Abstract
The inflammatory response initiated upon burn injury is also associated with extensive metabolic adjustments. While there is a significant body of literature on the characterization of these changes at the metabolite level, little is known on the mechanisms of induction, especially with respect to the role of gene expression. We have comprehensively analyzed changes in gene expression in rat livers during the first 7 d after 20% total body surface area burn injury using Affymetrix microarrays. A total of 740 genes were significantly altered in expression at 1, 2, 4, and 7 d after burn injury compared to sham-burn controls. Functional classification based on gene ontology terms indicated that metabolism, transport, signaling, and defense/inflammation response accounted for more than 70% of the significantly altered genes. Fisher least-significant difference post-hoc testing of the 740 differentially expressed genes indicated that over 60% of the genes demonstrated significant changes in expression either on d 1 or on d 7 postburn. The gene expression trends were corroborated by biochemical measurements of triglycerides and fatty acids 24 h postburn but not at later time points. This suggests that fatty acids are used, at least in part, in the liver as energy substrates for the first 4 d after injury. Our data also suggest that long-term regulation of energy substrate utilization in the liver following burn injury is primarily at the posttranscriptional level. Last, relevance networks of significantly expressed genes indicate the involvement of key small molecules in the hepatic response to 20% total body surface area burn injury.
Keywords: liver gene expression, 20% burn injury, microarrays, energy expenditure
INTRODUCTION
The body’s response to burn injury is characterized by an increase in overall energy expenditure. Metabolic processes, such as gluconeogenesis, ureagenesis, futile cycles (fatty acid synthesis, fatty acid breakdown, and the Cori cycle), the necessity to compensate for the increased loss of body heat through injured skin [1], and up-regulation of liver-synthesized plasma proteins [2], all contribute to this increase in energy requirement [3]. Several studies have demonstrated that an unabated increase in the overall energy expenditure and a state of high metabolic activity arising from severe burn injury or burn injury followed by infection correlates positively with progressive deterioration of major organ systems, including the liver, ultimately leading to multiple organ dysfunction syndrome [4–6]. Therefore, there is significant interest in understanding the contributions of the different postburn physiological processes to the increase energy expenditure in the liver after burn injury.
Metabolic alterations following burn injury have been primarily studied in whole body systems as well as by using isolated perfused organ systems in combination with an array of biochemical techniques [7–9]. The latter studies involve perfusing the organ after burn injury for determining net rates of production or uptake of various metabolites [7, 9]. This approach has been used to describe changes in metabolic processes and establish a biochemical basis for the liver response to burn injury [7, 10, 11]. However, few studies have established the gene expression basis for these changes [12, 13]. Our laboratory [13] has recently investigated the gene expression basis underlying the liver’s short-term (first 24 h) response to burn injury. In conjunction with metabolite measurements, we developed a comprehensive picture of changes in the energy-generating processes in the liver following burn injury that suggest fatty acids are increasingly imported into the liver and oxidized to meet the enhanced energy demands during the first 24 h following burn injury [13]. In addition, alterations in gene expression were sufficient to describe the alterations in the different metabolic processes initiated in the liver after burn injury. However, the role that gene expression plays in the long-term response to burn injury has not been thoroughly investigated and forms the basis of this study.
The objective of this study was to characterize the long-term changes in gene expression during the first 7 d after burn injury. Our data indicate that significant changes in gene expression are observed only on d 1 and 7 postburn, with minimal alterations in gene expression on d 2 and 4. However, changes in triglyceride (TG) and free fatty acids (FFA) are observed on all days, indicating that long-term responses are governed at both the gene expression and the metabolite levels.
MATERIALS AND METHODS
In Vivo Experiments
Animal experiments were performed with male Sprague Dawley rats (Charles River Laboratories, Boston, MA) weighing 150 to 200 g using experimental protocols approved by the Subcommittee on Research Animal Care, Massachusetts General Hospital. Rats were individually housed in a temperature-controlled (25°C) and light-controlled room (12 h light–dark cycle) and allowed to adjust to their new surroundings for at least 5 d prior to the experiment. Water and rat chow were provided ad libitum. Although prior studies suggest that food intake is decreased in the first 24 h after burns [14, 15], pair-feeding was not adopted as unpublished data (not shown) from our laboratory show similar food intake for sham and scald-burn-injured animals. On the day of the treatment, the animals were randomly divided into 2 groups: burn and sham-burn. Three animals were used in each experimental group, based on the minimum number of arrays required for performing meaningful statistical analysis of the gene expression data. For the burn-injury group, a standardized thermal injury was performed on the rats according to methods originally described by Walker and Mason [16]. Briefly, a scald burn of the dorsum, calculated to be ∼20% of the rat’s total body surface area (TBSA), was induced by immersing the designated area in boiling water for 10 s, leading to a third-degree (full-thickness) burn. Prior studies suggest that this burn size is largely nonlethal and induces a systemic inflammatory response as well as hypermetabolic features in the rats [7–9, 17]. Rats were resuscitated with an intraperitoneal injection of sterile saline solution (1.5 mL/kg body weight/% TBSA) immediately after burn. At each time point (1, 2, 4, and 7 d), 3 animals belonging to each group were sacrificed; the liver and serum were collected and stored at –80°C after being inflicted with 20% TBSA burn injury. Control sham-burn rats (n = 3) were treated identically except that they were immersed into a 37°C water bath and immediately sacrificed to collect the livers.
RNA Extraction and Microarray Hybridization
Total RNA was isolated from homogenized liver tissue from each time point (1, 2, 4, and 7 d) using the Nucleospin II RNA isolation kit from Clontech (Palo Alto, CA). Briefly, 50 mg of liver tissue was homogenized in the presence of buffer RA1 (supplied by the manufacturer) and beta-mercaptoethanol. To the clear homogenate, 70% ethanol was added and loaded onto the column to retain the RNA on the membrane. After DNase I treatment, the RNA was eluted into 30 µL of RNAse-free water. Biotinylated c-RNA was prepared from 20 µg of total RNA using protocols provided by Affymetrix and 15 µg of labeled cRNA used to hybridize Affymerix Rat RAE230A arrays. Arrays were washed and scanned according to protocols provided by Affymetrix at the Bauer Center for Genomics, Harvard University, and analyzed using the Affymetrix GENECHIP MAS V.5.0 software. Labeled cRNA from each liver sample was hybridized to a single microarray, for a total of 24 microarrays (3 arrays for 2 experimental conditions at 4 time points).
Data Analysis
All intensity values were scaled to a target intensity of 500 using the Affymetrix GENECHIP MAS V.0 software to account for differences between the replicate chips and their hybridization efficiencies. The entire dataset is available at the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) with the Accession Number GSE5647. The gene expression data were initially filtered to include only genes that were present in all 3 replicate arrays for any experimental condition. A 1-way analysis of variance (ANOVA) filter was used to test each gene independently for a statistical difference in the fold-change in expression (i.e., ratio of expression in burn liver to sham-burn) between the different time points (1, 2, 4, and 7 d). The output of the analysis is the probability (P value) that a difference in expression is observed by chance, i.e., probability of getting a false positive. Genes with P values less than 0.05 were selected as demonstrating a statistically significant difference in expression in the burn-injured liver as compared to the sham-burn liver. The occurrence of false positives was minimized using the false discovery rate (FDR) method [18], which adjusts the ANOVA P value (P < 0.05) to reduce the occurrence of the false positives. Post-hoc analysis was performed using the Fisher least-significant difference test [19] to determine the specific days on which each gene shows a significant change in expression.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
The mRNA sequences for 8 selected genes and 18S rRNA (housekeeping gene) were retrieved from the GenBank database and gene-specific primers were designed for each transcript. RNA was extracted from ∼50 mg of whole liver tissue and RT-PCR was performed with ∼100 ng of total RNA using the Superscript II 1-step RT-PCR kit (Invitrogen, San Diego, CA) on a ICycler real-time PCR machine (Bio-Rad, Hercules, CA). The cycle number at which the fluorescence in each amplification reaction increased beyond a threshold (in the exponential phase of amplification) was determined using the MyiQ software (Bio-Rad). Threshold cycle numbers for each gene were normalized to that of 18S rRNA as described earlier [20]. All RT-PCR experiments were done in triplicate.
TG and FFA Assays
TG levels in liver tissue were measured using a commercially available TG kit (TR0100; Sigma, St. Louis, MO), based on the release of glycerol from TG by lipoprotein lipase. Replacing the lipase with water permitted the quantification of free glycerol in the supernatant. FFA were measured based on the release of hydrogen peroxide in the presence of acyl-CoA synthetase and acyl-CoA oxidase that catalyzes the conversion of FFA into enoyl-coA, using a kit from Roche Molecular Biochemicals (Indianapolis, IN).
Protein Interaction Networks
Protein interaction networks were developed using the Pathway-Architect software (Stratagene, La Jolla, CA) and used to establish connections between the significantly expressed genes in the liver on d 1, 2, 4, and 7 after burn injury. PathwayArchitect utilizes a natural language selection algorithm (MedScan) to establish connections between genes based on information contained in a curated database of biological objects, interactions, and regulations generated from published literature. In this study, we used connections based on regulation (e.g., transcriptional, posttranscriptional), promoter binding, molecular transport (e.g., secretion, trafficking), metabolism, protein modification, binding, expression, and small molecule regulation. Two major protein interaction types were generated using this approach: (1) direct interaction networks wherein connections are only identified using the statistically significant gene list submitted to PathwayArchitect; (2) relevance networks wherein additional genes or small molecules are added [21] by PathwayArchitect to create a denser interaction network. In both cases the output from PathwayArchitect is a map of significantly expressed genes that are linked to one another either directly or indirectly through the expression of other genes or small molecules.
RESULTS
Male Sprague Dawley rats were subjected to a 20% TBSA scald-burn injury and liver tissue was collected 1, 2, 4, and 7 d after burn injury. Changes in the expression of liver genes following burn injury were determined using high-density Affymetrix RG230A arrays.
