Abstract
Here we report the first example of using β-galactosidase to trigger the formation of cell compatible, supramolecular nanofibers, which ultimately may lead to a new approach for the development of soft nanotechnology.
This article reports the use of β-galactosidase (β-gal) for triggering molecular self-assembly in water to form ordered nanostructures and to result in a hydrogel as potential nanobiomaterials. In the past two and half decades, nanotechnology has evolved into a subfield of science. In the meantime, the nanometer sizes and surface characters shared between the nanostructures in physical science and biological science have led to the recognition that “future developments in nanoscience could provide the basis for artificial life”1 because living cells consist of arrays of dynamically self-assembled nanostructures (e.g., enzymes, receptors, microtubules, ribosomes, or molecular motors) for carrying out metabolic processes, proliferation, differentiation, and other biological functions. These conceptual advancements have resulted in the development of soft nanotechnology, the branch of nanotechnology focuses on the synthesis, properties, and functions of organic nanostructures for mimicking and interacting with naturally existing nanostructures.2 Based on this notion, supramolecular hydrogels,3 consisting of water and self-assembled nanofibers formed via supramolecular interactions, are becoming attractive candidates for the development of soft nanotechnology. Supramolecular nanofibers/hydrogels have been offering new scaffolds for regenerative medicine,4 dressings for wound healing,5 templates for biomineralization,6 vehicles for controlled drug release,7 matrices for protein microarrays,8 a low-cost platform for screening enzyme inhibitors,9 and components for enzyme mimetics.10 Recently, the supramolecular hydrogel of lanreotide has received regulatory approval as subcutaneous long-acting implants for acromegaly treatment, which certainly underscores the potentials of supramolecular nanofibers /hydrogels in biomedicine.11
Generally, the change of temperature, pH, or ionic strength can initiate molecular self-assembly in water, thus resulting in hydrogelation. Although this method has led to the formation of numerous nanofibers/hydrogels, it still has some inherent disadvantages. For example, because it may cause irreversible damages to cells, the change of temperature, pH, or ionic strength has limited use for self-assembly/hydrogelation in vivo. Because supramolecular interaction and enzymatic reaction are ubiquitous in nature for the formation of cellular nanostructures via self-assembly,12 it is reasonable to use an enzyme to trigger the molecular self-assembly for generating supramolecular nanofibers/hydrogels for interacting and regulating biological systems.13 Using phosphatase,14 carboxylesterase,15 β-lactamase,16 and matrix metalloprotease (MMP),17 we have demonstrated that enzymes can trigger the formation of supramolecular nanofibers/hydrogels in vitro, in vivo, even inside cells.14c, 15b These results not only validate the enzyme-based approach13a for soft nanotechnology, but also encourage us to examine other important enzymes for generating supramolecular nanofibers/hydrogels.
Catalyzing the hydrolysis of the glycosidic linkages in saccharides and glycoconjugates, glycosidases are a class of enzymes fundamental to the existence of life as well as invaluable drug targets.18 Among glycosidases, β-gal has received the most extensive study as a model for enzymology and a useful tool for biomedical researches.19 Given the importance of β-gal, we choose it to trigger molecular self-assembly and hydrogelation. Upon the treatment of β-gal, the precursor 4 (i.e., a β-gal substrate) undergoes hydrolysis to form aglycone 5, which self-assembles into supramolecular nanofibers and gels water. At the concentration of 1.0 mg/mL, 5 form mechanical stable hydrogel containing 3D nanofiber networks. In addition, 4 shows little inhibition towards Escherichia coli (E. coli), with and without active β-gal, suggesting that 4 is cell compatible. As the first example of using β-gal to trigger the formation of supramolecular nanofibers/hydrogels, this work ultimately may lead to a new approach that utilizes metabolic enzymes/processes for the development of soft nanotechnology.
