol-labeled PtdIns (Figure 6C). These data suggested that the PtdIns pools can be drained both via the ER and the PM, consistent with the movement of the lipid between these membranes. When we looked at the localization of the DAG sensor under these conditions, it did not show ER localization even under conditions when the PI-PLC was targeted to the ER but DAG could be detected in the PM when PI-PLC was PM-targeted (Figure 6A). Notably, the number of small DAG positive motile particles in the cytoplasm was greatly reduced in cells expressing either the ER- or PM-targeted PI-PLC (Figure 6A).
Figure 6. Depletion of different pools of PtdIns by membrane targeted PI-PLC enzymes.
(A) The GFP-tagged DAG sensor was expressed with the cytoplasmic mRFP-tagged PI-PLC (left), its targeted versions directed to the PM (middle) or the cytoplasmic surface of the ER (right) in HEK293-AT1 cells. Scale bars, 10 µm. (B, C) HEK293-AT1 cells were transfected with the cytoplasmic or membrane targeted versions of PI-PLC and labeled with [32P]phosphate for 3 hrs at the end of one day transfection or myo-[3H]inositol for 24 hrs as described under Methods. Labeled lipids were extracted from the cell pellets, separated by TLC, and quantified either by a PhosphorImager for [32P] phosphate labeling or densitometry of films for myo-[3H]inositol samples. Error bars indicate SEM (from 4 independent experiments, each performed in duplicates). Also see Figure S4 for the effects of PtdIns depletion on the level of [3H]PtdIns4P and distribution of cellular PtdIns4P and PtdIns3P reporters.
Although expression of cytoplasmic PI-PLC had the smallest impact on overall PtdIns levels, it already substantially diminished the endosomal localization of the PtdIns3P reporter, FYVE domain (Gillooly et al., 2000) and induced enlarged vesicles, especially in cells expressing the FYVE domain (Figure S4A). (The residual FYVE domain localization to the enlarged vesicles was resistant to PI 3-kinase inhibition.) Expression of cytoplasmic PI-PLC also reduced the Golgi localization of PtdIns4P-recognizing PH domains (FAPP1, OSH1, OSBP) (Figure S4B). Importantly, none of these effects were observed when the catalytically inactive H86A mutant PLC was used (Figures S4A and S4B). Assessment of the PtdIns4P depletion in myo-[3H]inositol-labeled cells also showed that PtdIns4P closely followed the myo-[3H]inositol labeling of PtdIns showing the largest decrease when the PI-PLC was targeted to the PM (Figure S4C). This result indicated that a large fraction of PtdIns4P is found in the PM in agreement with recent immunostaining studies (Hammond et al., 2009), even if most PtdIns4P live biosensors primarily decorate the Golgi (Balla and Varnai, 2009).