Temporal Changes in Gene Expression
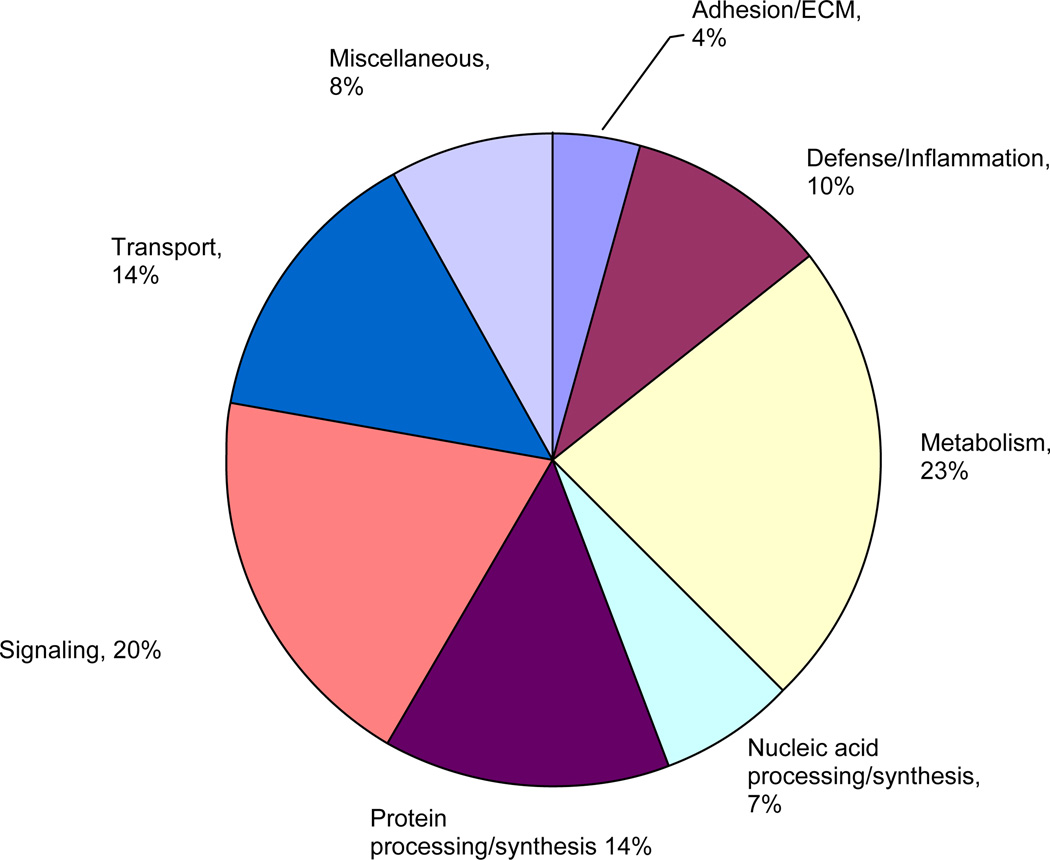
Of the 15,923 genes and ESTs represented on the RG230A array, 13,820 were present in all 3 arrays at each time point and were subjected to 1-way ANOVA to test each gene independently for a statistically significant difference in expression in burn liver compared to sham-burn controls between the different time points (1, 2, 4, and 7 d). Seven hundred forty-two probe sets passed the ANOVA filter and were further corrected for a FDR of 0.05, as described in Materials and Methods. The 740 significantly altered probe sets were annotated using resources in the Affymetrix Netaffx analysis center (http://www.affymetrix.com/analysis/ index.affx), Unigene database, and scientific literature available at PubMed (http://www.ncbi.nlm.nih.gov/ entrez/query.fcgi). A total of 414 of the 740 genes were annotated and sorted into 8 functional categories (Fig. 1).
FIG. 1.
Functional classification of statistically significant genes. Genes that passed the FDR statistical filter were categorized on the basis of their biological function using definitions in the Affymetrix NeAffx center (http://www.affymetrix.com/analysis/index.affx). (Color version of figure is available online.)
Genes involved in metabolic processes (e.g., carbohydrate, cholesterol, lipid, and nucleic acid metabolism) accounted for nearly 23% of all differentially expressed genes. Other categories that were significantly altered include signaling (20%; genes involved in transcriptional regulation and signal transduction), transport (14%; genes involved in mobilizing electrons, ions, lipids, and sterols), protein synthesis and metabolism (14%; genes involved in the processing of polypeptides), and defense/inflammatory response (10%; genes involved in cytokine signaling, complement, and clotting systems). Together, these 5 categories accounted for 80% of all significantly altered genes and reflect changes that are typical following burn injury.
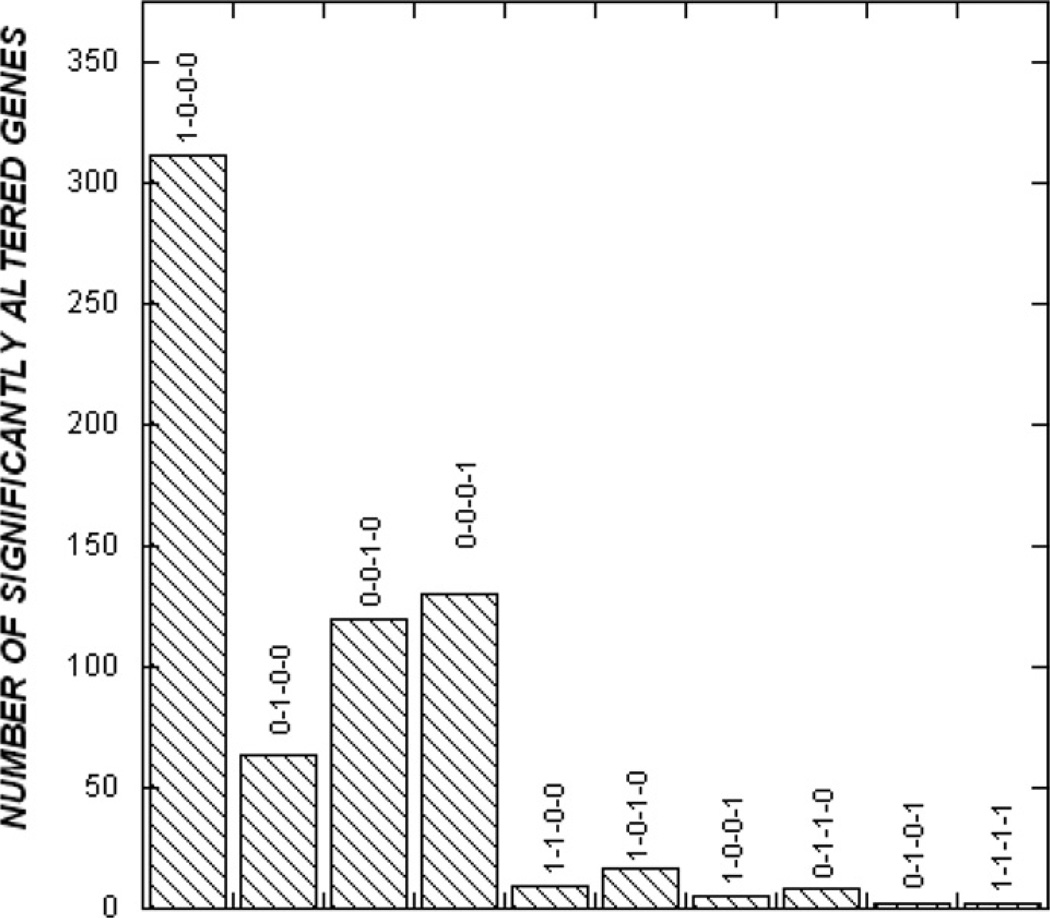
The probe sets that passed the FDR filter were also further analyzed using the Fisher least-significant difference post-hoc test to identify the time point(s) at which the statistically significant changes in expression were observed. Two dominant temporal trends were observed in the entire data set of 740 genes and ESTs (Fig. 2). First, more than 90% of all changes in expression were transient (i.e., on any 1 d following burn injury) and not sustained over several days. In fact, only 3 genes (MAD homologue 2, glutamyl-prolyl-tRNA synthetase, and ribosomal protein L32) demonstrated a change in gene expression on all time points (d 1, 2, 4, and 7 after burn injury). Second, nearly 42% of the observed changes in expression (310 probe sets) were observed on d 1 after burn injury (Fig. 2). The number of significantly altered genes in liver tissue was markedly lower on d 2 (8.6% or 64 probe sets) and increased slightly on d 4 (16.2% or 120 probe sets) and 7 (17.6% or 130 probe sets) as compared to sham-burn controls.
FIG. 2.
Temporal analysis of gene expression. The Fisher least-significant difference test was used to determine the time points at which a statistically significant fold-change in expression between burn and sham-burn liver was observed.
Genes in the major ontology categories also exhibited markedly different temporal patterns over the duration of the experiment. Genes involved in metabolic processes accounted for 20 to 28% of all significantly expressed genes at any time point and was consistently one of the largest ontology groups whose expression was altered. On the other hand, genes involved in defense/inflammatory response were dominant in the list of significantly expressed genes only on d 1 (13% of total significantly altered genes) and gradually decreased over time. Similarly, the fraction of significantly altered genes in the protein synthesis and transport ontologies was highest on d 4 after burn injury. Genes classified as belonging to the signaling ontology group exhibited biphasic behavior, with the maximum contribution on d 2 (35%), followed by a decrease to 10% on d 4 and an increase to 25% on d 7 after burn injury. The expression changes of selected genes were independently confirmed using quantitative reverse transcription-PCR (Table 1) and are in good agreement with the microarray data. Salient changes in 3 of these categories—metabolism, defense/inflammatory response, and signaling—are discussed below.