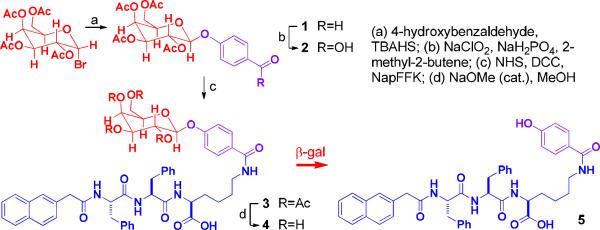
Scheme 1 shows the design of the precursor, which consists a galactose (in red color), as the relatively hydrophilic segment, and 2-naphthylacetic acid-Phe-Phe (NapFF, in blue color), as an effective self-assembly motif.20 To allow the cleavage of the precursor by β-gal at the active site, we use 4-hydroxybenzoic acid19b (in purple color) to connect galactose and the self-assembly motif via the four-carbon side chain of lysine (in blue color). A molecular docking model based on the crystal structure of the complex of β-gal and galactose19c (Fig. 1) suggests the design of the precursor should permit expected hydrolysis to generate the hydrogelator for self-assembly. Based on a procedure reported by Roy,21 phase-transfer catalysis glycosylates 4-hydroxylbenzaldehyde to afford the aryl β-d-galactopyranoside (1) using acetobromo-α-d-galactose, which undergoes a Kraus oxidation22 to 7-O-(β-d-galactopyranosyl) benzoic acid (2) in a quantitative yield. N,N'-dicyclohexylcarbodiimide (DCC) and N-hydroxysuccinimide (NHS) activate the carboxylic acid group of 2 to react with the amine group on the side chain of lysine of NapFF-Lys (NapFFK).20b Finally, Zemplén de-O-acylation gives 4 in a good yield (96 %).
Scheme 1.
The synthetic route for making the precursor 4 and the transformation from 4 to 5 by β-gal.
Fig.1.
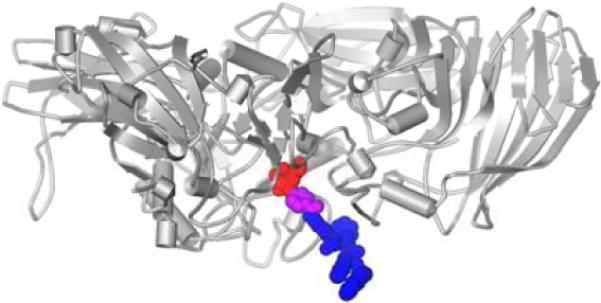
The binding of one subunit of β-galactosidase (in gray) to the precursor 4 based on the crystal structure of β-gal.
After obtaining 4, we test its ability to form supramolecular nanofibers upon the catalysis of β-gal. At the concentration of 1.0 mg/mL, 4 only forms a solution (Fig. 2A), and the variation of pH of the solution from 5 to 11 causes no hydrolysis and leads to neither hydrogelation nor the formation of the nanofibers. After the addition of 2.0 U β-gal (8 μL, 0.25 U/μL), 4 completely transforms to 5 in 20 hours according to LC-MS analysis, and the solution turns into a transparent hydrogel (300 μL, 1.0 mg/mL) (Fig. 2B). Interestingly, at a higher concentration (8.0 mg/mL), the solution of 4 becomes viscous in 4 hours after being dissolved in water, and the solution finally turns into a translucent hydrogel after the pH of the solution being increased to 11 (Fig. 2C), even without the addition of β-gal. The gel remains stable after the pH is changed from 11 back to 7.5. This result suggests that the addition of NaOH causes hydrogelation by changing ionic strength instead of pH. Not surprisingly, the addition of β-gal to the fresh prepared or aged solution of 4 (pH = 7.5) at 8.0 mg/mL can easily induce hydrogelation.
Fig.2.