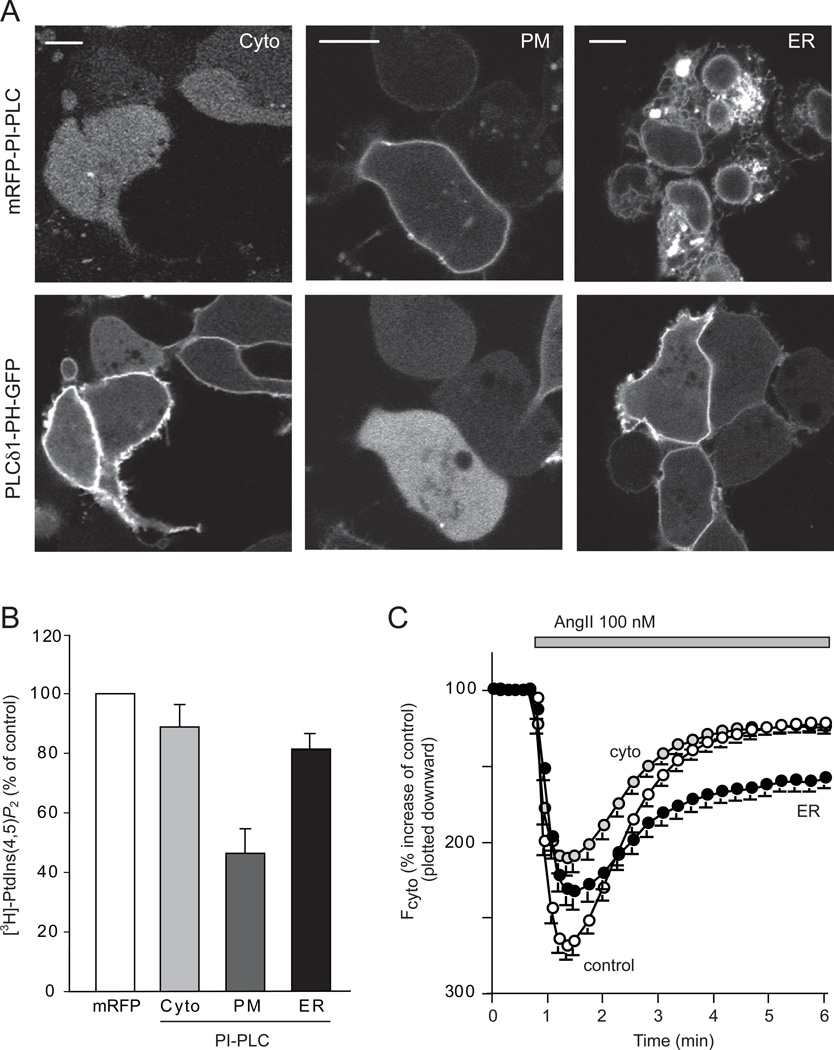
PtdIns(4,5)P2 changes during depletion of PtdIns pools
Next we assessed the PtdIns(4,5)P2 changes during PtdIns depletion in the different membrane compartments. Expression of cytosolic PI-PLC had only a minimal impact (a ~10% decrease) on the level of myo-[3H]inositol labeled PtdIns(4,5)P2 and it was without a noticeable effect on the PM localization of the PtdIns(4,5)P2 reporter, PLCδ1PH-GFP (Figures 7A and 7B). Surprisingly, the ER-targeted PI-PLC only minimally affected the myo-[3H]inositol labeled PtdIns(4,5)P2 (~20% decrease) in sharp contrast to the ~50% decrease in both PtdIns and PtdIns4P (Figure 7B). Consistent with this finding, the PM localization of the PLCδ1PH-GFP was not noticeably affected by the expression of the ER-targeted PI-PLC (Figure 7A). Massive PtdIns(4,5)P2 depletion was only achieved through depletion of PtdIns by the PM-targeted PI-PLC. In this case the reduction (~50%) was comparable to those in the levels of PtdIns and PtdIns4P. PtdIns(4,5)P2 depletion in the PM was confirmed by the greatly reduced or completely lacking PM localization of the PLCδ1PH-GFP (Figures 7A and 7B).
Figure 7. Effects of PtdIns depletion by expression of PI-PLC enzymes on the plasma membrane PtdIns(4,5)P2.
(A) The PtdIns(4,5)P2 sensor, PLCδ1-PH-GFP was expressed with mRFP-conjugated cytoplasmic, PM- or ER- targeted PI-PLC in HEK293-AT1 cells. Scale bars, 10 µm. (B) HEK293-AT1 cells were transfected with the various forms of PI-PLC and labeled with myo-[3H]inositol for 24 hrs as described under Methods. Labeled lipids were extracted from the cell pellets, separated by TLC, and quantified by densitometry of exposed films to measure total myo-[3H]inositol labeled PtdIns(4,5)P2 levels. Error bars indicate SEM (from 4 independent experiments, each performed in duplicates). (C) PLCδ1-PH-GFP was expressed together with mRFP only, cytoplasmic-, PM- or ER- targeted PI-PLC in HEK293-AT1 cells. After 24 hrs, cells were analyzed by confocal microscopy and time-lapse images were recorded after AngII stimulation. The average responses of the cytoplasmic fluorescence intensity of 80–100 cells (mean ± S.E.M) are shown. After normalization to pre-stimulatory levels, these intensity increases were plotted downward to better conceptualize that they reflect the PtdIns(4,5)P2 decreases in the membrane. Note the PH domain was only partially re-localized to the membrane in cells expressing ER-PI-PLC.