TABLE 1.
RT-PCR Analysis of Selected Genes from the Microarray Data
| Gene name | Affymetrix ID | Significant expression on |
Microarray fold-change |
RT-PCR fold-change |
|---|---|---|---|---|
| Coagulation factor X | 1369852_at | D1 | 1.82 ± 0.49 | 2.47 ± 0.29 |
| IL-6 signal transducer | 1370957_at | D1 | − 1.35 ± 0.07 | − 1.04 ± 0.14 |
| Solute carrier family 6 | 1368778_at | D1 | 1.89 ± 0.63 | 2.32 ± 0.31 |
| CD36 antigen | 1367689_at | D2 | 3.95 ± 1.17 | 3.69 ± 0.48 |
| Kininogen | 137114_a_at | D2 | 2.77 ± 0.96 | 3.01 ± 0.18 |
| Onecut 1 | 1387760_a_at | D4 | −4.0 ± 0.2 | − 3.22 ± 0.09 |
| Protooncogene Met | 1369218_at | D7 | 1.83 ± 0.54 | 2.52 ± 0.26 |
| Signal tranducer and activator of transcription 3 | 1371781_at | D7 | 1.76 ± 0.45 | 2.75 ± 0.33 |
Metabolism Genes
Genes classified as metabolism-related constituted the single largest ontology category altered in expression at all time points where gene expression was profiled (Table 2). Genes involved in the regulation of fatty acid oxidation (propionyl coenzyme A carboxylase beta), cholesterol synthesis (farnesyl diphosphate synthase), cholesterol transport (sterol carrier protein 2), and bile acid conjugation (taurine transporter) were all significantly increased in expression on d 1 after burn injury. In addition, genes involved in the TCA cycle (pyruvate dehydrogenase E1) were also up-regulated on d 1.
TABLE 2.
Metabolism Genes Altered by 20% Burn Injury
| Affymetrix ID | P | Gene | Symbol | Significant change in expression on d |
|---|---|---|---|---|
| 1367667_at | 0.00297956 | Farensyl diphosphate synthase | fdps | 1 |
| 1388361_at | 0.00019917 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 10 | 1 | |
| 1370151_at | 0.000564898 | Carbamoyl-phosphate synthetase 1, murine thymoma viral (v-akt) oncogene |
Cps1 | 1 |
| 1368832_at | 0.013284629 | Homolog 2 | Akt2 | 1 |
| 1388324_at | 0.000261272 | Nitrilase 1 | Nit1 | 1 |
| 1370370_at | 0.000642335 | Hyaluronidase | Hyal2 | 1 |
| 1368435_at | 0.000774279 | Cytochrome P450, 8b1, sterol 12 alpha-hydrolase | Cyp8b1 | 1 |
| 1371380_at | 0.001045579 | Pyruvate dehydrogenase E1 alpha 1 | Pdha1 | 1 |
| 1367906_at | 0.001385744 | Acid phosphatase 2, lysozymal | Acp2 | 1 |
| 1369061_at | 0.001396486 | glutathione reductase | Gsr | 1 |
| 1377060_at | 0.002422737 | Methylcrotonoyl-Coenzyme A carboxylase 2 | Mccc2 | 1 |
| 1368165_at | 0.002695078 | Phosphoribosyl pyrophosphate synthetase 1 | Prps1 | 1 |
| 1369074_at | 0.004335644 | Amino acid transport system A3 | Slc38a4 | 1 |
| 1368178_at | 0.00455661 | PDZ domain containing 1 | Pdzk1 | 1 |
| 1370187_at | 0.005333347 | Propionyl coenzyme A carboxylase, beta | Pccb | 1 |
| 1371388_at | 0.005740121 | Pyruvate dehydrogenase (lipoamide) beta | Pdhb | 1 |
| 1372101_at | 0.006610444 | ER transmembrane protein Dri 42 | Ppap2b | 1 |
| 1368331_at | 0.007263767 | Chitobiase, di-N-acetyl- | Ctbs | 1 |
| 1370563_at | 0.010115802 | 3-alpha-hydroxysteroid dehydrogenase | 1 | |
| 1367612_at | 0.010725964 | Microsomal glutathione-S-transferase 1 | Mgst1 | 1 |
| 1371966_at | 0.010736311 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 | Pcmt1 | 1 |
| 1367617_at | 0.010969807 | Aldolase A | AldoA | 1 |
| 1387790_at | 0.013292224 | Phosphoribosylaminoimidazole carboxylase | Paics | 1 |
| 1387376_at | 0.013455723 | Aldehyde oxidase 1 | Aox1 | 1 |
| 1386893_at | 0.0151628 | Granulin | Grn | 1 |
| 1371482_at | 0.016301613 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | Ndufs2 | 1 |
| 1370879_at | 0.016746012 | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
1 | |
| 1367575_at | 0.018391572 | Enolase 1, alpha | Eno1 | 1 |
| 1398898_at | 0.019612191 | Endosulfine alpha | Ensa | 1 |
| 1371894_at | 0.021821892 | Glucosamine (N-acetyl)-6-sulfatase | Gns | 1 |
| 1371034_at | 0.022442353 | One cut domain, family member 1 | Onecut1 | 1 |
| 1372471_at | 0.027579584 | UDP-glucose ceramide glucosyltransferase-like 1 | Ugcgl1 | 1 |
| 1398286_at | 0.029644819 | Cysteine sulfinic acid decarboxylase | Csad | 1 |
| 1387896_at | 0.030345056 | sterol carrier protein 2 | Scp2 | 1 |
| 1367642_at | 0.03062091 | Succinate-CoA ligase, GDP-forming, alpha subunit | Suclg1 | 1 |
| 1367917_at | 0.035257827 | Cytochrome P450, family 2, subfamily d, polypeptide 26 | Cyp2d26 | 1 |
| 1373337_at | 0.038129876 | Glyoxylate reductase/hydroxypyruvate reductase | Grhpr | 1 |
| 1373362_at | 0.04022897 | Ubiquinol-cytochrome c reductase core protein II | Uqcrc2 | 1 |
| 1372297_at | 0.046419782 | Glutathione-S-transferase, alpha 4 | Gsta4 | 1 |
| 1368092_at | 0.047029507 | Fumarylacetoacetate hydrolase | Fah | 1 |
| 1374959_at | 0.049248511 | NAD(P)H dehydrogenase, quinone 2 | 1 | |
| 1370284_at | 0.00014788 | ATPase epsilon subunit | ATP5e | 2 |
| 1373041_at | 0.003020817 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 3 | Ndufb3 | 2 |
| 1386871_at | 0.03682998 | Glutathione peroxidase 4 | Gpx4 | 2 |
| 1387630_at | 0.001082576 | ELOVL family member 5, elongation of long chain fatty acids | Elovl5 | 2 |
| 1367622_at | 0.002946455 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | Atp5h | 2 |
| 1370445_at | 0.003936523 | phosphatidylserine-specific phospholipase A1 | Pspla1 | 2 |
| 1368270_at | 0.005692623 | Apolipoprotein B editing complex 1 | ApoBec1 | 2 |
| 1367689_a_at | 0.021897134 | cd36 antigen | Cd36 | 2 |
| 1392604_at | 0.022606882 | NAD(P)-dependent steroid dehydrogenase-like | Nsdh1 | 2 |
| 1367855_at | 0.035360287 | Scavenger receptor class B, member 1 | Scarb1 | 2 |
| 1388414_at | 0.000354217 | NADH dehydrogenase (ubiquinone) Fe-S protein 5b, 15 kDa (NADH- coenzyme Q reductase) |
Ndufs5b | 4 |
| 1388304_at | 0.001786453 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | Ndufb5 | 4 |
| 1371912_at | 0.002073967 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | Ndufs7 | 4 |
| 1367777_at | 0.002208063 | 4-dienoyl CoA reductase 1, | Decr1 | 4 |
| 1368409_at | 0.002582013 | Glutathione-S-transferase, theta 2 | Gstt2 | 4 |
| 1369275_s_at | 0.004015466 | Cytochrome P450 IIA1 | Cyp2a1 | 4 |
| 1388489_at | 0.004797647 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | 4 | |
| 1387240_at | 0.008292462 | Retinol dehydrogenase type III | Rdh3 | 4 |
| 1368275_at | 0.009227855 | Sterol-C4-methyl oxidase-like | Sc4mol | 4 |
| 1389906_at | 0.010398835 | Farnesyl diphosphate farnesyl transferase 1 | Fdft1 | 4 |
| 1369546_at | 0.012943779 | Butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma- butyrobetaine hydroxylase) |
Bbox1 | 4 |
| 1375906_at | 0.01525469 | Nudix (nucleotide diphosphate linked moiety X)-type motif 3 | 4 | |
| 1389334_at | 0.017602044 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | 4 | |
| 1367705_at | 0.023270459 | Glutaredoxin 1 (thioltransferase) | Glrx1 | 4 |
| 1376592_at | 0.02564911 | Methylmalonyl CoA epimerase | Mcee | 4 |
| 1367909_at | 0.02613897 | Dicarbonyl-L-xylulose reductase | Dcxr | 4 |
| 1367720_at | 0.026742304 | Aminolevulinate, delta-, dehydratase | Alad | 4 |
| 1374265_at | 0.031520387 | Similar to arylacetamide deacetylase (esterase) | 4 | |
| 1372004_at | 0.037134398 | Heme binding protein 1 | Hebp1 | 4 |
| 1387296_at | 0.043136174 | Cytochrome P450, family 2, subfamily J, polypeptide 4 | Cyp2j4 | 4 |
| 1375964_at | 0.047951776 | Phosphoserine phosphatase | Psph | 4 |
| 1375412_at | 0.000525169 | Arylsulfatase B | Arsb | 7 |
| 1367674_at | 0.001123933 | Phosphatidylinositol transfer protein, beta | Pitpnb | 7 |
| 1367568_a_at | 0.001740532 | Matrix gamma carboxyglutamic acid | Mgp | 7 |
| 1370952_at | 0.003298992 | Glutathione-S-transferase, mu 2 | Gstm2 | 7 |
| 1373874_at | 0.003529545 | Sphingosine-1-phosphate phosphatase 1 | Sgpp1 | 7 |
| 1369560_at | 0.003554545 | Glycerol-3-phosphate dehydrogenase 1 | Gpd1 | 7 |
| 1379243_at | 0.008376253 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) | Ndufa6 | 7 |
| 1387284_at | 0.008499174 | Dihydropyrimidinase | Dpys | 7 |
| 1398772_at | 0.011159968 | NSFL1 (p97) | Nsfl1c | 7 |
| 1398353_at | 0.013274267 | SAR1a gene homolog 1 | Sara1 | 7 |
| 1369785_at | 0.020692991 | Phosphoribosyl pyrophosphate amidotransferase | Ppat | 7 |
| 1371415_at | 0.021285045 | Ubiquinol-cytochrome c reductase hinge protein | Uqcrb | 7 |
| 1368188_at | 0.02916268 | 4-Hydroxyphenylpyruvic acid dioxygenase | Hpd | 7 |
| 1368121_at | 0.030787215 | Aldo-keto reductase family 7, member A3 | Akr7a3 | 7 |
| 1368180_s_at | 0.036416989 | Glutathione-S-transferase, alpha type2 | Gsta2 | 7 |
| 1388586_at | 0.037280773 | Synaptojanin 1 | Synj1 | 7 |
| 1367824_at | 0.037492574 | Farnesyltransferase, CAAX box, alpha | Fnta | 7 |
| 1387725_at | 0.039647225 | L-gulonolactone oxidase | Gulo | 7 |
| 1374426_at | 0.048203726 | Ubiquinol-cytochrome c reductase binding protein | Uqcrb | 7 |
The metabolic processes down-regulated on d 1 include glycolysis (aldolase A, enolase 1), urea cycle (carbomoyl-phosphate synthase 1), and amino acid transport/catabolism (amino acid transport system 3, methylcrotonoyl-Coenzyme A carboxylase 2). A significant category of genes that were down-regulated on d 1 belonged to the antioxidant family of enzymes. Glutathione reductase and peroxiredoxin 4 were significantly less in burn liver as compared to sham liver, indicating that antioxidant defense systems were down-regulated at 24 h.
