Abstract
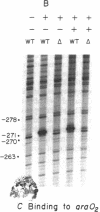
Two sets of experiments have been performed to test the DNA loop model of repression of the araBAD operon of Escherichia coli. First, dimethyl sulfate methylation protection measurements on normally growing cells show that the AraC regulatory protein occupies the araI site in the presence and absence of the inducer arabinose. Similarly, the araO2 site is shown to be occupied by AraC protein in the presence and absence of arabinose; however, its occupancy by AraC is greatly reduced when araI and adjacent sequences are deleted. Thus, AraC protein binds to araO2 cooperatively with some other component of the ara system located at least 60 base pairs away. Second, the mutational analysis presented here shows that the DNA components required for repression of araBAD are araI, araO2, and perhaps the araBAD operon RNA polymerase binding site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Brent R. Repression of transcription in yeast. Cell. 1985 Aug;42(1):3–4. doi: 10.1016/s0092-8674(85)80091-x. [DOI] [PubMed] [Google Scholar]
- Busby S., Irani M., Crombrugghe B. Isolation of mutant promoters in the Escherichia coli galactose operon using local mutagenesis on cloned DNA fragments. J Mol Biol. 1982 Jan 15;154(2):197–209. doi: 10.1016/0022-2836(82)90060-2. [DOI] [PubMed] [Google Scholar]
- Colomé J., Wilcox G., Englesberg E. Constitutive mutations in the controlling site region of the araBAD operon of Escherichia coli B/r that decrease sensitivity to catabolite repression. J Bacteriol. 1977 Feb;129(2):948–958. doi: 10.1128/jb.129.2.948-958.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Schleif R. Deletion analysis of the Escherichia coli ara PC and PBAD promoters. J Mol Biol. 1984 Nov 25;180(1):201–204. doi: 10.1016/0022-2836(84)90437-6. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Squires C., Meronk F., Jr The L-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1100–1107. doi: 10.1073/pnas.62.4.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielow L., Largen M., Englesberg E. Initiator constitutive mutants of the L-arabinose operon (OIBAD) of Escherichia coli B/r. Genetics. 1971 Nov;69(3):289–302. doi: 10.1093/genetics/69.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Schleif R. Arabinose C protein: regulation of the arabinose operon in vitro. Nat New Biol. 1971 Oct 6;233(40):166–170. doi: 10.1038/newbio233166a0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J Mol Biol. 1984 Sep 25;178(3):611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani M. H., Orosz L., Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983 Mar;32(3):783–788. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. Identification of araC protein and two-dimensional gels, its in vivo instability and normal level. J Mol Biol. 1981 Jun 15;149(1):133–139. doi: 10.1016/0022-2836(81)90265-5. [DOI] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R., Lis J. T. The regulatory region of the L-arabinose operon: a physical, genetic and physiological study. J Mol Biol. 1975 Jul 5;95(3):417–431. doi: 10.1016/0022-2836(75)90200-4. [DOI] [PubMed] [Google Scholar]
- Sheppard D. E., Englesberg E. Further evidence for positive control of the L-arabinose system by gene araC. J Mol Biol. 1967 May 14;25(3):443–454. doi: 10.1016/0022-2836(67)90197-0. [DOI] [PubMed] [Google Scholar]
- Wilcox G., Meuris P., Bass R., Englesberg E. Regulation of the L-arabinose operon BAD in vitro. J Biol Chem. 1974 May 10;249(9):2946–2952. [PubMed] [Google Scholar]