The relative preservation of the PM PtdIns(4,5)P2 pool in cells expressing the ER targeted PI-PLC raised the possibility that a significant fraction of ER-PtdIns pool is not used for PtdIns(4,5)P2 synthesis, or that the maintenance of PtdIns(4,5)P2 in the PM is a priority for the cells and that the remaining PtdIns is diverted toward the PM. To test whether the relatively preserved PtdIns(4,5)P2 levels in cells expressing ER-targeted PI-PLC is still maintained against an increased PLC activity, we followed PtdIns(4,5)P2 levels during stimulation with AngII. As shown in Figure 7C, AngII stimulation rapidly released the PLCδ1 PH-domain from the membrane in both the control cells an the cells expressing the ER-targeted PI-PLC, but the re-localization of the probe was strongly impaired in cells expressing ER-targeted PI-PLC. (The smaller increase in the cytoplasmic intensity of the PH-domain probe upon stimulation of the cells expressing the PI-PLC either in the cytosol or the ER was consistent with an already reduced PtdIns(4,5)P2 level and hence membrane localization of the probe before stimulation). PLCδ1-PH domain translocation response was not performed with the PM-targeted PI-PLC, since it has already mostly eliminated the membrane localization of this PH domain (Figure 7A). These data collectively suggested that cells expressing the ER-targeted PI-PLC are able to maintain their basal PtdIns(4,5)P2 at a slightly reduced level but they are unable to re-supply this pool during massive agonist-induced PLC activation.
DISCUSSION
The present experiments were designed to obtain information on the distribution and dynamics of the cellular PtdIns pools in living mammalian cells. Since there is no known protein domain that would specifically recognize PtdIns, we approached this question from two directions. First, we used a PtdIns-specific bacterial PLC to hydrolyze PtdIns and captured the reaction product, DAG with a sensitive DAG sensor. Second, we used GFP-tagged enzymes of PtdIns synthesis, PIS and the CDS1 and -2. Both of these approaches identified highly mobile small particles in the cytoplasm as the sites of PtdIns synthesis and the source of DAG when a PtdIns-specific bacterial PI-PLC is expressed in the cytosol. The PIS positive membranes originate from the ER and use an exit pathway that requires the cycling of the small GTPase Sar1. These organelles have lighter density than the bulk of ER and they contain little if any CDS1/2, the enzyme that supplies CDP-DAG for PtdIns synthesis.
The relationship between the DAG positive structures in PI-PLC expressing cells and the PIS positive organelles is not clear at present. Co-localization studies in live cells were uninformative because of the speed of movements of these structures relative to the speed of image acquisition and because fixation distorted the appearance of DAG positive particles. The DAG vesicles are more numerous and many of them are smaller than the PIS organelles and hence, likely to be at least partially distinct from the latter. However, this could simply be due to higher number of DAG sensors being associated with these objects making their detection more efficient. Nevertheless, the connection between these structures is strongly indicated by the ability of the ER- or PM-targeted PI-PLC to decrease the number of DAG particles. We also cannot rule out the possibility that the tiny DAG positive particles originate from the PIS positive membranes formed by the action of the expressed PI-PLC enzyme.
A few observations suggest that the mobile fraction of the PIS enzyme is functionally different from the one associated with the tubular ER. First, the highest specific activity of the PIS enzyme was associated with light membrane fractions separated from the bulk ER. Second, a catalytically inactive PIS enzyme did not get sorted in the mobile compartment. Third, membrane fractionation from cells expressing both PIS-HA and Sar1-H79G, which prevents the formation of the dynamic PIS pool, greatly reduced the PIS activity associated with the light membrane fractions. Notably, Imai and Gershengorn (1987) found significantly higher Km values for myo-inositol and CDP-DAG for the PIS activity present in the ER membranes than in the membrane fractions that they attributed to the PM. It is not unlikely that the PIS in their “PM” fraction corresponded to the mobile compartment described in the present study. More studies are needed to understand the functional difference between the two pools of PIS enzymes.