Fewer metabolic genes were altered in expression on d 2, 4, and 7 after burn injury as compared to d 1. Genes involved in lipid catabolism (scavenger receptor class B1, phosphatidylserine-specific phospholipase A1, fatty acid translocase) were all down-regulated on d 2, indicating that fatty acids were no longer used as the primary energy-yielding substrate. Similarly, ATP biosynthesis was also reduced on d 2, as inferred from the significant reduction in the expression of the ATPase epsilon subunit gene. On d 4, the increase in expression of genes involved in fatty acid biosynthesis (gamma-butyrobetaine hydroxylase, farnesyl diphosphate farnesyl transferase 1, sterol-C4-methyl oxidase-like) and the decrease of genes involved in fatty acid oxidation (4-dienoyl CoA reductase 1) indicated the liver replenishing its TG reserves that were used during the response to burn injury. However, the expression of other genes involved in energy generation such as those in amino acid uptake were also not significantly expressed on d 2 after burn injury.
Several genes involved in mitochondrial electron transport were also significantly altered in expression on d 4, including multiple subunits of the NADH ubiquinone dehydrogenase (NADH-coenzyme Q reductase, NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, NADH dehydrogenase (ubiquinone) Fe-S protein 7, NADH dehydrogenase (ubiquinone) 1-alpha subcomplex, 3, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10).
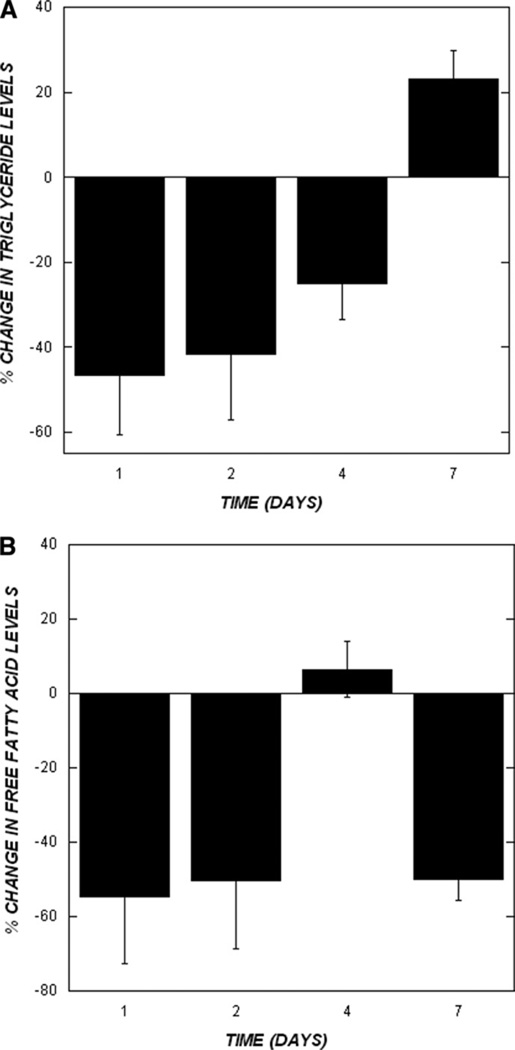
We also used biochemical assays to measure the levels of TG and FFA in the liver after burn injury to determine the extent of correlation between changes in the expression of metabolic genes and the levels of TG and FFA in the liver. TG (46 and 41%) and FFA (54 and 51%) levels in liver tissue from burn-injured rats were less than sham-burn controls at d 1 and 2, respectively (Fig. 3A, B). While TG and FFA levels in burn liver tissue were higher on d 4 compared to d 1 and 2, TG levels were still 25% less than the sham-burn control. On the other hand, FFA levels in liver tissue 4 d after burn injury were comparable to sham-burn controls. TG levels continued to increase and were 23% more in burn-injured livers on d 7 as compared to sham-burn controls (Fig. 3A), while FFA levels were approximately 50% of that seen in liver tissue from sham-burn controls (Fig. 3B).
FIG. 3.
Biochemical measurement of TG and FFA. The levels of (A) TG and (B) FFA in rat livers after burn injury were measured using commercially available kits from Sigma Chemical Co. and Roche Biochemicals, respectively. Data shown are from 3 burn and sham-burn livers at each time point and are the percentage change in burn livers compared to the sham-burn control. Statistical analysis was performed using the Student’s t-test and significance relative to the previous time point is indicated by an asterisk (P < 0.05).
Inflammatory Response Genes
Several genes belonging to the inflammation ontology group were significantly altered in expression only on d 1 and reflect the important role of these genes in the early acute phase response to burn injury (Table 3). These include genes belonging to the complement system (mannan-binding lectin serine protease 2, complement component 4 binding protein alpha), protease inhibitors (alpha 1 anti-trypsin, serine protease inhibitor), antioxidant systems (heme oxygenase 2, superoxide dismutase 1), coagulation factors (factor X and 2), and acute-phase proteins (major urinary protein 2, haptoglobin). The expression of constitutive hepatic proteins such as albumin (down-regulated on d 1) was also decreased, while the expression of acute phase genes such as kininogen (up-regulated on d 2) and serum amyloid P (up-regulated on d 4) was increased. Interestingly, the expression of acute-phase genes (fibrinogen, Fga, and ceruloplasmin, Cp) and anti-apoptosis (baculoviral IAP repeat-containing 4, Birc4) were also significantly up-regulated on d 7 after burn injury.
TABLE 3.