The optical images of (A) the solution of 4 (1.0 mg/mL, 300 μL, pH = 7.5); (B) the gel of 5 (1.0 mg/mL, 300 μL, pH = 7.5) formed by the addition of β-gal (2.0 U) to the solution of 4; and (C) the gel of 4 (8.0 mg/mL, 400 μL, pH = 11)
Rheology measurement also confirms that the solution of 4 at the concentration of 1.0 mg/mL (Fig. 3) behaves as a liquid before the addition of β-gal. The solution turns to mechanical stable gel after the addition of β-gal, evidenced by that the ratio of G' and G” increases from almost 1 to 2.95 and the critical strain becomes 20% from almost undetectable in the strain sweep. According to the frequency sweep, the formed gel shows a little dependence to the frequency at low frequency range, which confirms the formation of the hydrogel, albeit a relatively weak one.
Fig.3.
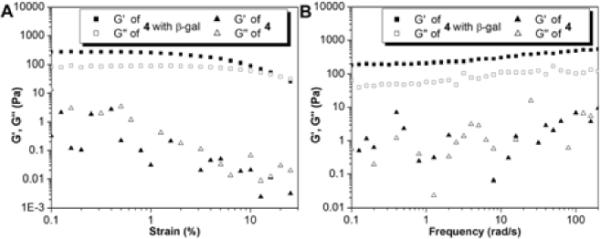
The (A) strain sweep and (B) frequency sweep of the solutions of 4 before and after adding β-gal (2.0 U) at the concentration of 1.0 mg/mL.
The transmission electron microscopy (TEM) images of the hydrogels reveal that nanofibers of 5 entangle into 3D networks (Fig. 4A). The diameter of the nanofibers is around 12 nm. When the concentration of 5 is at the 0.5 mg/mL, which is half of the gelation concentration, the nanofibers of 5 emerge to form the bundles with the narrowest width of 60 nm upon the treatment of β-gal (Fig. 4B). Because the bundles do not interact to form networks, they fail to result in a hydrogel. This result agrees with that a certain density of crosslinking is necessary for hydrogelation.
Fig.4.
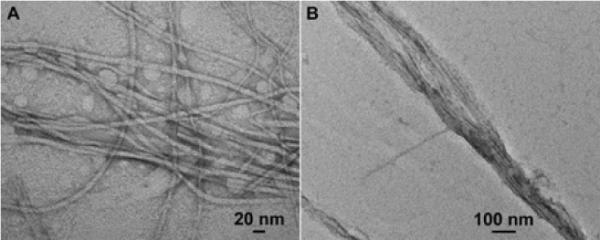
The negative stain TEM images of (A) the nanofibers from gel of 5 at the concentration of 1.0 mg/mL and (B) the nanostructures in the solution of 5 at the concentration of 0.5 mg/mL.
After characterizing the nanofibers of 5, we test the biocompatibility of the nanofibers towards E. coli. Because the Luria-Bertani (LB) media used to culture the bacteria contains enough ionic strength (ca. 170 mM NaCl) to induce nanofiber formation of 4 at the high concentration of 8.0 mg/mL, we use 4 at the concentration of 0.5 mg/mL for the in vitro assay. We have demonstrated at this concentration that the molecular aggregation of 5 is dependent on β-gal (see Fig. 4B), which is expressed in the bacteria. We use two strains of E. coli, wild type MG1655 (lacZ+)23 and a lacZ− isogenic strain, in which the active site of β-gal (E462, S463) has been deleted rendering it inactive. The activity of β-gal is confirmed by the blue-white assay. We treat the two strains with 4 at concentrations of 0, 1, 100 and 500 μg/mL in the presence of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), which relieves transcriptional repression of lacZ by LacI. After an extensive treatment (ca. 20 hours), we serial dilute the cultures to 10−5 before plating on LB agar and LB agar supplemented with 1 mM IPTG. We observe no difference in cell viability between the two strains (Fig. 5), suggesting that 4 is not toxic to E. coli with active β-gal, at least at a concentration of 0.5 mg/mL. Besides, we quantify the concentration of 4 and 5 in E. coli after incubation with 4 at the concentration of 0.5 mg/mL by LC-MS. The concentration of 4 is almost identical inside and outside E. coli, suggesting 4 can cross the two membranes. The concentration of 5 is 28% of that of 4 in the lacZ+ strain, but undetectable in the lacZ− strain.