Although the nature of these objects is still being evaluated, these findings have altered our views on PtdIns synthesis and distribution. These dynamic, mobile PtdIns distribution platforms would represent a very efficient way of making multiple contacts and deliver PtdIns to the various membranes. Dynamic contact zones between the ER and PM have been increasingly recognized as special sites for lipid transfer and metabolism (Stefan et al., 2011). The current findings indicate that a special PtdIns rich dynamic organelle potentially multiplies the probability of making contacts and exchange lipids with a variety of membranes. A possible way of delivering lipids from the mobile platforms to the target membranes would be a simple “fusion” event but we were unable to detect fusions with the PM or other organelles, although such fusions likely occur with the ER. We also ruled out by photoactivation experiments that the PIS positive membranes would simple be ER-derived transport vesicles directed to the Golgi. While these experiments did not rule out that PtdIns is distributed by simple vesicular transport, it is equally possible that lipid transfer proteins such as the PITPs participate in lipid exchange during the brief contacts between the PIS organelle and other membranes. We have not yet been able to identify the PITP(s) that would serve in that capacity. Since CDP-DAG synthesis is mostly confined to the ER proper, the PIS positive organelles must also return to the ER to resupply the PIS enzyme with its substrate and the multiple contacts and likely fusion events observed between the tubular ER and the PIS organelle could provide the basis for such an exchange.
The PtdIns specific PI-PLC also allowed us to selectively deplete PtdIns in the PM and the ER after targeting the enzyme to these membranes. Both of these manipulations depleted the small cytoplasmic DAG positive particles and also decreased the size of myo-[3H]-labeled PtdIns4P pools and eliminated the localization of PtdIns4P and PtdIns3P recognizing protein modules. These results suggested that PtdIns equilibrates between membranes during the prolonged times (16–24 hrs) of transfection. However, there was a significant difference between the effects of ER- or PM-targeted PI-PLC on PtdIns(4,5)P2. Not surprisingly, PtdIns(4,5)P2 was largely depleted when PtdIns was consumed at the PM. However, somewhat unexpectedly, PtdIns depletion via the ER only moderately decreased PM PtdIns(4,5)P2 under unstimulated conditions. This result suggested that a significant fraction of PtdIns in the ER PtdIns pools is not directed toward the PM and that PM PtdIns were likely originated directly from the PIS organelle. In this context it is important to note that during PIS knockdown there was a parallel decrease in PtdIns, PtdIns4P and PtdIns(4,5)P2 indicating that once PtdIns synthesis is intercepted at the source, it will equally affect all phosphoinositide pools.
There are several open questions that remain to be answered in future studies. How is the generation of the PIS organelle regulated and how does the cells sense the PtdIns status of its membranes? Our preliminary studies showed no obvious change in PIS distribution after agonist stimulation when PtdIns resynthesis is increased, or after loading cells with DAG analogues. It is most likely that we need a thorough quantitative analysis of the statistical behavior of these organelles. The frequency and resident times of contacts probably hold the key to answer these questions, but clearly more studies are needed to fully explore every dimension of these processes.
In summary, the present studies introduced several approaches to determine the localization of PtdIns in living cells and to manipulate PtdIns levels in distinct cellular compartments. Our results revealed that a significant part of PtdIns synthesis is associated with a dynamic ER-derived structure that is capable of making multiple contacts with a variety of organelles and is highly suitable to supply PtdIns lipids to the different membranes. We propose a separate name of PIPEROsome (PI Producing ER-derived Organelle) to designate this structure if it stands the scrutiny of further studies. These findings open new research directions to understand how cells generate and maintain their membrane lipid compositions and to explore the exact molecular events that take place during the contacts between these organelles and their target membranes.