Inflammation Genes Altered by 20% Burn Injury
| Affymetrix ID | P | Gene | Symbol | Significant change in expression on d |
|---|---|---|---|---|
| 1387891_at | 0.001371 | Peroxiredoxin 4 | Prdx4 | 1 |
| 1370148_at | 0.011171 | Haptoglobin | Hp | 1 |
| 1371098_a_at | 6.38E-05 | Mannan-binding lectin serine protease 2 | Masp2 | 1 |
| 1367641_at | 0.000136 | Superoxide dismutase 1 | Sod1 | 1 |
| 1374798_at | 0.000584 | Interferon alpha responsive gene | 1 | |
| 1398850_at | 0.000741 | Peptidylprolyl isomerase A (cyclophilin A) | PpiA | 1 |
| 1369726_at | 0.001599 | TAP binding protein | Tapbp | 1 |
| 1371002_at | 0.001615 | Programmed cell death 2 | Pdcd2 | 1 |
| 1367609_at | 0.003306 | Macrophage migration inhibitory factor | Mif | 1 |
| 1367903_at | 0.003381 | Heme oxygenase (decycling) 2 | Hmox2 | 1 |
| 1371560_at | 0.003587 | Interferon regulatory factor 3 | Irf3 | 1 |
| 1371783_at | 0.00505 | Heat shock protein | 1 | |
| 1368707_at | 0.006201 | Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 | 1 |
| 1373818_at | 0.006734 | Heat shock protein 1, alpha | 1 | |
| 1371734_at | 0.016254 | Macrophage erythroblast attacher | Maea | 1 |
| 1369852_at | 0.017431 | Coagulation factor X | F10 | 1 |
| 1367555_at | 0.017548 | Albumin | Alb | 1 |
| 1389270_x_at | 0.018361 | Urinary protein 3 | Mup3 | 1 |
| 1367647_at | 0.018529 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
Serpina1 | 1 |
| 1388362_at | 0.021721 | Similar to novel cell death-regulatory protein GRIM19 | 1 | |
| 1368442_at | 0.024012 | Coagulation factor 2 | F2 | 1 |
| 1368446_at | 0.024705 | Serine protease inhibitor, Kazal type 1 | Spink1 | 1 |
| 1388809_at | 0.027976 | acid sphingomyelinase-like Phosphodiesterase 3A | Smpdl3a | 1 |
| 1372440_at | 0.03398 | Serine (or cysteine) proteinase inhibitor, clade E, member 2 | Serpine2 | 1 |
| 1368492_at | 0.034784 | Prostaglandin D2 synthase 2 | Ptgds2 | 1 |
| 1367696_at | 0.036135 | Interferon-induced transmembrane protein 2 | Ifit2m | 1 |
| 1370243_a_at | 0.036981 | Prothymosin alpha | Ptma | 1 |
| 1368118_at | 0.049492 | B-cell CLL/lymphoma 10 | Bcl10 | 1 |
| 1369764_at | 5.89E-05 | Complement component 4 binding protein, alpha | C4bpa | 1 |
| 1374785_at | 0.000471 | p60, early T-cell activation antigen | 2 | |
| 1388792_at | 0.041642 | Growth arrest and DNA-damage-inducible 45 gamma | Gadd45g | 2 |
| 1374334_at | 0.046284 | Immunoglobulin heavy chain (alpha polypeptide) | Igha | 2 |
| 1371124_a_at | 0.047002 | Kininogen 1 | Kng1 | 2 |
| 1367804_at | 0.004404 | Serum amyloid P-component | Sap | 4 |
| 1367850_at | 0.011204 | Fc receptor, IgG, low affinity III | Fcgr3 | 4 |
| 1373868_at | 0.0181 | BCL2-associated transcription factor 1 | Bclaf1 | 4 |
| 1367677_at | 0.035321 | Peroxiredoxin 5 | Prdx5 | 4 |
| 1374551_at | 0.048959 | Interferon-induced protein 35 | Ifi35 | 4 |
| 1367793_at | 0.016016 | D-dopachrome tautomerase | Ddt | 7 |
| 1371258_at | 0.021047 | Fibrinogen, alpha polypeptide | Fga | 7 |
| 1368420_at | 0.021348 | Ceruloplasmin | Cp | 7 |
| 1369248_a_at | 0.030362 | Baculoviral IAP repeat-containing 4 | Birc4 | 7 |
| 1388102_at | 0.040721 | Leukotriene B4 12-hydroxydehydrogenase | Ltb4dh | 7 |
Signal Transduction Genes
Genes that function as signaling intermediates (receptors, transcriptional regulators, kinases, and phosphatases) in the inflammatory response to burn injury were also significantly altered in expression on d 1 (Table 4). The observed changes in the expression of interleukin 6 signal transducer gp130 and SMAD suggest that interleukin-6 (IL-6) and transforming growth factor-beta are some of the inflammatory molecules that are actively involved in the early liver response to burn injury. Similarly, changes in the expression of mitogen-activated protein kinase-interacting kinase 2a, mitogen-activated protein kinase kinase kinase kinase 3, and inhibitor of kappaB kinase beta were also observed. The expression of several signaling-related genes was also significantly altered on d 7 after burn injury, including signal transducer and activator of transcription 3 (STAT3), leptin receptor, hepatocyte nuclear factor 4 alpha (HNF4α), the protein tyrosine phosphatase, non-receptor-type substrate 1 (Ptpns1), and hepatocyte growth factor receptor. These changes in expression are consistent with the changes in the expression of defense/inflammatory response genes observed on d 7 after burn injury.
TABLE 4.
Signaling Genes Altered by 20% Burn Injury
| Affymetrix ID | P | Gene | Symbol | Significant change in expression on d |
|---|---|---|---|---|
| 1368214_at | 1.64E-05 | MAD homolog 2 | Smad2 | 1 |
| 1374697_at | 0.000757 | Neuronal RhoA GEF protein | 1 | |
| 1375231_a_at | 0.001007 | CXXC finger 5 | 1 | |
| 1367833_at | 0.009637 | For proteasomal ATPase | 1 | |
| 1398814_at | 4.12E-05 | RAB11a | RAB11a | 1 |
| 1367736_at | 0.043696 | Ras-related GTP-binding protein ragA | Rraga | 1 |
| 1374468_at | 4.38E-05 | Myeloid differentiation primary response gene | Myd88 | 1 |
| 1398352_at | 0.000782 | Protein inhibitor of activated STAT, 4 | PIAS4 | 1 |
| 1373449_at | 0.001662 | TAF5-like RNA polymerase II, p300/CBP-associated factor- | PCAF | 1 |
| 1372460_at | 0.00233 | Protein kinase N3/SET translocation | 1 | |
| 1368424_at | 0.002381 | Inhibitor of kappaB kinase beta | Ikbkb | 1 |
| 1372485_at | 0.002484 | 6-Pyruvoyl-tetrahydropterin synthase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
Pcbd | 1 |
| 1372617_at | 0.003014 | Mitogen-activated protein kinase-interacting kinase 2a | MKNK2 | 1 |
| 1375760_at | 0.003019 | Transducer of ERBB2, 2 | Tob2 | 1 |
| 1373719_at | 0.003413 | Mitogen-activated protein kinase kinase kinase kinase 3 | MAP4K3 | 1 |
| 1375374_at | 0.003747 | Sequestosome 1 | Sqstm1 | 1 |
| 1373479_at | 0.004298 | Protein phosphatase 3, catalytic subunit, alpha isoform | Ppp3ca | 1 |
| 1372077_at | 0.007063 | Serine/threonine kinase receptor associated protein | Strap | 1 |
| 1367948_a_at | 0.007175 | Kinase insert domain protein receptor | kdr | 1 |
| 1386905_at | 0.007993 | Protein kinase, cAMP dependent regulatory, type I, alpha | Prkar1a | 1 |
| 1377013_at | 0.008106 | Williams–Beuren syndrome chromosome region 14 homolog | Wbscr14 | 1 |
| 1368076_at | 0.008124 | von Hippel–Lindau syndrome homolog | Vhl | 1 |
| 1373658_at | 0.009277 | Rac GTPase-activating protein 1 | 1 | |
| 1368220_at | 0.009881 | General transcription factor IIB | Gtf2b | 1 |
| 1372187_at | 0.01097 | Protein kinase C, nu | 1 | |
| 1369559_a_at | 0.011219 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
Cd47 | 1 |
| 1375882_at | 0.022334 | Protein phosphatase 3, catalytic subunit, beta isoform | Ppp3cb | 1 |
| 1370990_at | 0.023947 | Similar to cytokine receptor related protein 4 | 1 | |
| 1369174_at | 0.030888 | SMAD, mothers against DPP homolog 1 | Smad1 | 1 |
| 1371926_at | 0.033214 | Interleukin 6 signal transducer | Il6st | 1 |
| 1371491_at | 0.034935 | Notch gene homolog 1 (Drosophila) | Notch1 | 1 |
| 1375745_at | 0.036368 | Guanine nucleotide binding protein, alpha q polypeptide | Gnaq | 1 |
| 1367624_at | 0.040828 | Activating transcription factor 4 | Atf4 | 1 |
| 1367766_at | 0.044906 | Expressed in nonmetastatic cells 2 | Nme2 | 1 |
| 1385236_at | 0.044925 | Phosphatidylinositol 4-kinase, catalytic, beta polypeptide | Pik4cb | 1 |
| 1388686_at | 0.001482 | Down syndrome critical region homolog 1 | Dscr1 | 2 |
| 1370968_at | 0.02047 | Nuclear factor of kappa light chain gene enhancer in B-cells 1, p105 |
NfkB1 | 2 |
| 1367802_at | 0.001572 | Serum/glucocorticoid regulated kinase | Sgk | 2 |
| 1374404_at | 0.002935 | v-jun sarcoma virus 17 oncogene homolog | Jun | 2 |
| 1368089_at | 0.003712 | Phosphodiesterase 2A, cGMP-stimulated | Pde2A | 2 |
| 1389479_at | 0.006647 | Kruppel-like factor 3 | Klf3 | 2 |
| 1368146_at | 0.025522 | Dual specificity phosphatase 1 | Dusp1 | 2 |
| 1368134_a_at | 0.025522 | Interleukin 4 receptor | Il4r | 2 |
| 1390201_at | 0.025965 | Ras-related protein RAP-1A | RAP-1a | 2 |
| 1375985_at | 0.026917 | RAB2B, member RAS oncogene family | 2 | |
| 1389381_at | 0.040311 | sequestosome 1 | Sqstm1 | 2 |
| 1367618_a_at | 0.044889 | Guanine nucleotide binding protein, beta polypeptide 2-like 1 | Gnb2l1 | 2 |
| 1373421_at | 0.047408 | TG interacting factor | Tgif | 2 |
| 1371718_at | 0.000993 | Steroid receptor RNA activator 1 | Sra1 | 4 |
| 1374408_at | 0.005115 | Similar to CBF1 interacting corepressor | 4 | |
| 1373400_at | 0.014566 | Protein kinase, cAMP-dependent, regulatory, type 2, alpha | Prkar2a | 4 |
| 1368216_at | 0.016246 | RAB28, member RAS oncogene family | Rab28 | 4 |
| 1369658_at | 0.0259 | CCAAT/enhancer binding protein, alpha | Ceebpa | 4 |
| 1369177_at | 0.000304 | Phosphatidylinositol 4-kinase type II | Pi4K2a | 7 |