Fig.5.
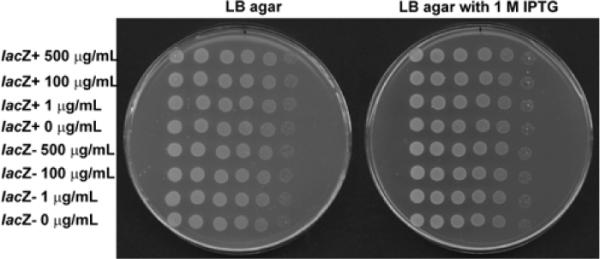
Treatment of isogenic MG1655 lacZ+ and lacZ− with 4. Strains were treated with 0, 1, 100, and 500 μg/mL of 4 in the presence of 1 mM IPTG at 37 °C for ca. 20 hours. Cultures were then serial diluted to 10−5 and plated from left to right on LB agar and LB agar with 1 mM IPTG.
In conclusion, we have demonstrated that β-galactosidase is able to trigger the formation of supramolecular nanofibers in water. Given the prevalence of the glycosidases in living organisms, this work opens a new venue to generate nanostructures via the action of enzymes. Despite the promises, several questions remain to be addressed in future study. Because the glycosidase are indispensable metabolic enzymes to cell growth and proliferation, how to utilize the distribution and expression level of these metabolic enzymes to selectively trigger intercellular nanofiber formation at certain cells,13a especially cancer cells with a fundamentally different metabolic profile,24 remains a challenge. Besides, the solubility of the glycopeptide as the hydrogelator precursor remains to be improved to have distinctive process of the formation of the nanofibers at high concentration. The future study of trigger control by addition of β-gal alone under variable pH, temperature, and concentration may provide useful insights for optimizing the β-gal instructed gelation at high concentration and high ionic strength.
Supplementary Material
Acknowledgments
‡We are grateful for financial support from the NIH (R01CA142746), a start-up grant from Brandeis University, and an HFSP grant (RGP0056/2008). We thank Dr. Isaac J. Krauss, Sebastian Temme and Quan Zhou for the suggestions on the glycoside synthesis and the EM facility at Brandeis University for assistance.
Footnotes
† Electronic Supplementary Information (ESI) available: [details of any DOI: 10.1039/b000000x/
Notes and references
- 1.Mann S. Angew. Chem. Int. Ed. 2008;47:5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 2.(a) Whitesides GM, Lipomi DJ. Faraday Discuss. 2009;143:373–384. doi: 10.1039/b917540g. [DOI] [PubMed] [Google Scholar]; (b) Zelzer M, Ulijn RV. Chem. Soc. Rev. 2010;39:3351–3357. doi: 10.1039/c0cs00035c. [DOI] [PubMed] [Google Scholar]; (c) de Loos M, Feringa BL, van Esch JH. Eur. J. Org. Chem. 2005;2005:3615–3631. [Google Scholar]
- 3.(a) Estroff LA, Hamilton AD. Chem. Rev. 2004;104:1201–1218. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]; (b) Zhao F, Ma ML, Xu B. Chem. Soc. Rev. 2009;38:883–891. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]
- 4.(a) Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]; (b) Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough JE, Ulijn RV. Adv. Mater. 2006;18:611–614. [Google Scholar]; (c) Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Liang G, Ma M, Abbah AS, Lu WW, Xu B. Chem. Commun. 2007:843–845. doi: 10.1039/b616563j. [DOI] [PubMed] [Google Scholar]
- 6.Schnepp ZAC, Gonzalez-McQuire R, Mann S. Adv. Mater. 2006;18:1869–1872. [Google Scholar]