EXPERIMENTAL PROCEDURES
The list of Materials and information on the DNA constructs can be found in the Supplemental Experimental Procedures
Transfection of cells for microscopy
COS-7 cells or HEK293-AT1 cells (a HEK293 cell line stably expressing the rat AT1a angiotensin receptor) were used. Cells (50,000 cells/ well) were plated onto 25-mm-diameter circular glass cover slips in 6 well plates and plasmid DNAs (0.5–1 µg/well) were transfected using the Lipofectamine2000 reagent (Invitrogen) and OPTI-MEM (Invitrogen) following the manufacturer’s instructions.
Live-cell imaging
After 20–24 hrs of transfection, cells were washed on the glass cover slips with a modified Krebs-Ringer solution, containing 120 mM NaCl, 4.7 mM KCl, 1.2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, 10 mM Na-Hepes, pH 7.4 and the coverslip was placed into a metal chamber (Atto, Invitrogen) that was mounted on a heated stage (35 °C, or room temperature for HEK293-AT1 cells). Cells were incubated in 1 ml of the Krebs-Ringer buffer and the stimuli were dissolved and added in 200 µl warm buffer removed from the cells. Cells were examined in inverted microscopes. Confocal images were obtained with a Zeiss LSM510-META laser confocal microscope (Carl Zeiss MicroImaging, Inc.) using a 63× oil-immersion objective equipped with an objective heater (Bioptech). Total internal reflection fluorescent microscopy (TIRF) analysis was performed at 35 °C in an Olympus dual launch TIRF microscope system equipped with a Photometrics Cascade II camera and a PlanApo 60×/1.45 objective.
Measurements of receptor-stimulated PtdIns(4,5)P2 kinetics using PLCδ1 PH domain translocation
HEK293-AT1 cells were transfected with the PLCδ1-PH-GFP construct together with cytoplasmic-, PM- or ER-PI-PLC constructs. After 24 h, cells were imaged in a Zeiss LSM 510-META confocal microscope at room temperature. After stimulation with 100 nM AngII, the translocation of the GFP-probe was monitored in time-lapse imaging. The translocation of the construct from the membrane to the cytosol was quantified by measuring cytosolic GFP intensity in ROIs outside the nucleus and plotted against time using the Zeiss image processing software. Intensity curves were normalized to prestimulatory values and they were averaged from a large number of cells in recordings obtained in several dishes in multiple independent experiments.
Analysis of myo-[3H]Inositol- or [32P]Phosphate-labeled lipids
HEK293-AT1 cells plated on 12-well plates were labeled with myo-[3H]inositol (20 µCi/ml) in 1 ml of inositol-free DMEM supplemented with 2% dialyzed FBS for 24 h or with 2 µCi/ml o-[32P]phosphate for 3 h in phosphate-free DMEM supplemented with 2% dialyzed FBS. The labeling was terminated by the addition of ice-cold perchloric acid (5% final concentration), and cells were kept on ice for 30 min. After scraping and freezing/thawing, the cells were centrifuged and the cell pellet was processed to extract the phosphoinositides by an acidic chloroform/methanol extraction followed by thin layer chromatography (TLC) essentially as described previously (Nakanishi et al., 1995).
RNA interference
The target sequence for PIS siRNA was 5’-TCCGGTGCTTCGGATCTACTA-3’ and the oligonucleotide duplexes were obtained from Qiagen. Cells were cultured in either 6-well dishes (for confocal microscopy) or 12-well plates (for metabolic labeling studies) and treated with 100 nM siRNA using the oligofectamine reagent, twice in consecutive days, and they were analyzed after 3 days incubation of siRNA.