| 1375151_at | 0.002215 | Similar to RAP2A, member of RAS oncogene family | Rap2b | 7 |
| 1374092_at | 0.003946 | Similar to WW domain containing transcription regulator 1 | 7 | |
| 1369511_at | 0.00417 | Endothelin receptor type A | EdnrA | 7 |
| 1387474_at | 0.004476 | Tachykinin receptor 1 | Tacr1 | 7 |
| 1383426_at | 0.005603 | Proline-serine-threonine phosphatase-interacting protein 1 | Pstpip1 | 7 |
| 1368924_at | 0.007385 | Growth hormone receptor | Ghr | 7 |
| 1389974_at | 0.010412 | Casein kinase II, alpha 1 polypeptide | Csnk2a1 | 7 |
| 1369218_at | 0.013213 | Met proto-oncogene | Met | 7 |
| 1371781_at | 0.014111 | Signal transducer and activator of transcription 3 | STAT3 | 7 |
| 1369289_at | 0.017492 | Hepatocyte nuclear factor 4, alpha | HNF4a | 7 |
| 1369323_at | 0.019408 | Leptin receptor | Lepr | 7 |
| 1370136_at | 0.022218 | Lamin B receptor | Lbr | 7 |
| 1373817_at | 0.024551 | Inhibitor of growth family, member 4 | Ing4 | 7 |
| 1367881_at | 0.032642 | Protein tyrosine phosphatase, non-receptor type substrate 1 | Ptpns1 | 7 |
| 1369004_at | 0.041529 | RAB26, member RAS oncogene family | Rab26 | 7 |
Protein Interaction Network Analysis
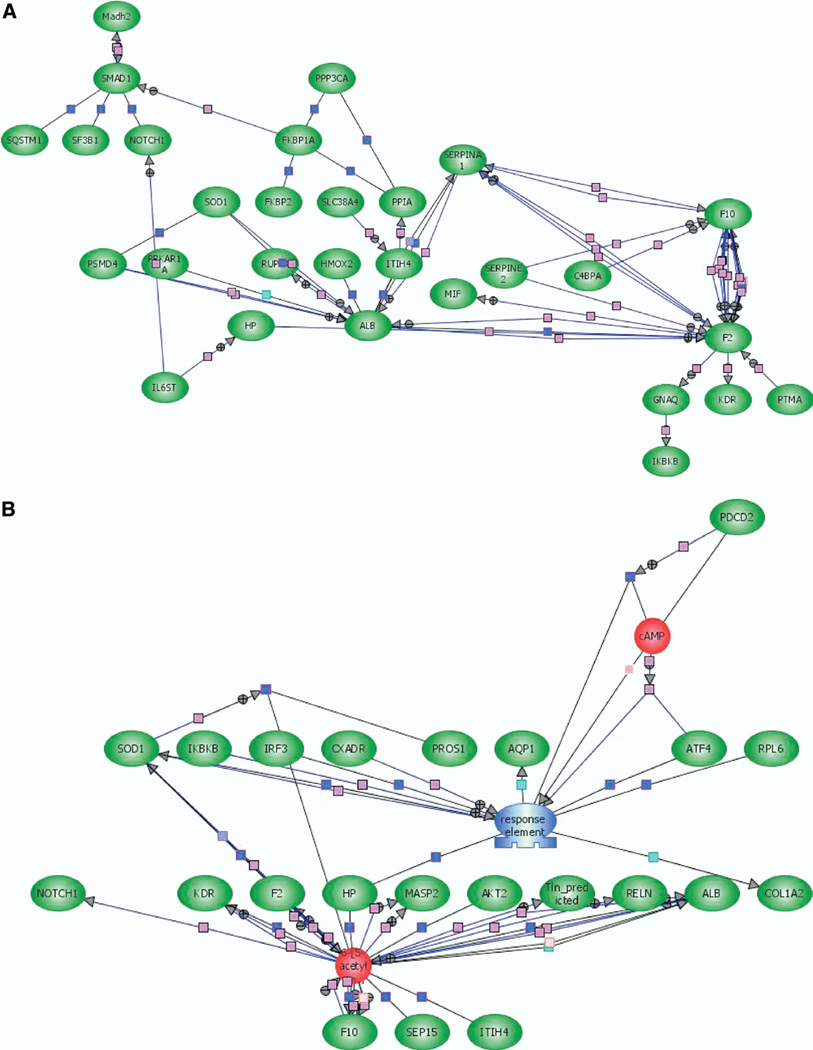
Protein interaction networks were used to identify relationships between the significantly expressed genes in the liver on d 1, 2, 4, and 7 after burn injury. Three types of networks were constructed using the Pathway Assist software. The simplest form of an interaction network was constructed to identify direct relationships between the significantly expressed genes, without introducing any additional genes or small molecules into the network. The direct relevance network between the significantly expressed genes on d 1 is shown in Fig. 4A and direct networks developed for d 2 and 7 are included in Supplemental Fig S1–S2. The network of interacting genes shows that albumin, the coagulation factors F2 and X, and serpinA1 are key to the early response to burn injury. In addition, the network also indicates that the transcription factor Onecut1 (hepatocyte nuclear factor 6A) is a key regulator as it has direct expression links to 6 other significantly expressed genes. Similarly, the direct interaction network on d 7 (see supplemental Figure S2) indicates that the transcription factor HNF4α plays an important role in regulating the expression of other significantly expressed genes. Last, the magnitude of changes in expression (down-regulation of HNF4α and up-regulation of target genes such as Slca25) indicates that HNF4α is a negative regulator of these genes.
FIG. 4.
Protein interaction networks for significantly expressed genes in rat liver after burn injury. Protein interaction networks were constructed using the PathwayArchitect software (Stratagene). (A) A direct interaction network between the different significantly expressed genes on d 1. Pink connections represent regulation, and blue connections represent direct protein–protein binding. (B) A relevance network based on various regulatory and expression links (described in Materials and Methods) for statistically significant genes on d 1. Red objects represent small metabolites, and green objects represent proteins. Blue connections represent protein–protein binding, and pink interactions designate regulation. (Color version of figure is available online.)
A second class of interaction maps was developed using a relevance network, based on a range of connections (e.g., protein–protein binding, small molecule binding, common transcriptional regulation, etc) between the significantly expressed genes. To construct these networks, additional genes or nodes were also introduced (identified by PathwayArchitect as relevant to the overall network) even though these genes did not demonstrate significant changes in expression after burn injury. Using this approach, specific genes or molecules were identified to be key nodes in the relevance network. In the network shown in Fig. 4B, Fig. 2 molecules—an unknown transcription factor and the small molecule certoparin—are important nodes, with the expression of haptoglobin and SOD1 serving as links between the 2 key nodes. Relevance networks for the significantly expressed genes on all other days are shown in Supplemental Fig S3–S5.
DISCUSSION
The objective of this study was to characterize the long-term changes in gene expression during the first 7 d after burn injury. Transcriptional changes in response to burn injury have been studied in several tissues and organs, including skin [22], skeletal muscle [23, 24], intestinal epithelium [25], and liver [12, 13]. In our prior work [13], we focused exclusively on characterizing the short-term changes (i.e., those occurring within 24 h) in liver gene expression after burn injury. The 20% burn injury model used in this study induces a significant, nonlethal injury response in rats that is different from more severe burn injury models (i.e., 40% TBSA burn injury or burn injury followed by infection). The 20% TBSA model has been extensively used to characterize burn-injury-induced insulin resistance as well as hypermetabolism [7, 26, 27]. This being a nonlethal model, the changes in expression observed in this study reflect dynamics of the normal and successful systemic response to burn injury where there is virtually no mortality. Furthermore, the changes reported herein are intrinsic to the liver and not likely contributed by inflammatory cell recruitment, since prior studies with this model indicate that neutrophil sequestration in liver is a very transient phenomenon lasting less than 6 h, well before the first time point of this study [17].
To date, long-term changes in liver gene expression following burn injury have not been comprehensively studied at the gene expression level. It should be noted that while Dasu et al. [12] monitored changes in liver gene expression for up to 10 d after burn injury, the time points that were used (2 h, 3 d, and 10 d) were not sufficient to develop a systematic understanding of the temporal evolution of postburn liver gene expression. Despite the differences in the time points at which gene expression was monitored, there is a high degree of similarity between the genes identified as significantly different in this study and that presented by Dasu et al. (Table 1 and Table 2) in [12], including genes such as mast cell protease 9, α2-macroglobulin, lipopolysaccharide-binding protein, stearyl-coA desaturase, lysyl oxidase, tyrosine aminotransferase, early growth response-1, and cholesterol hydroxylase. This similarity validates our approach and, along with the other significantly expressed genes, provides a comprehensive overview of changes in liver gene expression after burn injury.
An important feature of our analysis is that genes demonstrating significant changes in expression between burn and sham-burn livers were not selected based on a predetermined threshold (e.g., 2-fold). Instead, we chose to determine statistically significant changes in expression between the different conditions based on the variations in expression observed in the entire data set of 24 microarrays. The advantage with this approach is that it accounts for statistically significant and potentially meaningful small changes in expression (e.g., a 50% increase in expression) that would have been otherwise not identified by an arbitrarily chosen threshold value.
Our data show that the significantly major ontology categories (metabolism, signaling, protein synthesis and processing, transport, and defense/inflammation) were impacted to varying degrees in the liver at different time points after 20% burn injury. These results suggest that a majority of changes in gene expression in the liver after 20% TBSA burn injury are transient in nature and suggest temporal coordination between the different post-burn physiological responses in the liver. The distinct temporal trends observed also likely reflect the roles that the different processes play in the liver’s response to 20% TBSA burn injury. While metabolism genes were altered at all days on which gene expression was monitored, the processes that these genes encoded for were different. For example, fatty acid oxidation genes were up-regulated on d 1, whereas genes involved in fatty acid biosynthesis demonstrated an increase in expression on d 4 after injury. These changes likely underlie the fact that fatty acids are used as energy sources during the first 24 h and are replenished at 4 d after injury. Similarly, the increase in the expression of cytochrome-c oxidase subunits (subunits VIa and VIIa) are indicative of an increase in ATP synthesis in the liver on d 4 after burn injury, presumably to meet the increased energy demands after burn injury.