- 7.Vemula PK, Cruikshank GA, Karp JM, John G. Biomaterials. 2009;30:383–393. doi: 10.1016/j.biomaterials.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 8.Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Nat. Mater. 2004;3:58–64. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Xu B. Chem. Commun. 2004:2424–2425. doi: 10.1039/b408897b. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Zhao F, Wang Q, Zhang Y, Xu B. Chem. Soc. Rev. 2010;39:3425–3433. doi: 10.1039/b919450a. [DOI] [PubMed] [Google Scholar]
- 11.Cherif-Cheikh R, Bismuth F, Torres ML, Alloza R, Bosch MT, Montes M, Fuster E, Valles J, Cordero JA, Peraire C, Obach R, Antonijoan R. Proc. Int. Symp. Control. Release Bioact. Mater. 1998;25:798–799. [Google Scholar]
- 12.Lodish HF, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular cell biology. W.H. Freeman; New York: 2008. [Google Scholar]
- 13.(a) Yang Z, Liang G, Xu B. Acc. Chem. Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]; (b) Toledano S, Williams RJ, Jayawarna V, Ulijn RV. J. Am. Chem. Soc. 2006;128:1070–1071. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 14.(a) Yang ZM, Gu HW, Fu DG, Gao P, Lam JK, Xu B. Adv. Mater. 2004;16:1440–1444. [Google Scholar]; (b) Yang ZM, Liang GL, Wang L, Xu B. J. Am. Chem. Soc. 2006;128:3038–3043. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]; (c) Yang Z, Liang G, Guo Z, Xu B. Angew. Chem. Int. Ed. 2007;46:8216–8219. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- 15.(a) Zhao F, Gao Y, Shi J, Browdy HM, Xu B. Langmuir. 2010;27:1510–1512. doi: 10.1021/la103982e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B. Adv. Mater. 2007;19:3152–3156. [Google Scholar]
- 16.Yang ZM, Ho PL, Liang GL, Chow KH, Wang QG, Cao Y, Guo ZH, Xu B. J. Am. Chem. Soc. 2007;129:266–267. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- 17.Yang ZM, Ma ML, Xu B. Soft Matter. 2009;5:2546–2548. [Google Scholar]
- 18.(a) Henrissat B, Davies G. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]; (b) Asano N. Glycobiology. 2003;13:93R–104R. doi: 10.1093/glycob/cwg090. [DOI] [PubMed] [Google Scholar]
- 19.(a) Craven GR, Steers E, Anfinsen CB. J. Biol. Chem. 1965;240:2468–2477. [PubMed] [Google Scholar]; (b) Sinnott ML, Souchard IJ. Biochem. J. 1973;133:89–98. doi: 10.1042/bj1330089. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Juers DH, Heightman TD, Vasella A, McCarter JD, Mackenzie L, Withers SG, Matthews BW. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]; (d) Monod J. Science. 1966;154:475–483. doi: 10.1126/science.154.3748.475. [DOI] [PubMed] [Google Scholar]; (e) Bullock WO, Fernandez JM, Short JM. BioTechniques. 1987;5:376–379. [Google Scholar]; (f) Bondi A, Chieregatti G, Eusebi V, Fulcheri E, Bussolati G. Histochemistry. 1982;76:153–158. doi: 10.1007/BF00501918. [DOI] [PubMed] [Google Scholar]; (g) Lin W-C, Pretlow TP, Pretlow TG, Culp LA. Cancer Res. 1990;50:2808–2817. [PubMed] [Google Scholar]
- 20.(a) Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2010;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gao Y, Kuang Y, Guo Z-F, Guo Z, Krauss IJ, Xu B. J. Am. Chem. Soc. 2009;131:13576–13577. doi: 10.1021/ja904411z. [DOI] [PubMed] [Google Scholar]
- 21.(a) Roy R, Tropper FD. Can. J. Chem. 1991;69:817–821. [Google Scholar]; (b) Carrière D, Meunier SJ, Tropper FD, Cao S, Roy R. J. Mol. Catal. A: Chem. 2000;154:9–22. [Google Scholar]
- 22.Lindgren BO, Nilsson T. Acta Chem. Scand. 1973;27:888–890. [Google Scholar]
- 23.Bachmann BJ. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.