Density gradient centrifugation
COS-7 cells were grown on 10 cm culture dishes and transfected with PIS-GFP or PIS-HA (1 µg plasmid DNA /plate). Cells were harvested 24 h later by scraping into ice-cold 2.0 ml lysis medium (0.25 M sucrose, 10 mM Tris/HCl, pH 7.4, 1 mM EDTA, 10 µg/ml Aprotinin, 10 µg/ml Leupeptin, 1mM AEBSF). Cells were passed through 25 gauge needles 15 times and the membranes centrifuged at 2,000 × g for 5 min. The supernatant was then layered on top of a 10 ml linear OPTIPREP gradient made by a gradient maker using 10 and 25% OPTIPREP diluted from 60% OPTIPREP (Sigma) with 0.25 M sucrose, 60 mM Tris/HCl, pH 7.4, 6 mM EDTA. After centrifugation (17 hrs, 200,000 × g in an SW40 Ti rotor) the gradients were fractionated from the top into 0.5 ml fractions and an aliquot was used freshly to measure PIS activity. Proteins were precipitated by TCA and separated by PAGE. Western blotting was performed with antibodies against proteins representing the various organelles.
PtdIns synthase assay
The PIS-clone obtained in the pSport6 mammalian expression vector (Open Biosystem) or the PIS-GFP construct (see above) were transfected into COS-7 cells using Lipofectamine 2000. After 1 day transfection, cells in 6-well dishes were washed and scraped into 0.2 ml of 50 mM Tris-HCl pH8.0 and 0.1 mM EGTA and ruptured by freezing and thawing three times. The crude membranes were centrifuged in a benchtop centrifuge at 13,000 rpm for 10 min. For the gradient separated membranes, 100 µl of the 0.5 ml fractions were used. The assay was performed essentially as described by Carman and Fischl (Carman and Fischl, 1992). Briefly, the reaction mixture contained 50 mM Tris-HCl pH 8.0, 100 mM KCl, 20 mM MgCl2, 0.15% Triton X-100, 2 mM MnCl2, 5 µCi myo-[3H]inositol, 0.05 mM myo-inositol, 0.2 mM CDP-DAG (Avanti Polar lipids) and an aliquot of the crude membrane, all in a total volume of 0.1 ml. For the gradient samples the reaction volume was increased to two fold. After 30 min incubation at 37 °C, the reaction was stopped by adding of 0.35 ml of acidic methanol (0.1N HCl), followed by 0.5 ml of chloroform and 0.5 ml of 1M MgCl2. After vortexing, two phases were separated by centrifugation at 2000 rpm for 5min. The lower phase containing the chloroform fraction was transferred to scintillation vials, dried and counted for radioactivity to determine the incorporation of myo-[3H]inositol into PtdIns.
Highlights.
-
-
PtdIns synthesis takes place in a highly mobile organelle
-
-
The PtdIns-synthesizing organelle is ER-derived
-
-
The PtdIns synthesizing organelle is the ultimate source of polyphosphoinositides
-
-
PtdIns depletion at the plasma membrane but not the ER impacts PtdIns(4,5)P2 pools
Supplementary Material
ACKNOWLEDGEMENT
Confocal imaging was performed at the Microscopy & Imaging Core of the National Institute of Child Health and Human Development, NIH with the kind assistance of Drs. Vincent Schram and James T. Russell. The enthusiastic contribution of Miss Anna Mezey-Brownstein to this project during her Summer Internship is greatly appreciated. We are also grateful to Dr. Howard Goldfine (University of Pennsylvania) for Listeria strains. This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFRENCES
- Acharya JK, Labarca P, Delgado R, Jalink K, Zuker CS. Synaptic defects and compensatory regulation of inositol metabolism in inositol polyphosphate 1-phosphatase mutants. Neuron. 1998;20:1219–1229. doi: 10.1016/s0896-6273(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Bradley RM, Brady RO. The enzymatic synthesis of inositol phosphatide. J.Biol.Chem. 1958;233:1077–1083. [PubMed] [Google Scholar]
- Balla T, Baukal AJ, Guillemette G, Catt KJ. Multiple pathways of inositol polyphosphate metabolism in angiotensin-stimulated adrenal glomerulosa cells. J.Biol.Chem. 1988;263:4083–4091. [PubMed] [Google Scholar]
- Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Varnai P. Visualization of cellular phosphoinositide pools with GFP-fused protein-domains. Chapter 24, Unit 24 24. Curr Protoc Cell Biol. 2009 doi: 10.1002/0471143030.cb2404s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannam T, Goldfine H. Mutagenesis of active-site histidines of Listeria monocytogenes phosphatidylinositol-specific phospholipase C: effects on enzyme activity and biological function. Infect Immun. 1999;67:182–186. doi: 10.1128/iai.67.1.182-186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Downes CP. The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary, selective, consequences of inositol depletion in 1321N1 cells. Biochem.J. 1994;297:529–537. doi: 10.1042/bj2970529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Fischl AS. Phosphatidylinositol synthase from yeast. Methods Enzymol. 1992;209:305–312. doi: 10.1016/0076-6879(92)09038-5. [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Garner K. Function of the phosphatidylinositol transfer protein gene family: is phosphatidylinositol transfer the mechanism of action? Crit Rev Biochem Mol Biol. 2011;46:89–117. doi: 10.3109/10409238.2010.538664. [DOI] [PubMed] [Google Scholar]
- De Camilli P, Meldolesi J. Subcellular distribution of the PI effect in the pancreas of the guinea pig. Life Sci. 1974;15:711–721. doi: 10.1016/0024-3205(74)90509-8. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem. 2007;282:826–830. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier LM, Parton GP, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OH, Ryan M. Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim Biophys Acta. 1999;1441:237–254. doi: 10.1016/s1388-1981(99)00153-5. [DOI] [PubMed] [Google Scholar]
- Halet G. Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol Cell. 2005;97:501–518. doi: 10.1042/BC20040080. [DOI] [PubMed] [Google Scholar]
- Halford S, Dulai KS, Daw SC, Fitzgibbon J, Hunt DM. Isolation and chromosomal localization of two human CDP-diacylglycerol synthase (CDS) genes. Genomics. 1998;54:140–144. doi: 10.1006/geno.1998.5547. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock AM, Uhler MD, Agranoff BW. Cloning of CDP-diacylglycerol synthase from a human neuronal cell line. J Neurochem. 1996;67:2200–2203. doi: 10.1046/j.1471-4159.1996.67052200.x. [DOI] [PubMed] [Google Scholar]
- Hokin LE, Hokin MR. Effects of acetylcholine on the turnover of phosphoryl units in individual phospholipids of pancreas slices and brain cortex slices. Biochim.Biophys.Acta. 1955;18:102–110. doi: 10.1016/0006-3002(55)90013-5. [DOI] [PubMed] [Google Scholar]
- Hokin LE, Hokin MR. The incorporation of 32P from[32P]ATp into polyphosphoinositides and phosphatidic acid in erythrocyte membranes. Biochim.Biophys.Acta. 1964;84:563–575. doi: 10.1016/0926-6542(64)90126-x. [DOI] [PubMed] [Google Scholar]
- Ile KE, Schaaf G, Bankaitis VA. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- Imai A, Gershengorn MC. Independent phosphatidylinositol synthesis in pituitary plasma membrane and endoplasmic reticulum. Nature. 1987;325:726–728. doi: 10.1038/325726a0. [DOI] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson S, Nahorski SR, Challiss RA. Disruption by lithium of phosphatidylinositol-4,5-bisphosphate supply and inositol-1,4,5-trisphosphate generation in Chinese hamster ovary cells expressing human recombinant m1 muscarinic receptors. Mol.Pharmacol. 1994;46:1138–1148. [PubMed] [Google Scholar]
- Kumano Y, Nikawa J. Functional analysis of mutations in the PIS gene, which encodes Saccharomyces cerevisiae phosphatidylinositol synthase. FEMS Microbiol Lett. 1995;126:81–84. doi: 10.1111/j.1574-6968.1995.tb07394.x. [DOI] [PubMed] [Google Scholar]
- Lapetina EG, Michell RH. Stimulation by acetylcholine of phosphatidylinositol labelling: subcellular distribution in rat cerebral-cortex slices. Biochem.J. 1972;126:1141–1147. doi: 10.1042/bj1261141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RS. The molecular choreography of a store-operated calcium channel. Nature. 2007;446:284–287. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- Lykidis A, Jackson PD, Rock CO, Jackowski S. The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. J Biol Chem. 1997;272:33402–33409. doi: 10.1074/jbc.272.52.33402. [DOI] [PubMed] [Google Scholar]