Based on coordinated short-term (up to 24 h) changes in both gene expression data and metabolite measurements, our prior work revealed that fatty acids are important energy-yielding substrates during the first 24 h after 20% burn injury [13]. Moreover, we also observed that the changes in gene expression accounted for a majority of metabolic alterations up to 24 h after injury. The data presented here on the absence of genes involved in fatty acid oxidation on d 2, 4, and 7 and the persistent down-regulation of fatty acid levels (Fig. 3B) suggest that fatty acid utilization is not controlled at the transcrip-tional level in the liver beyond 24 h in response to 20% burn injury. Moreover, the lack of significant changes in the expression of genes involved in the utilization of alternate substrates (i.e., amino acids) for glucose production [28] indicates that transcriptional regulation is not the primary mode of regulation for amino acid utilization in the liver after burn injury. Together with our previous work [13], these data suggest that the early hepatic response to 20% TBSA burn injury is controlled, at least in part, at the transcriptional level, whereas long-term responses do not involve significant contributions from transcriptional control. It is likely that other regulatory mechanisms such as protein stability, enzyme activity, and substrate availability regulate the long-term responses.
The up-regulation in inflammatory genes after 24 h, but not after 48 and 72 h, is consistent with previous reports describing the hepatic acute phase response to injury [2]. The activation of inflammatory signaling pathways at 24 h postburn is consistent with our data on the changes in the expression of genes involved in inflammation as well as prior reports on changes in gene expression during the hepatic acute phase response [29–31]. For example, the increase in mitogen-activated protein kinase and nuclear factor-kappa B signaling pathway intermediates in the liver during the first 24 h after burn injury is consistent with the observations of Jeschke et al. [32], who observed nearly a 2-fold increase on d 1 after 40% TBSA burn injury in rats. However, the down-regulation of antioxidant defense systems at 24 h is surprising as large quantities of reactive oxygen species are associated with cellular energetics during the inflammatory response [33, 34].
The up-regulation of several acute-phase genes and signaling intermediates (e.g., STAT3, leptin receptor, HNF4α) that are known to be involved in the response to infection on d 7 suggests bacterial colonization of the wound in burn-injured animals. The up-regulation of Birc4 is particularly interesting, as Deveraux et al. [35] have shown that this protein directly inhibits caspase-3 and caspase-7 that are key mediators of apoptosis postburn [36]. Jeschke et al. [32] have also reported the decrease in caspase 3 at d 7 in rats upon 40% burn injury, which would also be consistent with the increase in the expression of the anti-caspase 3 molecule Birc4. Together, these results suggest the presence of an inflammatory response on d 7 after burn injury. Specifically, the change in the expression of Ptpns1, a signal regulatory protein that regulates innate immune function [37] and has been linked to infections [38], and hepatocyte growth factor, which is involved in sepsis [39, 40], support our hypothesis. In addition, the increase in the expression of STAT3 (the signal-transducing molecule for signaling through the gp130 receptor that is activated a broad range of molecules family of cytokines including IL-6, leukemia inhibitory factor, and epidermal growth factor) [41] and the leptin receptor (also a member of the gp130 family and used for signaling through the Jak/STAT pathway [42]), indicates enhanced cytokine signaling on d 7. However, no cultivatable bacteria were detected in the serum samples (not shown). This result is consistent with previous work from our laboratory demonstrating that, while bacterial counts (colony forming units/g tissue) were extremely high (∼108) in the wound site of 20% TBSA burn-injured animals [17], no bacteria were detected in the circulation. Moreover, the 20% TBSA burn injury model also has a negligible mortality rate [13, 17], which precludes the onset of sepsis and systemic infection. Therefore, it is plausible that the changes in gene expression in the liver on d 7 underlie the liver’s response to dead bacteria in circulation or bacterial cellular material and is primarily involved in preventing systemic infections.
We used PathwayArchitect to construct protein interaction networks and determine the biological connections between the significantly expressed genes. In this study, we used PathwayArchitect to identify genes that had direct interactions with other significantly expressed genes, and therefore, likely play a critical function in the response to burn injury. An example is the identification of the coagulation factors 2 (F2) and X (F10) as key elements in the d 1 direct interaction relevance network (Fig. 4A), which is consistent with the notion that coagulation and inflammation are interconnected processes [43, 44]. Specifically, inflammation and coagulation are thought to function in an auto-amplification loop where the onset of inflammation promotes the coagulation/ clotting pathways, which in turn increases inflammation [43]. F10 has been shown to play an active role in endothelial cell inflammation through synergistic interactions with pro-inflammatory cytokines such as tumor necrosis factor and IL-1β [45] and supports the molecular connection between coagulation processes and the inflammatory response.
In addition, the incorporation of relevance network analysis also enabled the identification of small molecules that likely regulate the response to burn injury. The identification of certoparin, a low molecular weight heparin that is an anticoagulant, is significant as low molecular weight heparins have also been reported to have anti-inflammatory properties [46, 47], which provides a further link between inflammation and coagulation interactions. We speculate that molecules like certoparin play important roles in the modulation of the inflammatory response in the liver on postburn d 1. Other key nodes identified through relevance networks, such as matrix metalloproteinase-2 (MMP-2) and Met on postburn d 7, also provide insights into the physiological responses in the liver after burn injury. MMP-2 specifically cleaves Type 4 collagen, which is a major component of basement membranes and has been suggested to cause liver injury [48]. On the other hand, the proto-oncogene Met is the cell surface receptor for hepatocyte growth factor, which is involved in promoting hepatocyte proliferation [49]. Therefore, the presence of both MMP-2 and Met in the relevance network on d 7 suggests the presence of liver injury on d 7 after injury and the up-regulation of processes that are geared toward countering its adverse effects through hepatocyte proliferation.
It should be noted that the data in the present study are primarily at the level of transcription and are being used to infer phenotypic changes in the liver (i.e., lipid metabolism). The modest correlation between gene expression and functional changes suggest that at least some of these processes are regulated at the level of transcription. However, since gene expression can be regulated at additional levels, including translational and posttranslational regulation, changes in gene expression need not always correlate to changes in function. Therefore, it is necessary to assess changes in expression at other (i.e., proteomic and metabolomic) levels to develop an integrated picture of postburn changes in the liver.
In summary, our data show significant differences in the gene expression changes during the short-term (<24 h) and long-term liver responses to 20% TBSA burn injury. In addition, biochemical measurements indicate that fatty acids are used, at least in part, in the liver as energy substrates for 4 d after injury. Together, these data suggest that regulation of energy substrate utilization is primarily at the posttranscriptional level during the long-term response. Last, relevance networks of significantly expressed genes indicate interactions between different genes and the involvement of key small molecules in hepatic response to 20% TBSA burn injury. Current work in our laboratory focuses on correlating the extent of injury to the dynamics of expression changes, by exploring if a more severe injury model (e.g., 40% TBSA burn injury or 20% TBSA burn injury with infection) will lead to persistent changes in metabolism genes or more pronounced alterations in gene expression [50, 51]. Additionally, we are investigating the correlation between the dynamics of gene expression to proteome and metabolomic level changes so that specific aspects of the hypermetabolic response can be related to changes in gene expression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Shriners Hospitals for Children (SHC 8560). The use of facilities at the Special Shared Facility for Functional Genomics and Proteomics is acknowledged.