- Ma L, Bindokas VP, Kuznetsov A, Rhodes C, Hays L, Edwardson JM, Ueda K, Steiner DF, Philipson LH. Direct imaging shows that insulin granule exocytosis occurs by complete vesicle fusion. Proc Natl Acad Sci U S A. 2004;101:9266–9271. doi: 10.1073/pnas.0403201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH. Inositol phospholipids and cell surface receptor function. Biochim.Biophys.Acta. 1975;415:81–147. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- Michell RH, Harwood JL, Coleman R, Hawthorne JN. Characteristics of rat liver phosphatidylinositol kinase and its presence in the plasma membrane. Biochim.Biophys.Acta. 1967;144:649–658. doi: 10.1016/0005-2760(67)90053-7. [DOI] [PubMed] [Google Scholar]
- Monaco ME, Cassai ND, Sidhu GS. Subcellular localization of phosphatidylinositol synthesis. Biochem Biophys Res Commun. 2006;348:1200–1204. doi: 10.1016/j.bbrc.2006.07.196. [DOI] [PubMed] [Google Scholar]
- Murphy TR, Vihtelic TS, Ile KE, Watson CT, Willer GB, Gregg RG, Bankaitis VA, Hyde DR. Phosphatidylinositol synthase is required for lens structural integrity and photoreceptor cell survival in the zebrafish eye. Exp Eye Res. 2011 doi: 10.1016/j.exer.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Catt KJ, Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc.Natl.Acad.Sci.U.S.A. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. Yeast mutant defective in synthesis of phosphatidylinositol. Isolation and characterization of a CDPdiacylglycerol--inositol 3-phosphatidyltransferase Km mutant. Eur J Biochem. 1982;125:445–451. doi: 10.1111/j.1432-1033.1982.tb06703.x. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. Phosphatidylinositol synthase from yeast. Biochim Biophys Acta. 1997;1348:173–178. doi: 10.1016/s0005-2760(97)00103-3. [DOI] [PubMed] [Google Scholar]
- Oancea E, Teruel MN, Quest AFG, Meyer T. Green fluorescent protein (GFP)-tagged cystein-rich domains from protein kinase C as a fluorescent indicators for diacylglycerol signaling in living cells. J. Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A. Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res. 2009;48:307–343. doi: 10.1016/j.plipres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Sillence DJ, Downes CP. Subcellular distribution of agonist-stimulated phosphatidylinositol synthesis in 1321 N1 astrocytoma cells. Biochem.J. 1993;290:381–387. doi: 10.1042/bj2900381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh Proteins Regulate Phosphoinositide Metabolism at ER-Plasma Membrane Contact Sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nikawa J, Imai H, Yamashita S, Hosaka K. Molecular cloning of rat phosphatidylinositol synthase cDNA by functional complementation of the yeast Saccharomyces cerevisiae pis mutation. FEBS Lett. 1996;393:89–92. doi: 10.1016/0014-5793(96)00858-7. [DOI] [PubMed] [Google Scholar]
- Thakur PC, Stuckenholz C, Rivera MR, Davison JM, Yao JK, Amsterdam A, Sadler KC, Bahary N. Lack of De novo phosphatidylinositol synthesis leads to endoplasmic reticulum stress and hepatic steatosis in cdipt-deficient zebrafish. Hepatology. 2011 doi: 10.1002/hep.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- Wei Z, Zenewicz LA, Goldfine H. Listeria monocytogenes phosphatidylinositol-specific phospholipase C has evolved for virulence by greatly reduced activity on GPI anchors. Proc Natl Acad Sci U S A. 2005;102:12927–12931. doi: 10.1073/pnas.0501725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS. Regulation of PLC-mediated signalling in vivo by CDP-diacylglycerol synthase. Nature. 1995;373:216–222. doi: 10.1038/373216a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.