REFERENCES
- 1.Kien CL, Young VR, Rohrbaugh DK, et al. Increased rates of whole body protein synthesis and breakdown in children recovering from burns. Ann Surg. 1978;187:383. doi: 10.1097/00000658-197804000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann H, Gauldie J. The acute phase response. Immunol Today. 1994;15:74. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 3.Tredget EE, Yu YM, Zhong S, et al. Role of interleukin 1 and tumor necrosis factor on energy metabolism in rabbits. Am J Physiol. 1988;255:E760. doi: 10.1152/ajpendo.1988.255.6.E760. [DOI] [PubMed] [Google Scholar]
- 4.Baue AE, Durham R, Faist E. Systemic inflammatory response syndrome (SIRS), multiple organ dysfunction syndrome (MODS), multiple organ failure (MOF): Are we winning the battle? Shock. 1998;10:79. doi: 10.1097/00024382-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Cerra F, West M, Keller GJM, et al. Hypermetabolism/organ failure: the role of the activated macrophage as a metabolic regulator. Prog Clin Biol Med. 1988;264:27. [PubMed] [Google Scholar]
- 6.Herndon D, Curreri P, Abston S, et al. Treatment of burns. Curr Probl Surg. 1987;24:341. doi: 10.1016/0011-3840(87)90010-4. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Berthiaume F, Stephanopoulos GN, et al. Metabolic flux analysis of postburn hepatic hypermetabolism. Metab Eng. 2000;2:312. doi: 10.1006/mben.2000.0160. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Berthiaume F, Stephanopoulos GN, et al. Metabolic flux analysis: a powerful tool for monitoring tissue function. Tissue Eng. 1999;5:347. doi: 10.1089/ten.1999.5.347. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi Y, Yu YM, Zupke C, et al. Effect of burn injury on glucose and nitrogen metabolism in the liver: preliminary studies in a perfused liver system. Surgery. 1997;121:295. doi: 10.1016/s0039-6060(97)90358-5. [DOI] [PubMed] [Google Scholar]
- 10.Scholmerich J, Kremer B, Richter IE, et al. Effect of cutaneous human or mouse burn toxin on the metabolic function of isolated liver cells. Scand J Plast Reconstr Surg. 1979;13:223. doi: 10.3109/02844317909013061. [DOI] [PubMed] [Google Scholar]
- 11.Chen CL, Fei Z, Carter EA, et al. Metabolic fate of extrahepatic arginine in liver after burn injury. Metabolism. 2003;52:1232. doi: 10.1016/s0026-0495(03)00282-8. [DOI] [PubMed] [Google Scholar]
- 12.Dasu MR, Cobb JP, Laramie JM, et al. Gene expression profiles of livers from thermally injured rats. Gene. 2004;327:51. doi: 10.1016/j.gene.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Vemula M, Berthiaume F, Jayaraman A, et al. Expression profiling analysis of the metabolic and inflammatory changes following burn injury in rats. Physiol Genom. 2004;18:87. doi: 10.1152/physiolgenomics.00189.2003. [DOI] [PubMed] [Google Scholar]
- 14.Fong Y, Minei J, Marano M, et al. Skeletal muscle amino acid and myofibrillar protein mRNA response to thermal injury and infection. Am J Physiol. 1991;261:R536. doi: 10.1152/ajpregu.1991.261.3.R536. [DOI] [PubMed] [Google Scholar]
- 15.Rothwell N, Little R, Rose J. Brown adipose tissue activity and oxygen consumption after scald injury in the rat. Circ Shock. 1991;33:33. [PubMed] [Google Scholar]
- 16.Walker HL, Mason ADJ. A standard animal burn. J Trauma. 1968;8:1049. doi: 10.1097/00005373-196811000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Baskaran H, Yarmush ML, Berthiaume F. Dynamics of tissue neutrophil sequestration after cutaneous burns in rats. J Surg Res. 2000;93:89. doi: 10.1006/jsre.2000.5955. [DOI] [PubMed] [Google Scholar]
- 18.Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2001;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott LR, Longnecker M. An Introduction to Statistical Methods and Data Analysis. Pacific Grove, CA: Duxbury; 2001. [Google Scholar]
- 20.Jayaraman A, Yarmush ML, Roth CM. Dynamics of gene expression in rat hepatocytes under stress. Metab Eng. 2000;2:1. doi: 10.1006/mben.2000.0153. [DOI] [PubMed] [Google Scholar]
- 21.Pradines J, Farutin V, Rowley S, et al. Analyzing protein lists with large networks: edge-count probabilities in random graphs with given expected degrees. J Comput Biol. 2005;12:113. doi: 10.1089/cmb.2005.12.113. [DOI] [PubMed] [Google Scholar]
- 22.Feezor RJ, Paddock HN, Baker HV, et al. Temporal patterns of gene expression in murine cutaneous burn wound healing. Physiol Genom. 2004;16:341. doi: 10.1152/physiolgenomics.00101.2003. [DOI] [PubMed] [Google Scholar]
- 23.Barrow RE, Dasu MR, Ferrando AA, et al. Gene expression patterns in skeletal muscle of thermally injured children treated with oxandrolone. Ann Surg. 2003;237:422. doi: 10.1097/01.SLA.0000055276.10357.FB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herndon DN, Dasu MR, Wolfe RR, et al. Gene expression profiles and protein balance in skeletal muscle of burned children after beta-adrenergic blockade. Am J Physiol Endocrinol Metab. 2003;285:E783. doi: 10.1152/ajpendo.00508.2002. [DOI] [PubMed] [Google Scholar]
- 25.Varedi M, Lee HM, Greeley GHJ, et al. Gene expression in intestinal epithelial cells, IEC-6, is altered by burn injury-induced circulating factors. Shock. 2001;16:259. doi: 10.1097/00024382-200116040-00004. [DOI] [PubMed] [Google Scholar]
- 26.Carter EA, Derojas-Walker T, Tamir S, et al. Nitric oxide production is intensely and persistently increased in tissue by thermal injury. Biochem J. 1994;304:201. doi: 10.1042/bj3040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Fei Z, Carter E, et al. Metabolic fate of extrahepatic arginine in liver after burn injury. Metabolism. 2003;52:1232. doi: 10.1016/s0026-0495(03)00282-8. [DOI] [PubMed] [Google Scholar]
- 28.Pawlik TM, Lohmann R, Souba WW, et al. Hepatic glutamine transporter activation in burn injury: Role of amino acids and phosphatidylinositol-3-kinase. Am J Physiol Gastrointest Liver Physiol. 2000;278:G532. doi: 10.1152/ajpgi.2000.278.4.G532. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Baumann H. Dual signaling role of the protein tyrosine phosphatase SHP-2 in regulating expression of acute-phase plasma proteins by interleukin-6 cytokine receptors in hepatic cells. Mol Cell Biol. 1999;19:5326. doi: 10.1128/mcb.19.8.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray A, Yu GY, Ray BK. Cytokine-responsive induction of SAF-1 activity is mediated by a mitogen-activated protein kinase signaling pathway. Mol Cell Biol. 2002;22:1027. doi: 10.1128/MCB.22.4.1027-1035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia C, Cheshire JK, Patel H, et al. Cross-talk between transcription factors NF-kappa B and C/EBP in the transcriptional regulation of genes. Int J Biochem Cell Biol. 1997;29:1525. doi: 10.1016/s1357-2725(97)00083-6. [DOI] [PubMed] [Google Scholar]
- 32.Jeschke MG, Low JF, Spies M, et al. Cell proliferation, apoptosis, NF-kappaB expression, enzyme, protein, and weight changes in livers of burned rats. Am J Physiol Gastrointest Liver Physiol. 2001;280:G1314. doi: 10.1152/ajpgi.2001.280.6.G1314. [DOI] [PubMed] [Google Scholar]
- 33.Horton JW. Free radicals and lipid peroxidation mediated injury in burn trauma: The role of antioxidant therapy. Toxicology. 2003;189:75. doi: 10.1016/s0300-483x(03)00154-9. [DOI] [PubMed] [Google Scholar]
- 34.Hosnuter M, Gurel A, Babuccu O, et al. The effect of CAPE on lipid peroxidation and nitric oxide levels in the plasma of rats following thermal injury. Burns. 2004;30:121. doi: 10.1016/j.burns.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 35.Deveraux QL, Takahashi R, Salvesen GS, et al. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 36.Klein D, Schubert T, Horch RE, et al. Insulin treatment improves hepatic morphology and function through modulation of hepatic signals after severe trauma. Ann Surg. 2004;240:340. doi: 10.1097/01.sla.0000133353.57674.cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Beek EM, Cochrane F, Barclay AN, et al. Signal regulatory proteins in the immune system. J Immunol. 2005;175:7781. doi: 10.4049/jimmunol.175.12.7781. [DOI] [PubMed] [Google Scholar]
- 38.Manca C, Tsenova L, Freeman S, Hypervirulent M, et al. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J Interferon Cytokine Res. 2005;25:694. doi: 10.1089/jir.2005.25.694. [DOI] [PubMed] [Google Scholar]
- 39.Masson S, Daveau M, Francois A, et al. Up-regulated expression of HGF in rat liver cells after experimental endotoxemia: a potential pathway for enhancement of liver regeneration. Growth Factors. 2001;18:237. doi: 10.3109/08977190109029113. [DOI] [PubMed] [Google Scholar]
- 40.Sekine K, Fujishima S, Aikawa N. Plasma hepatocyte growth factor is increased in early-phase sepsis. J Infect Chemother. 2004;10:110. doi: 10.1007/s10156-004-0301-y. [DOI] [PubMed] [Google Scholar]
- 41.Kuropatwinski KK, De Imus C, Gearing D, et al. Influence of subunit combinations on signaling by receptors for oncostatin M, leukemia inhibitory factor, and interleukin-6. J Biol Chem. 1997;272:15135. doi: 10.1074/jbc.272.24.15135. [DOI] [PubMed] [Google Scholar]
- 42.Fruhbeck G. Intracellular signalling pathways activated by leptin. Biochem J. 2006;393:7. doi: 10.1042/BJ20051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esmon CT, Fukudome K, Mather T, et al. Inflammation, sepsis, and coagulation. Haematologica. 1999;84:254. [PubMed] [Google Scholar]
- 44.Koller M, Kutscha-Lissberg F, Brom J, et al. Influence of low molecular weight heparin (certoparin) and unfractionated heparin on the release of cytokines from human leukocytes. Inflammation. 2001;25:531. doi: 10.1023/a:1012883916991. [DOI] [PubMed] [Google Scholar]
- 45.Hezi-Yamit A, Wong PW, Bien-Ly N, et al. Synergistic induction of tissue factor by coagulation factor Xa and TNF: evidence for involvement of negative regulatory signaling cascades. Proc Natl Acad Sci USA. 2005;102:12077. doi: 10.1073/pnas.0504526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deepa PR, Varalakshmi P. Favourable modulation of the inflammatory changes in hypercholesterolemic atherogenesis by a low-molecular-weight heparin derivative. Int J Cardiol. 2006;106:338. doi: 10.1016/j.ijcard.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Hochart H, Jenkins PV, Smith OP, et al. Low-molecular weight and unfractionated heparins induce a downregulation of inflammation: Decreased levels of proinflammatory cytokines and nuclear factor-kappaB in LPS-stimulated human monocytes. Br J Haematol. 2006;133:62. doi: 10.1111/j.1365-2141.2006.05959.x. [DOI] [PubMed] [Google Scholar]
- 48.Bansal MB, Kovalovich K, Gupta R, et al. Interleukin-6 protects hepatocytes from CCl4-mediated necrosis and apoptosis in mice by reducing MMP-2 expression. J Hepatol. 2005;42:548. doi: 10.1016/j.jhep.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond DE, Carter S, Clague MJ. Met receptor dynamics and signalling. Curr Top Microbiol Immunol. 2004;286:21. doi: 10.1007/978-3-540-69494-6_2. [DOI] [PubMed] [Google Scholar]
- 50.Jeschke M, Finnerty C, Norbury W, et al. Burn size determines the inflammatory and hypermetabolic response. Shock. 2006;25:87. doi: 10.1186/cc6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrow RE, Meyer NA, Jeschke MG. Effect of varying burn sizes and ambient temperature on the hypermetabolic rate in thermally injured rats. J Surg Res. 2001;99:253. doi: 10.1006/jsre.2001.6183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.